Abstract
The Lyme disease spirochete, Borrelia burgdorferi, encodes a novel type of DNA-binding protein named EbfC. Orthologs of EbfC are encoded by a wide range of bacterial species, so characterization of the borrelial protein has implications that span the eubacterial kingdom. The present work defines the DNA sequence required for high-affinity binding by EbfC to be the 4 bp broken palindrome GTnAC, where ‘n’ can be any nucleotide. Two high-affinity EbfC-binding sites are located immediately 5′ of B. burgdorferi erp transcriptional promoters, and binding of EbfC was found to alter the conformation of erp promoter DNA. Consensus EbfC-binding sites are abundantly distributed throughout the B. burgdorferi genome, occurring approximately once every 1 kb. These and other features of EbfC suggest that this small protein and its orthologs may represent a distinctive type of bacterial nucleoid-associated protein. EbfC was shown to bind DNA as a homodimer, and site-directed mutagenesis studies indicated that EbfC and its orthologs appear to bind DNA via a novel α-helical ‘tweezer’-like structure.
INTRODUCTION
The Lyme disease spirochete, Borrelia burgdorferi, is maintained in nature through cycles of alternating infections of vertebrate hosts and tick vectors. To facilitate infection of those distinctive types of animals, and to enable efficient transmission between hosts and vectors, B. burgdorferi controls production of a large number of bacterial proteins. Among these are the Erp proteins, a polymorphic family of surface-exposed, outer membrane lipoproteins that are expressed throughout mammalian infection but largely repressed during tick colonization (1). Known functions of Erp family members include binding of the host serum components complement factor H and plasminogen, and adherence to the extracellular matrix protein laminin (2–10). All Lyme disease spirochetes contain multiple variants of related prophages, known as cp32 elements, which replicate episomally as 32 kb circular plasmids (11–14). Each cp32 contains a mono- or bicistronic-erp locus. The B. burgdorferi type strain, B31, is known to harbor 10 different cp32 family members, and encodes 13 distinct Erp proteins (1,15). Despite the often extensive sequence variation found among erp open reading frames, all erp loci share a unifying feature in possessing highly conserved sequences immediately 5′ of the open reading frames. Within this DNA sequence is the transcriptional promoter and two operator sites that bind cytoplasmic proteins (16). Transcriptional fusion studies identified that the operator region closest to the transcriptional promoter, Operator 2, is involved in transcriptional repression (16). Members of our laboratories recently discovered that a chromosomally encoded protein of B. burgdorferi, named EbfC, specifically binds two sites within erp Operator 2 (17).
Many other species of bacteria encode orthologs of EbfC, suggesting that this protein performs a function(s) conserved throughout the kingdom Eubacteria. These homologous proteins have been variously classed as ‘domain of unknown function’ 149, Pfam 2575, COG-0718 and YbaB (18,19). The 3D structures of the Escherichia coli and Haemophilus influenzae orthologs have been determined (18) and (http://www.rcsb.org/pdb/explore.do?structureId=1PUG). Both proteins crystallized as dimers, consisting of a central β-folded dimer-interface region and protruding α-helices at either end, forming a distinctive structure that has been described as having a ‘tweezer-like’ shape (18). The present studies extended characterization of the B. burgdorferi EbfC protein, demonstrating that this small protein both specifically and nonspecifically binds DNA, can bind DNA independently of context and alters DNA conformation. Site-directed mutagenesis approaches were also employed to investigate the mode of interaction between EbfC and DNA.
MATERIALS AND METHODS
Site-directed mutagenesis
Site-directed mutagenesis of both the erpAB Operator 2 and ebfC were performed by sequence overlap extension PCR mutagenesis (20). Primers used in the PCR reactions are listed on Table 1. All mutagenized plasmids were sequenced on both strands to verify the desired mutations.
Table 1.
Oligonucleotide primers used for site-directed mutagenesis to produce mutant recombinant EbfC proteinsa
| Construct | Primer name | Sequence (from 5′ to 3′) |
|---|---|---|
| K16A | K16A-1 | TCT AGC GTT GCG AAT AAT ATT GAC AAT ATT AAA AAG G |
| K16A-2 | AAT ATT ATT CGC AAC GCT AGA CAT ATT TTT CAA AAA ATC TAA CGG | |
| D20A | D20A-1 | AAT AAT ATT GCC AAT ATT AAA AAG GAA ATT TCT AAA ATT ACG |
| D20A-2 | TTT AAT ATT GGC AAT ATT ATT CTT AAC GCT AG | |
| K23A | K23A-1 | GAC AAT ATT GCA AAG GAA ATT TCT AAA ATT ACG |
| K23A-2 | AAT TTC CTT TGC AAT ATT GTC AAT ATT ATT CTT AAC G | |
| N77A | N77A-1 | TCT GCT TTA GCT GAT GCT GTC TCT AAG GTT AAA G |
| N77A-2 | GAC AGC ATC AGC TAA AGC AGA TTT AAT CAT TTG | |
| D78A | D78A-1 | GCT TTA AAT GCT GCT GTC TCT AAG GTT AAA GAA G |
| D78A-2 | AGA GAC AGC AGC ATT TAA AGC AGA TTT AAT CAT TTG | |
| K82A | K82A-1 | GCT GTC TCT GCG GTT AAA GAA GAG ATA AAA TTA AAA ACC ATG G |
| K82A-2 | TTC TTT AAC CGC AGA GAC AGC ATC ATT TAA AGC AG | |
| K84A | K84A-1 | TCT AAG GTT GCA GAA GAG ATA AAA TTA AAA ACC ATG G |
| K84A-2 | TAT CTC TTC TGC AAC CTT AGA GAC AGC ATC ATT TAA AG | |
| E85A | E85A-1 | AAG GTT AAA GCA GAG ATA AAA TTA AAA ACC ATG G |
| E85A-2 | TTT TAT CTC TGC TTT AAC CTT AGA GAC AGC ATC | |
| K88A | K88A-1 | GAA GAG ATA GCA TTT AAA ACC ATG GGA GTT CTT CC |
| K88A-2 | GGT TTT TAA TGC TAT CTC TTC TTT AAC CTT AGA G |
aAll PCR used p462M5 as template (17).
