Abstract
The consensus sequence of p53 is repeated half sites of PuPuPuC(A/T)(A/T)GPyPyPy. GtAGCAttAGCCCAGACATGTCC is a 14-3-3σ promoter p53 regulation site; the first core sequence is CAttAG, and the second is CATG. Both mutants GtAGgAttAGCCCAGACATGTCC and GtAGCAttAGCCCAGACATcTCC can be activated by p53 as a 1.5-fold half site. The original p53 regulated site on the 14-3-3σ promoter is a whole site, and CATTAG is a functional core sequence. The p53-binding affinity and the activity of CATTAG were lower than for the mutant CATATG core sequence. Wild-type p53 acts as a tetramer to bind to the whole site; however, it also can bind to a half site by one of its dimers. Wild-type p53 can only bind to a half site with core sequence CATG but not to CATATG. The 1.5-fold half site or whole site with core sequence CATATG can be bound by wild-type p53. A p53 mutant, A344, forms dimeric p53; it can only bind to CATG, and not to CATATG. Therefore, tetrameric and dimeric p53 can bind to a two-base A/T gap core sequence, but only tetrameric p53 can bind to a four-base A/T gap core sequence.
INTRODUCTION
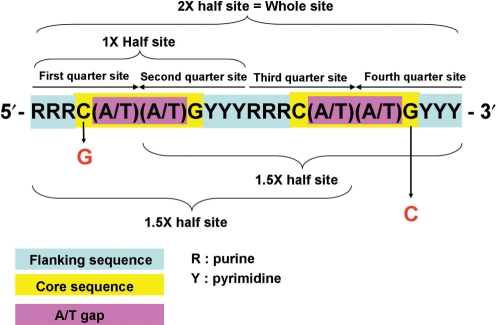
p53 is a key regulator of the cell cycle. After DNA damage, p53 can be activated to block cell-cycle progression and mediate multiple check points. The p53 target genes, p21 and 14-3-3σ, can be induced by activated p53 to arrest the G1-to-S phase and G2-to-M phase of the cell cycle, respectively (1,2). The p53 consensus sequence is 10-bp repeat of PuPuPuC(A/T)(A/T)GPyPyPy, separated by a spacer with up to 13 bases (3). C(A/T)(A/T)G is the core sequence, and the purine (pu) and pyrimidine (py) bases comprise the flanking sequence (Figure 1). The whole p53-binding sequence is the 10-base direct repeated sequence. Only 10 and 5 bases comprise the half site and quarter site of the p53-binding site, respectively. The p53-binding sequence with a half site plus a quarter site is equal to 1.5-fold of the p53 half site (4). A half site of the p53-binding site possesses p53-binding affinity (4); however, it can only be activated by p53 in a sequence-specific manner or with other required cofactors (5–7). About 1.5-fold of the p53 half site can be activated by p53, but its activity is much lower than for the whole p53-binding site (5,7). Both p21 and 14-3-3σ have two p53 consensus sequences on their promoter (8). The p21 promoter at 5′ site one (GAACATGTCCcAACATGTTg) and 3′ site two (GAagAAGaCTGGGCATGTCT) can be activated by p53. The 14-3-3σ promoter 3′ site two (GtAGCAttAGCCCAGACATGTCC) can be activated by p53 but not that at 5′ site one (AGGCATGTgCcAcCATGCCC) (9). However, the p21 and 14-3-3σ promoter p53-binding sites presented some mismatched bases (the lowercase letters note the mismatch bases). There are three mismatch bases in the flanking sequence of the p53-binding site in 14-3-3σ site one, but only one mismatch base in 14-3-3σ site two. There are two bases of the A/T gap in the core sequence of the p53-binding site in p21 site one and 14-3-3σ site one, but there are four bases in 14-3-3σ site two. Although the entire p53 consensus sequence comprises the whole p53-binding site, a 1.5-fold half site of the p53-binding site could be activated by p53, similar to p21 site two (7). In order to identify whether the p53-regulated site on the 14-3-3σ promoter is a whole site or a 1.5-fold half site, different mutant core sequences were cloned and tested for their functional activities. Interestingly, the p53-binding site on the 14-3-3σ promoter contained four bases of the A/T gap CATTAG core sequence. It differed from the p53 consensus by two bases of A/T gap CATG core sequence. To clarify how many bases of the A/T gap in the p53 core sequence are tolerant for p53-activating genes, basic promoter vectors with different A/T gap p53 response elements were constructed. The promoter activities and -binding affinities were measured in our experiments.
Figure 1.