Recombinant EbfC
Proteins were produced from either p462M5 (17) or mutant derivatives thereof (Table 1), using E. coli Rosetta (DE3) (pLysS) (Novagen). Following induction by addition of 1 mM isopropylthio-β-galactopyranoside, bacteria were harvested by centrifugation and lysed by sonication in 20 mM NaPO4, 0.5 M NaCl, 30 mM imidazole, pH 7.4. Lysates were cleared by centrifugation and injected onto 5 ml HisTrap-HP columns using an ÄKTA-FPLC with UPC-900 UV absorbance monitor and Frac920 fraction collector (GE Healthcare). Columns were subjected to the lysis buffer containing a linear gradient of imidazole ranging from 30 mM to 750 mM. Fractions (1 ml each) were collected, and aliquots subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie brilliant blue staining, to assess protein purity. Proteins were concentrated using YM-30 Centricon columns (Amicon), then dialyzed against 50 mM Tris–HCl, 1 mM phenylmethanesulfonyl fluoride, 1 mM dithiothreitol (DTT), 10% glycerol, pH 7.5. Recombinant protein concentrations were determined by bicinchoninic acid (BCA) protein assays (Pierce). Aliquots were snap frozen in liquid nitrogen, then stored at −80°C. Frozen protein preparations were thawed on ice immediately before use.
Electrophoretic mobility shift assays
Sequences of biotinylated DNAs used to define optimal EbfC-binding activities by electrophoretic mobility shift assays (EMSA) are illustrated in Figure 1. DNAs were produced by PCR amplification using appropriate primers, templates and LA Taq polymerase (Takara). Amplification consisted of 40 cycles of 94°C for 1 min, 55°C for 1 min and 68°C for 1 min. Reaction products were separated by agarose gel electrophoresis and visualized by staining with ethidium bromide. DNAs were extracted into nuclease-free water using Wizard SV gel purification system (Promega). DNA concentrations were determined spectrophotometrically by measuring absorbance at 260 nm, with an absorbance of 1.0 = 50 μg/ml DNA, then each diluted to a final concentration of 1 nM (10× stock).
Figure 1.
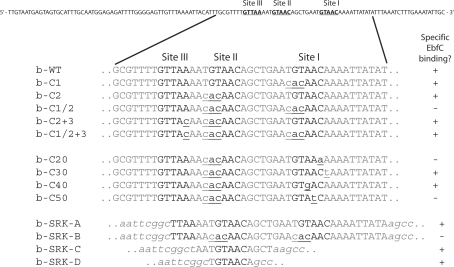
Sequences of DNA probes used for defining the sequence requirements for high-affinity binding by EbfC. All were based upon the sequence of the B. burgdorferi strain B31 erpAB Operator 2. The ability of each DNA to form high-affinity EbfC–DNA interactions is indicated. All probes were labeled with a biotin moiety at the right end. The uppermost sequence is the complete sequence of the wild-type probe b-WT. Other than the b-SRK series, all other EMSA probes were identical to b-WT except where indicated by underlining and lower case lettering. Sequences of EbfC-binding sites I, II and III are shown in bold black type. Parts of the pCR2.1-derived sequences of the b-SRK series are indicated by lower case italics.
EMSAs were performed using 100 pM biotin-labeled double-stranded DNA probes as previously described (16,17), with binding conditions of 50 mM Tris–HCl, 1 mM DTT, 8 μl/ml Protease inhibitor (Sigma. St. Louis, MO), 2 μl/ml Phosphatase Inhibitor Cocktail II (Sigma), 50 μg/ml bovine serum albumin, 10% glycerol, pH = 7.5 at room temperature. Binding reactions were allowed to proceed for 20 min at room temperature, before being subjected to electrophoresis through 6% DNA Retardation Gels (Invitrogen) for a total of 9000 V-min. Gels were transferred to nylon membranes, cross-linked by UV light and biotinylated DNAs were detected by Chemiluminescent Nucleic Acid Detection Modules (Pierce) and Kodak Biomax film.
Dissociation constant determination
Dissociation constants (Kd) for EbfC–DNA interactions in the absence of poly-dI-dC were determined by analyses of EMSA gel images (21). Exposed films were scanned in 8 bit depth at 1200 dpi resolution, analyzed using Image J 1.37v (22) and the ratios of bound:free DNA in each lane were determined. These values were transferred into Microsoft Excel for graphing and analyses. The quantity of DNA in each band or total lane was expressed as concentration of the known input DNA (23,24).
Binding was analyzed according to a model in which n molecules of protein (P) bind DNA (D):
| 1 |
with the equilibrium constant:
| 2 |
The Kd for the titration of DNA with EbfC were determined by graphing the ratio of bound to free DNA for each reaction. These ratios were graphed in relation to known concentration of free protein [P]. Since the ratios of free protein to DNA were extremely high, the binding of protein to DNA did not appreciatively change concentration of free protein (21). The Kd is therefore the concentration of EbfC where the ratio of bound to free DNA = 1.
Rearranging Equation (2) and taking natural logs gives:
Thus, for EbfC, a graph of ln ([D]/[EbfCnD]) as a function of ln [EbfC] will have a slope equal to the negative value of the stoichiometry, −n, and an x-intercept at which Kd = nln [EbfC].
For EMSAs with two DNA probes, the ratio of bound to free DNA cannot be determined for each specific DNA probe, because it is impossible to determine which DNA species is responsible for any single protein DNA complex. As such, this analysis was performed by measuring the disappearance of each individual free-DNA species. In these cases, the calculated binding activities were the ratios of protein concentrations at which each free-DNA species disappeared by 50%.
DNA-bending analyses
Five 150 bp DNA fragments, overlapping stepwise by 30 bp increments over a total of 250 bp, were produced by PCR, using the cloned erpAB insert of pBLS434a as template (13). The upstream oligonucleotide primer for each DNA fragment was modified with a biotin moiety at the 5′ end. Natural curvature of the DNAs was analyzed by electrophoresis through 15% polyacrylamide gels and staining with ethidium bromide. EMSAs using purified recombinant EbfC were performed as described above. Predictions of natural DNA-bending were calculated using bend.it, with default parameters (http://hydra.icgeb.trieste.it/dna/bend_it.html) (25).