The p53 consensus sequence. The purine (R) and pyrimidine (Y) bases comprise the flanking sequence. C(A/T)(A/T)G is the core sequence, and the (A/T)(A/T) within the core sequence is an A/T gap. The whole site is 20 bases with a ten-base direct repeat as a 2× half site, and each 10 bases are a 1× half site. Each five bases are quarter sites. A whole site with a core sequence C to G mutation within the first quarter site is a 1.5× half site, with a core sequence G to C mutation within the fourth quarter site is another 1.5× half site.
p53 binding to the whole p53 response element is tetrameric and it can bind to the p53 half site by one of its dimers. A dimeric form of p53 mutant p53A344 also can bind to the p53 half site (4). To understand the four-base A/T gap CATTAG p53 core sequence binding character to p53, a dimeric form of p53 mutant p53A344 and a tetrameric p53 were used in a DNA affinity purification assay (DAPA).
MATERIALS AND METHODS
Construction of reporter vectors with p53 response element sequence
Oligonucleotides (20–34 bp) with p53 response element sequences were synthesized and annealed to SmaI-digested pGL3 promoter vector (Promega). The vector and insert were ligated using a 1:4500 ratio at 22°C for 15 min then transformed at 37°C overnight. The positive clones were checked by DNA sequencing.
Luciferase activity assay
H1299 cells were grown in 12-well tissue culture dishes to 60% confluence and then cotransfected with 0.5 μg of pcDNA 3.0 (Invitrogen), pcDNA 3.0 p53, pcDNA 3.0 p53F46, or pcDNA 3.0 p53Δ364–393 in the presence of 0.5 μg of pGL3 promoter firefly luciferase (Promega) or pGL3 promoter with a p53 response element sequence and 10 ng of pCMV renilla luciferase plasmids (Promega). Twenty-four hours post-transfection, cells were harvested in 0.25 ml reporter lysis buffer and were subjected to a dual luciferase assay according to the manufacturer's protocol (Dual-Luciferase Reporter Assay System, Promega). Firefly luciferase activity was normalized to renilla luciferase activity and data are presented as the mean ± SD of three independent experiments, each performed in triplicate.
P53 mutants expression vector construct
The p53F46, p53Δ364–393, p53A344Δ364–393 and p53A344 point mutations or deletion clones were created by site-directed mutagenesis according to the manufacturer's instructions (Phusion Site-Directed Mutagenesis Kit, Finnzymes). p53F46 was created using the following primers: TCCCGGACGATATTGAACAATGG and ACAGCATCAAATCATCCATTGCTTGG. p53Δ364-393 was created using the following primers: TGAGGATCCACTAGTAACGGCCGC and CCTGCTCCCCCCTGGCTC. p53A344Δ364–393 and p53A344 were created using the following primers: CAAGGCCTCATTCGCCTCTCGGAACATC and GAACTCAAGGATGCCCAGGCTGG. All constructs were checked by DNA sequencing.
DNA affinity purification assay
Nuclear extract (100 μg) was prepared from 5 × 106 H1299 cells transfected with 16 μg of p53Δ364–393, p53A344Δ364–393 or p53A344 plasmid. The nuclear extracts were precleared by incubation with 40 μl streptavidin–agarose beads (4%) with a 50% slurry at 4°C for 1 h with rotation. They were then centrifuged at 4000 × g for 1 min at 4°C. The supernatant was collected as the precleared nuclear extract. The binding reaction was performed by mixing the precleared nuclear extract proteins with the annealing probe (0.3 nmol), 5× binding buffer [100 mM HEPES; pH 7.6, 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM DTT, 1% Tween 20 (w/v), 150 mM KCl], 10 μl sonicated salmon sperm DNA (1 μg/μl) and the 40 μl streptavidin–agarose beads (4%) with a 50% slurry in 1 ml. This was incubated at room temperature for 1 h with rotation. The beads were washed three times with 1× PBS. The binding proteins were eluted with 20 μl of 5× loading dye, then separated by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS–PAGE) followed by western blot analysis, and were probed with p53 antibody (p53 Ab-2, Lab Vision). Nuclear extract (10 μg) was mixed with 20 μl 5× loading dye as the 10% ‘input’ sample. To differentiate binding affinities of various p53 proteins to the p53 response element, either the low or high dosage of nuclear extract was used in the DAPA assay. The 30 or 300 μg of nuclear extract was used in each binding reaction, with 10 μg of nuclear extract used as an ‘input’ loading control, which was 33% or 3.3% of the sample, respectively.