Size fractionation chromatography
The abilities of the various recombinant EbfC preparations to form multimeric structures were analyzed by size fractionation chromatography as previously described (17). Proteins were individually loaded onto a Superdex 75 10/300 column (GE Healthcare) and separated using a Waters 600 pump and controller equipped with a Waters 996 UV/Vis detector.
EbfC model
The B. burgdorferi EbfC was modeled based on the solved structure of the orthologous protein YbaB of H. influenzae (PDB 1j8b) (18). The sequences of EbfC and YbaB were aligned with BlastP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid side-chain substitutions were made at positions of different sequences with the O-program (26). The alignment showed that compared with YbaB, EbfC contains an additional amino acid residue (Leu63) inserted into the loop before the C-terminal α-helix. This loop (residues 61–65) was modified to contain Leu63 so that its side chain would be in close proximity to Phe59 and Ala67 to form hydrophobic interactions. The resulting EbfC model was minimized with the CNS program (27). The figure of the EbfC model was generated with Molscript (28) and Raster3D (29,30).
Analysis of ebfC sequences of Lyme disease Borrelia spp.
The ebfC loci of Lyme disease borreliae were amplified by PCR using oligonucleotides EBFC-18 (5′-CATCTGGCTTAACAATACATAAAG-3′) and EBFC-19 (5′-GACAATGTTCTTTATTATAAGGTG-3′), which are specific for sequences bordering the B. burgdorferi strain B31 ebfC open reading frame on the 5′ and 3′ sides, respectively. Bacteria analyzed in this work were B. burgdorferi (sensu stricto) strains B31, N40, 297 and Sh-2-82, Borrelia afzelii strains PKo and VS461, Borrelia garinii strain Ip90 and Borrelia bissettii strains 25‱015 and DN127.
RESULTS
Sequence-specific binding of DNA by EbfC
Our earlier EMSA studies found that competitor DNAs containing the broken palindrome TGTA/TACA competed for EbfC binding to erp promoters (Perp), while DNAs that instead contained 5′-CACA/TACA-3′ did not compete (17). To more precisely define the sequence requirements for high-affinity interactions between EbfC and DNA, site-directed mutagenesis was employed to create a series of variants in the Operator 2 region of the strain B31 erpAB promoter (PerpAB).
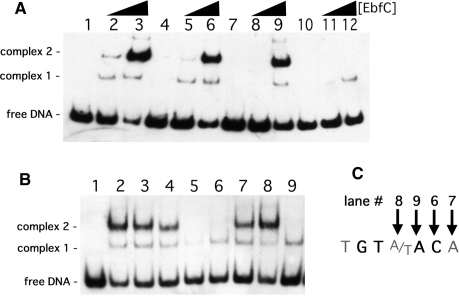
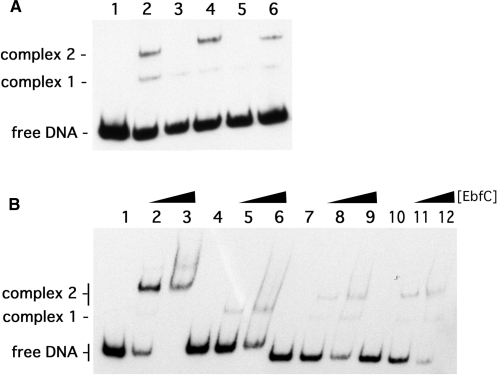
Initially, mutants of PerpAB were produced in which either site I, site II or both sites I and II were changed to 5′-CACAACA-3′, all in the context of otherwise normal PerpAB DNA sequences (Figure 1). EMSA using a probe with the wild-type PerpAB sequence, b-WT, yielded two observable DNA–EbfC complexes, with the slower mobility (upper) complex having a higher affinity interaction (Figure 2A). Probes that contain only one TGTA/TACA palindrome (b-C1 and b-C2) exhibited the same two EbfC–DNA complexes, but at decreased relative levels (Figure 3A). The signal strengths of the slower mobility complexes for EbfC binding to either probes b-C1 or b-C2 were approximately one half the value for b-WT. There were no apparent differences in EbfC binding to DNA probes that contained only wild type site I or site II, indicating relatively equal affinities for both sites. However, the mutation of both TGTA/TACA palindromes in probe b-C1/2 dramatically reduced EbfC binding (Figure 2A). These results confirm that EbfC preferentially binds the previously identified palindromic sequence and demonstrate that the apparent binding affinity when two sites are present is greater than when DNA contains only one site. This was likely due to the higher relative concentration of high-affinity binding sites when two adjacent sequences are present, as opposed to a single site alone.
Figure 2.
Definition of the EbfC preferred binding sequence. (A) Representative EMSA of DNA probes containing two, one or no EbfC-binding consensus sequences and titration of increasing amounts of EbfC. Lanes 1–3 contain probe b-WT, lanes 4–6 contain probe b-C1, lanes 7–9 contain probe b-C2 and lanes 10–12 contain probe b-C1/2. Lanes 1, 4, 7 and 10 lacked EbfC, lanes 2, 5, 8 and 11 contained 8 μM EbfC and lanes 3, 6, 9 and 12 contained 40 μM EbfC. (B) Association of 4 μM EbfC with probes b-WT (lane 2), b-C1 (lane 3), b-C2 (lane 4), b-C1/2 (lane 5) and b-C2 derivatives b-C20 (lane 6), b-C30 (lane 7), b-C40 (lane 8) and b-C50 (lane 9). (C) Definition of the EbfC high-affinity binding sequence. Lane numbers refer to the EMSA lanes shown in (B).
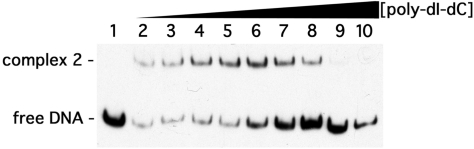
Figure 3.
Competition of EbfC binding to probe b-WT by the non-specific competitor poly-dI-dC. EMSA included 100 pM b-WT DNA, and lanes 2–10 contained 1 μM EbfC; also contained either 0, 1, 5, 10, 100, 500, 1000, 5000 or 25 000 pg/μl poly-dI-dC, respectively.
To identify the minimal DNA sequence required for specific EbfC binding, additional DNA probes based on PerpAB were produced in which site I was mutated to the non-binding sequence, 5′-CACAACA-3′, and nucleotides of site II were individually changed away from TGTA/TACA (Figure 1). Changing either the seventh nucleotide from A to T (b-C30) or the fourth nucleotide from A/T to G/C (b-C40) had no apparent affect on binding (Figure 2B, lanes 7 and 8, respectively). In contrast, the mutation of the fifth A to T (b-C50) or sixth C to A (b-C20) eliminated the specific EbfC–DNA primary complex 2 (Figure 2B, lanes 9 and 6, respectively). Thus the minimum DNA sequence necessary for specific interaction with EbfC is the four base broken palindrome GTnAC, where n may be any nucleotide (Figure 2A). With these results, EbfC is one of only two spirochetal DNA-binding proteins for which the consensus high-affinity binding sequence has been determined (31).
Specificity of EbfC binding to DNA was assessed using poly-dI-dC as a competitor for binding to PerpAB DNA. Poly-dI-dC DNA forms normal DNA helices but does not contain conventional DNA sequence due to the presence of the inosine nucleotide. Using this DNA permitted separation of sequence specific from structural EbfC–DNA interactions. Poly-dI-dC successfully competed only when present in concentrations >1 ng/μl (Figure 3). Based on the molar ratio of specific EbfC-binding sites in the erpAB Operator 2 probe relative to the number of non-specific 5 bp sequences in poly-dI-dC, EbfC has ∼40-fold lower affinity for poly-dI-dC than for PerpAB Operator 2.
EbfC binds DNA as a homodimer
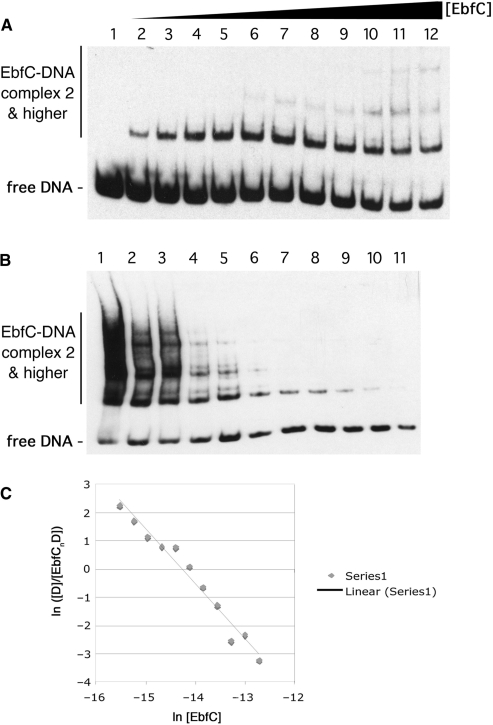
The Kd for EbfC binding to Perp DNA was determined by titrating increasing concentrations of recombinant EbfC to labeled DNA probe b-WT, followed by EMSA analysis of protein-DNA complex formation (Figure 4A). The protein:DNA ratios in all reactions were extremely high, such that binding of protein to DNA would not significantly affect the concentration of free protein. As described in the following paragraph, EbfC binds DNA as a homodimer. The calculated Kd for the reaction 2 EbfC + b-WT ↔ 2 EbfC/b-WT is 388 ± 108 nM.
Figure 4.
Determination of the Kd of EbfC binding to wild-type erpAB Operator 2 DNA. (A) Titration of labeled probe b-WT, a 124 bp segment including erpAB Operator 2 (Figure 1), with increasing concentrations of purified, recombinant EbfC. Each lane represents equally loaded reactions consisting of 100 pM b-WT plus 0, 68, 140, 210, 270, 340, 410, 480, 550, 620, 680 or 820 nM EbfC for lanes 1–12, respectively. (B) Serial dilution of non-equilibrated EbfC + DNA reactions. A mixture of 3 μM EbfC and 4 nM b-WT was produced and immediately serially diluted with three volumes of the previous tube and one volume of binding buffer. Final concentrations of EbfC were 3.0, 2.3, 1.7, 1.3, 0.98, 0.74, 0.56, 0.42, 0.32, 0.24, 0.18 μM and concentrations of probe b-WT were 4.0, 3.1, 2.4, 1.9, 1.4, 1.1, 0.88, 0.68, 0.52, 0.41, 0.31 nM for the mixtures in lanes 1–11, respectively. These reactions were allowed to equilibrate before loading equal volumes onto the gel to perform the mobility shift assay. (C) Band intensities from (B) were determined and ln([D]/[EbfCnD]) was graphed as a function of ln [EbfC]. The slope of the resulting line is equal to −n, the negative value of the binding stoichiometry and the x-intercept is the point at which n ln [EbfC] = lnKd.
Serial dilution was also performed to confirm that result and to determine binding stoichiometry (Figure 4B). This method allows estimation of the complex stoichiometry n and the Kd (32). These analyses yielded a binding stoichiometry of n = 2.0 EbfC monomers per DNA molecule, indicating that EbfC binds DNA as a dimer (Figure 4C). DNA-binding analyses of the E. coli and H. influenzae EbfC orthologs yielded that same result (Cooley,A.E. et al., submitted for publication). These data are also consistent with the observed formation of B. burgdorferi EbfC dimers in solution (17) and crystallization of the E. coli and H. influenzae orthologs as homodimers (18) and (http://www.rcsb.org/pdb/explore.do?structureId=1PUG). By this technique, the calculated Kd for dimeric recombinant EbfC binding to Perp Operator 2 DNA was 313 nM, consistent with the results obtained from our other analyses described above.
DNA-binding by EbfC is context independent
All of the probes used in the above studies contained EbfC binding site(s) in the context of PerpAB (Figure 1). As mentioned above, all erp promoters contain two EbfC binding sites. In the strain B31 erpHY locus, EbfC binding site I is identical to that of PerpAB and all other known erp promoters, but site II is altered to a non-binding 5′-GTAAT-3′ (16,17). However, other sequence changes in PerpHY created a new EbfC-binding consensus immediately 5′ of site II, which we designate site III (Figure 1). This alternative site is particularly noteworthy, because the different spacing of the two sites in PerpHY could influence EbfC–DNA interactions. To study that possibility, PerpAB was mutated to resemble PerpHY by disruption of site II and creation of site III, both with and without a consensus site I. As anticipated, in the absence of a consensus site I, changing the site II sequence from that of PerpAB to the non-consensus sequence of PerpHY eliminated specific binding (Figure 5A, lane 5 [b-C1/2]). However, in that same context, creation of a consensus sequence at site III was sufficient for EbfC binding (Figure 5A, lane 6 [b-C1/2+3]). Notably, mobilities of EbfC–DNA complexes formed with probes containing a consensus sequence at site III were comparable to those with wild type PerpAB sequences.