RESULTS
The original p53-regulated site on the 14-3-3σ promoter is a p53 whole site response element, and CATTAG is a functional core sequence
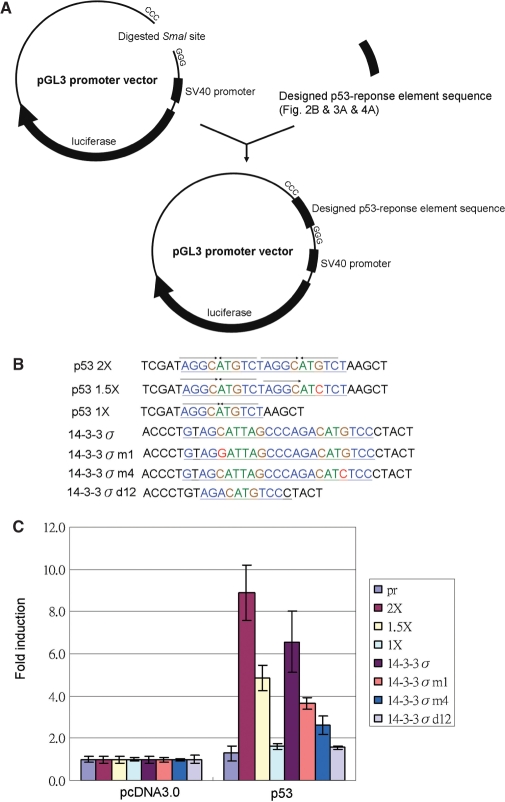
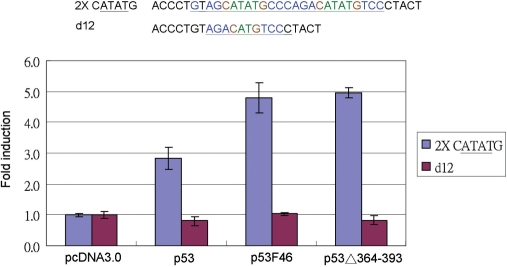
Designed p53 response element sequences were constructed in the pGL3 promoter digested by SmaI (Figure 2A). These designed p53 response element sequences with core sequence mutants are listed in Figure 2B. The whole p53 site is as a perfect match sequence for the p53 consensus sequence (p53 2×), and the fourth quarter with a TcTCT mutation is a 1.5-fold half site of the p53-binding site (p53 1.5×). Deletion of the first and second quarter produces a half site of the p53-binding site (p53 1×). After cotransfection with the p53 expression vector, the reporter activity of the whole site (p53 2×) is higher than that of the 1.5-fold half site (p53 1.5×), but there is no activity in a half site (p53 1×) construct compared to that for the pGL3 promoter vector only (pr) (Figure 2C). The p53 response element on the 14-3-3σ promoter, its first half site with a four-base A/T gap in the core sequence is different from the common p53 core sequence with a two-base A/T gap (2,7), may have no function. We supposed that the full 14-3-3σ p53 response element sequence is a 1.5-fold half site similar to that of p53 1.5×. Therefore, the first-quarter mutation (14-3-3 m1) activities were the same as for the full 14-3-3σ sequence, and the fourth-quarter mutation (14-3-3 m4) had no activity. The promoter activity of the full 14-3-3σ p53 response element sequence is higher than 14-3-3 m1. The 14-3-3 m1 is also higher than 14-3-3 m4 activity. The first- and second-quarter double-deleted mutants 14-3-3 d12 is no promoter activity at all (Figure 2C). Therefore, the original p53-regulated site on the 14-3-3σ promoter acts as a whole site, with CATTAG as a functional core sequence.
Figure 2.
The 14-3-3σ promoter p53 response element activity assay. (A) Double strands of oligonucleotides with designed p53 response element sequences were cloned to the SmaI site of the pGL3 promoter vector with a luciferase reporter gene. (B) Designed p53 response element sequences. p53 2×: p53 whole site; p53 1.5×: 1.5-fold p53 of half site with fourth-quarter site mutation; p53 1×: p53 half site with deletion of first and second quarters; 14-3-3σ: p53 response element of 14-3-3σ promoter; 14-3-3σm1: first-quarter mutation; 14-3-3σm4: fourth-quarter mutation; 14-3-3σd12: first- and second-quarter deletions. (C) Designed promoters were cotransfected with p53 expression vectors into H1299 cells, and the folds of luciferase promoter activity compared to the pcDNA3.0 as 1. The activity of p53 2× is higher than p53 1.5×, and the activity of p53 1× is similar to pGL3 promoter vector only (pr). The p53 induced activity pattern of 14-3-3σ promoter constructs is the highest from 14-3-3σ > 14-3-3σm1 > 14-3-3σm4. There was no activity in the 14-3-3σd12 mutant construct.