Figure 5.
Context-independent DNA-binding by EbfC. (A) Interactions between EbfC and erpAB variant DNAs: wild-type b-WT without added protein (lane 1); probe b-C2, which contains a consensus sequence at site I and non-consensus sequences at sites II and III (lane 2); probe b-C20, which lacks consensus sequences at sites I, II or III (lane 3); probe b-C2+3, which lacks a consensus sequence at site II but contains consensus sequences at site I and III (lane 4); probe b-C1/2, which lacks consensus sequences at sites I, II or III (lane 5); and probe b-C1/2+3, which contains a consensus binding sequence at site III but lacks consensus sequences at site I and II (lane 6). All lanes contained 2 μM EbfC and 1 ng/μl poly-dI-dC. (B) Binding of EbfC to DNA probes derived from insertion of small DNAs containing consensus EbfC-binding sites into the TA-cloning site of pCR2.1. Lanes 1–3 contained b-SRK-A, a 34 bp insert containing two EbfC-binding sites. Lanes 4–6 contained b-SRK-B, which contains the same insert as does b-SRK-A except with mutations in both binding sites such that EbfC does not specifically interact (negative control). Lanes 7–9 contained b-SRK-C, which consists of a 12 bp insert containing one consensus EbfC-binding sequence. Lanes 10–12 contained b-SRK-D, which consists of a 7 bp insert with one consensus binding sequence. Lanes 1, 4, 7 and 10 lacked EbfC protein, lanes 2, 5, 8 and 11 contained 4 μM EbfC, while lanes 3, 6, 9 and 12 contained 12 μM EbfC. All lanes contained 1 ng/μl poly-dI-dC.
The effects of surrounding DNA sequences on EbfC binding were also examined through use of constructs created by insertion of short DNA sequences into the TA-cloning site of plasmid pCR2.1. Labeled probes were produced from those constructs by PCR using the vector's M13F and M13R priming sites, and consisted of the DNA insert flanked by pCR2.1 DNA. That DNA is relatively rich in G + C content as compared with the high A + T content of PerpAB and the rest of the B. burgdorferi genome [www.invitrogen.com/content/sfs/vectors/pcr2_1topo_map.pdf and (15,33)]. Inclusion of a 29 bp sequence of PerpAB that contained two GTnAC palindromes yielded strong EbfC binding (Figure 5B, lanes 1–3 [b-SRK-A]). As a control, the same sequence in which both EbfC-binding sites were changed to 5′-ACAAC-3′ did not form the specific EbfC–DNA complex 2 (Figure 5B, lanes 4–6 [b-SRK-B]). A single EbfC-binding site on either a 13 or 7 bp insert yielded production of the specific EbfC–DNA complex 2 (Figure 5B, lanes 7–9 [b-SRK-C] and 10–12 [b-SRK-D], respectively). Thus, we conclude that EbfC is able to bind GTnAC sequences regardless of the composition of surrounding DNA, and is therefore likely to bind any consensus sequence within the B. burgdorferi genome.
Binding of EbfC affects DNA bending
DNA-binding proteins often change the conformation of targeted DNA. To examine whether or not EbfC-binding alters DNA conformation, its effects upon erpAB 5′ DNA were examined.
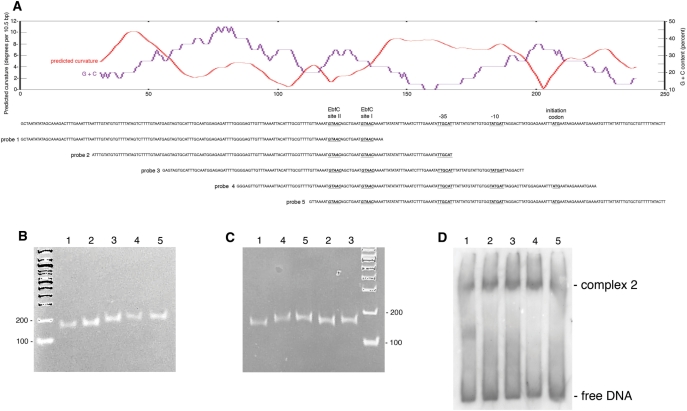
Five overlapping 150 bp fragments of PerpAB were produced, spanning a 250 bp region of erpAB centered on the two high-affinity EbfC-binding sites (Figure 6A). Modeling analyses of this 250 bp sequence predicted an extended region of DNA curvature that includes the promoter −35 and −10 regions (Figure 6A). Polyacrylamide electrophoresis of the five 150 bp fragments confirmed that prediction, probes 4 and 5, which contain the predicted bent region toward their centers, migrated more slowly than did probe 1, which does not contain the predicted bent region (Figure 6B and C). This is the first reported occurrence of naturally bent DNA in a spirochete, and its juxtaposition with PerpAB suggests that the bend may impact upon levels of erp transcription.
Figure 6.
PerpAB DNA is naturally bent, and binding of EbfC alters DNA conformation. (A) A 250 bp segment of the erpAB locus, spanning the two high-affinity EbfC-binding sequences and the transcriptional promoter, is illustrated in the center of the panel. Above the DNA sequence is the G + C content and predicted naturally occurring bending pattern of this DNA. Note that an extended bend of 8°–10° per 10.5 bp helical turn is predicted to occur over a 50 bp region that overlaps the −35 and −10 promoter sequences (40°−50° total curvature). Sequences of five overlapping 150 bp DNA probes are indicated underneath. (B) DNA probes 1–5 after polyacrylamide gel electrophoresis, stained with ethidium bromide. The number of each DNA is indicated above the lanes. DNA size markers (bp) were separated in the first lane. (C) DNA probes 1–5 in randomized order, subjected to polyacrylamide gel electrophoresis and ethidium bromide staining. The number of each DNA probe is indicated above the lanes. DNA size markers (bp) were separated in the last lane. (D) EMSA of probes 1–5 with EbfC. The number of each DNA probe is indicated above the lanes.
The effect of EbfC on the innate bend in PerpAB was next examined by EMSA. EbfC complexes with DNA probes 2, 3 and 4 all migrated at the same rates, while free probe 4 was retarded relative to probes 2 and 3, as above (Figure 6D). Thus, binding of EbfC to erp Operator 2 alters the conformation of PerpAB DNA.
The ‘tweezer’ domain of EbfC is vital for DNA binding
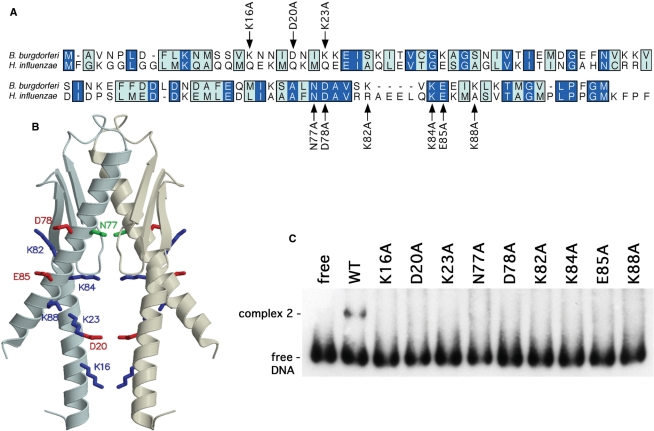
The EbfC ortholog of H. influenzae KW20 Rd (YbaB, HI_0442) shares 27.5% identity and 59.3% similarity with B. burgdorferi EbfC (Figure 7A). As noted in the Introduction section, both the H. influenzae and E. coli EbfC orthologs have been crystallized, with both forming nearly identical structures with a ‘tweezer’-like appearance (18). The predicted gap between the ‘tweezer’ arms of EbfC ranges between 15 and 22 Å, comparable with the diameter of B-DNA, suggesting that the protruding α-helices could form the DNA-binding domain (18,34). To address that hypothesis, site directed mutagenesis of B. burgdorferi EbfC was performed to individually replace nine of the amino acids in those α-helices (Figure 7A and B). Each recombinant protein was expressed and purified two separate times, to guard against preparation artifacts. None of the mutated variants of EbfC was able to detectably bind PerpAB, even when tested at protein concentrations 6.5-fold greater than the wild-type EbfC (Figure 7C).
Figure 7.
Identification of EbfC amino acid residues involved with binding DNA. (A) Alignment of B. burgdorferi EbfC and H. influenzae YbaB, with identical residues boxed in dark blue, and similar amino acids in light blue. Residues that were individually mutated to alanine are indicated. (B) Model of B. burgdorferi EbfC based on the solved structure of its H. influenzae ortholog. Residues specifically mutated to alanine in this work are indicated. (C) EMSA of DNA probe b-WT with wild-type (WT) and each mutant EbfC protein. The wild-type recombinant EbfC was analyzed at a final concentration of 3 mM, and each mutant at 19.5 mM.
Size-exclusion chromatography demonstrated that wild-type EbfC and the K16A, D20A, K23A, D78A, K82A, K84A, E85A and K88A mutants all preferentially formed dimers in solution (data not shown) (17). The N77A mutant protein did not form higher order structures, indicating that this residue, which is predicted to be located adjacent to the β-folded region of EbfC, plays a role in dimerization. These results indicate that the dimerization and DNA binding functions of EbfC are distinct, with the DNA-binding function residing in the α-helical ‘tweezer’ domains.
EbfC also forms tetramers and octamers in solution (17). The K16A, D20A, K23A, D78A, K82A, K84A, E85A and K88A mutants all retained ability to form those higher ordered multimers (data not shown), indicating that all aspects of EbfC multimerization are independent of its ability to bind DNA.
EbfC is conserved among Lyme disease spirochetes
The bacteria that cause Lyme disease have been divided into several genospecies, including B. burgdorferi (sensu stricto) and B. afzelii, B. garinii and B. bissettii, each of which is associated with different symptoms in humans and degrees of infectivity (35–37). To ascertain the degrees of EbfC sequence conservation among Lyme disease spirochetes, ebfC loci were sequenced from several strains of distinct borrelial genospecies, isolated from across North America and Eurasia. All of the examined bacteria encode an identical EbfC protein, suggestive of constraints upon sequence variation and indicating that the results obtained from the present work on the EbfC protein of B. burgdorferi strain B31 are applicable to all Lyme disease bacteria.
DISCUSSION
A previous study from our laboratories discovered that B. burgdorferi EbfC is a DNA-binding protein. Orthologs of EbfC are encoded by a large number of other bacterial species, suggesting a kingdom-wide, conserved function for this protein. Analyses of crystallized EbfC orthologs of H. influenzae and E. coli revealed a unique, ‘tweezer’ like structure. The present studies indicated that the extending α-helices that comprise the ‘tweezer’ arms are involved in binding DNA, a feature that suggests the EbfC family to constitute a new type of DNA-binding protein. EbfC bound to DNA as a dimer, consistent with crystallographic data from the H. influenzae and E. coli orthologs.
The present studies revealed that B. burgdorferi EbfC preferentially binds DNA containing the sequence GTnAC, where n = any nucleotide. This sequence occurs 833 times on the major chromosome of B. burgdorferi strain B31, a frequency of one site every 1093 base pairs. The B. burgdorferi EbfC can also bind DNA which lacks a GTnAC sequence. These features suggest that EbfC is not a gene-specific regulatory protein, as originally hypothesized (16,17). Instead, this protein's simple and abundant binding sequence, abilities to bind DNA nonspecifically and to alter DNA conformation, plus its small size, are characteristics shared with bacterial nucleoid-associated proteins (38,39). EbfC forms tetramers and octamers in solution, and mutations that prevent DNA-binding did not disrupt ability to form those higher ordered structures. Thus the multimerization and DNA-binding activities of EbfC are distinct, raising the possibility that EbfC might also multimerize when bound to DNA and thereby serve as a bridge to condense DNA. The genome of B. burgdorferi is the most complex bacterial genome yet to be analyzed. The sequenced genome of the type strain, B31, consists of a linear major chromosome and more than 20 other linear and circular smaller chromosomes/plasmids/episomal prophages (15,33,40). The mechanisms by which this bacterium facilitates maintenance, partitioning and compaction of this complex genome are largely unknown (41). The well studied E. coli compacts its chromosome through DNA super coiling and interactions with small DNA-binding proteins, the nucleoid-associated or ‘histone-like’ proteins (38,39,42,43). To date, 12 major nucleoid-associated proteins have been identified in E. coli (38,39,42). In addition to functioning in chromatin organization and compaction, nucleoid-associated proteins often play roles in a variety of other DNA processes, such as modulation of transcription, transposition, recombination, repair and DNA replication (38,44–46). The combination of structural and regulatory roles suggests that the global genome functions of the nucleoid are tightly linked to its organizational ability (47,48). Prior to this work, only two B. burgdorferi proteins had been identified which exhibit activities resembling those of nucleoid-associated proteins. The B. burgdorferi Hbb protein, which resembles E. coli HU and integration host factor (IHF), can bind and bend DNA, and is capable of complementing some functions of an E. coli HU/IHF mutant (49–51). A second known protein, Gac, is produced from independent synthesis of the carboxy-terminal domain of GyrA, and can complement an E. coli HU mutant to promote Mu DNA transposition (52). The roles of those two putative nucleoid-associated proteins in maintaining B. burgdorferi DNA have not yet been defined. The B. burgdorferi genome does not contain any recognizable homologs of known bacterial nucleoid-associated proteins such as H-NS or Fis (33). Thus, the current picture is that B. burgdorferi compacts its segmented genome, which consists largely of linear DNAs, using only Hbb, Gac and, we hypothesize, EbfC.
The B. burgdorferi ebfC is located immediately 3′ of dnaX, and the two genes are transcriptionally linked [our unpublished results and (17,33)]. Thus, EbfC protein levels are likely linked to those of the dnaX products, the γ and τ subunits of DNA polymerase III. If additional analyses confirm that EbfC is in fact a nucleoid-associated protein, then the connection between DNA replication and DNA conformation afforded by the dnaX-ebfC locus may prove central to the vitality of B. burgdorferi. Consistent with that hypothesis, repeated attempts by our laboritories and others to inactivate B. burgdorferi ebfC have been unsuccessful (P. Stewart, personal communication), suggesting that EbfC may be an essential borrelial protein.
Almost all components of the B. burgdorferi segmented genome contain the same frequency of EbfC consensus-binding sites, with a site approximately every 1100–1400 bp. The cp32 family members are distinctive in having an EbfC-binding sequence every 630–650 bp. The erp Operator 2 sites are unique in being the only locations in B. burgdorferi where two GTnAC sites are found in close proximity. The present studies demonstrated that EbfC exhibits a greater affinity for DNA containing two GTnAC sites than for DNA with only one site. Many bacterial nucleoid-associated proteins influence gene transcription levels as consequences of the locations of preferred binding sites relative to promoters (43,44,53,54). Noting that ebfC transcription is linked to that of DNA polymerase III subunits, and erp genes are maximally expressed during periods of rapid bacterial division, we hypothesize that cellular EbfC levels could serve as a signal of bacterial replication rates to influence expression of Erp lipoproteins. High-affinity EbfC-binding sequences are also scattered throughout the B. burgdorferi genome. Although some sites have been identified in close proximity to known transcriptional promoters (55,56), the effects of EbfC upon those genes remain to be investigated.
In conclusion, B. burgdorferi EbfC possesses both sequence-specific and sequence-independent DNA binding activities. Genes orthologous to ebfC are found in the genomes of a broad range of bacterial species, and the orthologs of E. coli and H. influenzae are also DNA-binding proteins (Cooley,A.E. et al., submitted for publiction). Thus, characterization of B. burgdorferi EbfC impacts upon the entire kingdom Eubacteria. It also appears that EbfC family members interact with DNA through a unique fold, and represent a novel type of DNA-binding protein.
FUNDING
US National Institutes of Health (grant R01-AI044‱254 to B.S. and R01-GM070‱662 to M.F.); National Institutes of Health Training Grant in Microbial Pathogenesis T32-AI49‱795 (to S.R.); University of Kentucky Graduate School Dissertation Year Fellowship (to S.R.). Funding for open access charge: US National Institutes of Health (grant R01-AI044254).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This work is dedicated to Prof. Martin Freundlich, in gratitude for his many contributions to the study of bacterial DNA-binding proteins. We thank Sherwood Casjens, Jeffery Ebersole, Osnat Herzberg, Charlotte Kaetzel, Richard Marconi, Karen Novak, Phillip Stewart, Susan Straley, Ashutosh Verma and Michael Woodman for assistance during this work.
REFERENCES
- 1.Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K, Riley SP. The Lyme disease spirochete Erp protein family: structure, function and regulation of expression. In: Cabello FC, Godfrey HP, Hulinska D, editors. Molecular Biology of Spirochetes. Amsterdam: IOS Press; 2006. pp. 354–372. [Google Scholar]
- 2.Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty PS, Akins D, Meri S. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 2002;169:3847–3853. doi: 10.4049/jimmunol.169.7.3847. [DOI] [PubMed] [Google Scholar]
- 3.Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, Bykowski T, Stevenson B. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 2008;298(Suppl. 1):257–267. doi: 10.1016/j.ijmm.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, Wallich R, Zipfel PF, Kraiczy P, Stevenson B. The Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 2009;77:300–306. doi: 10.1128/IAI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. Microbiology. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haupt K, Kraiczy P, Wallich R, Brade V, Skerka C, Zipfel PF. FHR-1, an additional human plasma protein, binds to complement regulator-acquiring surface proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 2008;298(Suppl. 1):287–291. [Google Scholar]
- 7.Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä IJT, Meri S. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- 8.Kraiczy P, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF, Wallich R. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 2003;33:697–707. doi: 10.1002/eji.200323571. [DOI] [PubMed] [Google Scholar]
- 9.Metts MS, McDowell JV, Theisen M, Hansen PR, Marconi RT. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 2003;71:3587–3596. doi: 10.1128/IAI.71.6.3587-3596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 2002;70:491–497. doi: 10.1128/IAI.70.2.491-497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggers CH, Samuels DS. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 1999;181:7308–7313. doi: 10.1128/jb.181.23.7308-7313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson B, Tilly K, Rosa PA. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevenson B, Zückert WR, Akins DR. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. In: Saier MH, García-Lara J, editors. The Spirochetes: Molecular and Cellular Biology. Oxford: Horizon Press; 2001. pp. 87–100. [Google Scholar]
- 15.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 16.Babb K, McAlister JD, Miller JC, Stevenson B. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 2004;186:2745–2756. doi: 10.1128/JB.186.9.2745-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babb K, Bykowski T, Riley SP, Miller MC, DeMoll E, Stevenson B. Borrelia burgdorferi EbfC, a novel, chromosomally-encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 2006;188:4331–4339. doi: 10.1128/JB.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim K, Tempczyk A, Parsons JF, Bonander N, Toedt J, Kelman Z, Howard A, Eisenstein E, Herzberg O. Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR. Proteins. 2003;50:375–379. doi: 10.1002/prot.10297. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagensis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 21.Adams CA, Fried MG. Analysis of protein-DNA equilibria by native gel electrophoresis. In: Schuck P, editor. Protein Interactions: Biophysical Approaches for the Study of Complex Reversible Systems. New York: Springer; 2007. pp. 417–446. [Google Scholar]
- 22.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 23.Fried MG, Crothers DM. Equilibria and kinetics of Lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fried MG, Kanugula S, Bromberg JL, Pegg AE. DNA binding mechanism of O6-alkylguanine-DNA alkyltransferase: stoichiometry and effects of DNA base composition and secondary structure on complex stability. Biochemistry. 1996;35:15295–15301. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 25.Munteanu MG, Vlahovicek K, Parthasaraty S, Simon I, Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci. 1998;23:341–346. doi: 10.1016/s0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA. Interactive electron-density map interpretation: from INTER to O. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2115–2125. doi: 10.1107/S0907444904023509. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Bacon DJ, Anderson WF. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 1988;6:219–220. [Google Scholar]
- 30.Merritt EA, Bacon DJ. Raster3D photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 31.Tourand Y, Korbryn K, Chaconas G. Sequence-specific recognition but position-dependent cleavage of two distinct telomeres by the Borrelia burgdorferi telomere resolvase, ResT. Mol. Microbiol. 2003;48:901–911. doi: 10.1046/j.1365-2958.2003.03485.x. [DOI] [PubMed] [Google Scholar]
- 32.Fried MG, Kanugula S, Bromberg JL, Pegg AE. DNA binding mechanism of O6-alkylguanine-DNA alkyltransferase: stoichiometry and effects of DNA base composition and secondary structure on complex stability. Biochemistry. 1996;35:15295–15301. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 33.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 34.Mandelkern M, Elias JG, Eden D, Crothers DM. The dimensions of DNA in solution. J. Mol. Biol. 1981;152:153–161. doi: 10.1016/0022-2836(81)90099-1. [DOI] [PubMed] [Google Scholar]
- 35.Anthonissen FM, De Kesel M, Hoet PP, Bigaignon GH. Evidence for the involvement of different genospecies of Borrelia in the clinical outcome of Lyme disease in Belgium. Res. Microbiol. 1994;145:327–331. doi: 10.1016/0923-2508(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 36.Balmelli T, Piffaretti JC. Association between different clinical manifestations of Lyme disease and different species of Borrelia burgdorferi sensu lato. Res. Microbiol. 1995;146:329–340. doi: 10.1016/0923-2508(96)81056-4. [DOI] [PubMed] [Google Scholar]
- 37.van Dam AP, Kuiper H, Vos K, Widjojokusumo A, de Jongh BM, Spanjaard L, Ramselaar ACP, Kramer MD, Dankert J. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin. Infect. Dis. 1993;17:708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- 38.Drlica K, Rouviére-Yaniv J. Histonelike proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavans J, Oppenheim A. DNA-protein interactions and bacterial chromosome architecture. Phys. Biol. 2006;3:R1–R10. doi: 10.1088/1478-3975/3/4/R01. [DOI] [PubMed] [Google Scholar]
- 40.Miller JC, Bono JL, Babb K, El-Hage N, Casjens S, Stevenson B. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 2000;182:6254–6258. doi: 10.1128/jb.182.21.6254-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch BJ, Bendich AJ. The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: comparison of Escherichia coli and spirochetes of the genus Borrelia. J. Bacteriol. 1997;179:2228–2237. doi: 10.1128/jb.179.7.2228-2237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luijsterburg MS, Noom MC, Wuite GJL, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Travers A, Muskhelishvili G. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J. Mol. Biol. 1998;279:1027–1043. doi: 10.1006/jmbi.1998.1834. [DOI] [PubMed] [Google Scholar]
- 44.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 45.Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;4:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 46.Jaffar Ali BM, Amit R, Braslavsky I, Oppenheim AB, Gileadi O, Stavans J. Compaction of single DNA molecules induced by binding of integration host factor (IHF) Proc. Natl Acad. Sci. USA. 2001;98:10658–10663. doi: 10.1073/pnas.181029198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Mouw KW, Rice PA. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol. Microbiol. 2007;63:1319–1330. doi: 10.1111/j.1365-2958.2007.05586.x. [DOI] [PubMed] [Google Scholar]
- 50.Kobryn K, Naigamwalla DZ, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 51.Tilly K, Fuhrman J, Campbell J, Samuels DS. Isolation of Borrelia burgdorferi genes encoding homologues of DNA-binding protein HU and ribosomal protein S20. Microbiology. 1996;142:2471–2479. doi: 10.1099/00221287-142-9-2471. [DOI] [PubMed] [Google Scholar]
- 52.Knight SW, Samuels DS. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J. 1999;18:4875–4881. doi: 10.1093/emboj/18.17.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freundlich M, Ramani N, Mathew E, Sirko A, Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol. Microbiol. 1992;6:2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- 54.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 55.von Lackum K, Ollison KM, Bykowski T, Nowalk AJ, Hughes JL, Carroll JA, Zückert WR, Stevenson B. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's mammal–tick infectious cycle. Microbiology. 2007;153:1361–1371. doi: 10.1099/mic.0.2006/003350-0. [DOI] [PubMed] [Google Scholar]
- 56.Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, Hu LT. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 2007;189:2653–2659. doi: 10.1128/JB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]