Four-base A/T gap acts as p53 core sequence response element
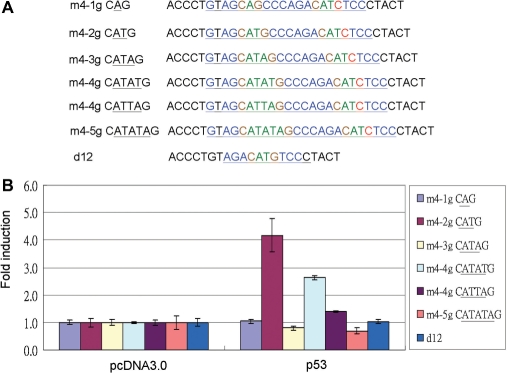
To evaluate if the A/T base gap in the p53 core sequence can be a functional core sequence of the p53 response element. One to five A/T base gaps were synthesized and cloned into 1.5-fold half sites (Figure 3A). The promoter activities were examined by luciferase activity assay. Only two- and four-base gaps can be activated by p53 (Figure 2B). When comparing two-base A/T gap core sequences with four-base A/T gaps, the activities were higher in two-base A/T gaps than in four-base A/T gaps. However, the 4 A/T base gap CATATG activity was higher than CATTAG (Figure 3B).
Figure 3.
The promoter activity in various A/T gap core sequences of p53 promoter response element. (A) Designed various A/T gap core sequence p53 response elements. m4-1g CAG: fourth quarter-site mutation and first half-site core sequence CAG. m4-2g CATG, m4-3g CATAG, m4-4g CATTAG, m4-4g CATATG, m4-5g CATATAG and d12 as negative control as in Figure 2B. (B) Designed various A/T gap core sequences of p53 promoter response element constructs were cotransfected with p53 expression vector into H1299 cells, and the luciferase promoter activity was compared to pcDNA3.0 as 1. The activity order is: m4-2g CATG > m4-4g CATATG > m4-4g CATTAG. No activity could be detected in m4-1g CAG, m4-3g CATAG, m4-5g CATATAG or d12.
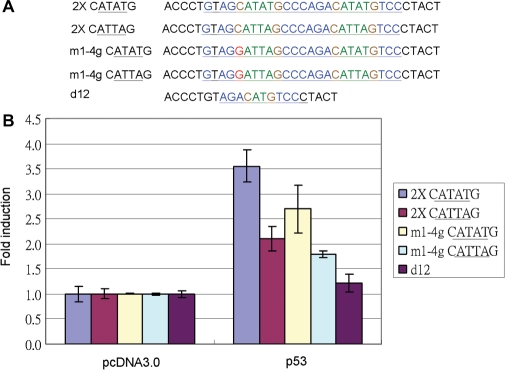
The whole p53 response element sequence activation by p53 was higher than for the 1.5-fold half site (5). Is the four-base A/T gap core sequence as functional in the whole site as in the 1.5-fold half site as previously demonstrated? The four-base A/T gap mutants in the whole p53 response element sequence were cloned, and the promoter activity was examined by a luciferase activity assay (Figure 4A). The results indicated that the core CATATG activity activated by p53 is higher than for CATTAG in both the whole site and the 1.5-fold half site (Figure 4B). Two super p53 mutants, p53F46 and p53Δ364–393 (10,11), also can activate the p53 whole site with the CATATG core sequence, even more so than wild-type p53 (Figure 5).
Figure 4.
Promoter activity assay for four-base A/T gap core sequence with p53 2X and 1.5X-fold p53 half site. (A) Designed four-base A/T gap core sequence p53 response element constructs and its mutants: 2× CATATG: whole site with CATATG core sequence; 2× CATTAG: whole site with CATTAG core sequence; m1-4g CATATG: mutation of first-quarter site with CATATG core sequence; m1-4g CATTAG: mutation of first-quarter site with CATTAG core sequence; and d12 is a negative control. (B) Designed four-base A/T gap core sequence p53 response element and mutant promoter constructs were co-transfected with p53 expression vectors into H1299 cells. The folds induction of luciferase promoter activity compared to the pcDNA3.0 as 1. Both 2- and 1.5-fold half-site core sequences of CATATG promoter activity were higher than CATTAG core sequence.
Figure 5.
Super p53 mutant activation of four-base A/T gap core sequences of p53 response element. Super p53 expression vector p53F46 or p53Δ364-393 was co-transfected with four-base A/T core sequence p53 promoter response elements into H1299 cells, and the folds of luciferase promoters activity induction compared to the pcDNA3.0 as 1. Both super p53 mutants can activate with higher activity than wild-type p53 at the four-base A/T gap core sequence of p53 promoter response element.
Tetrameric and dimeric p53 can bind to the two-base A/T gap core sequence, but only tetrameric p53 can bind to the four-base A/T gap core sequence
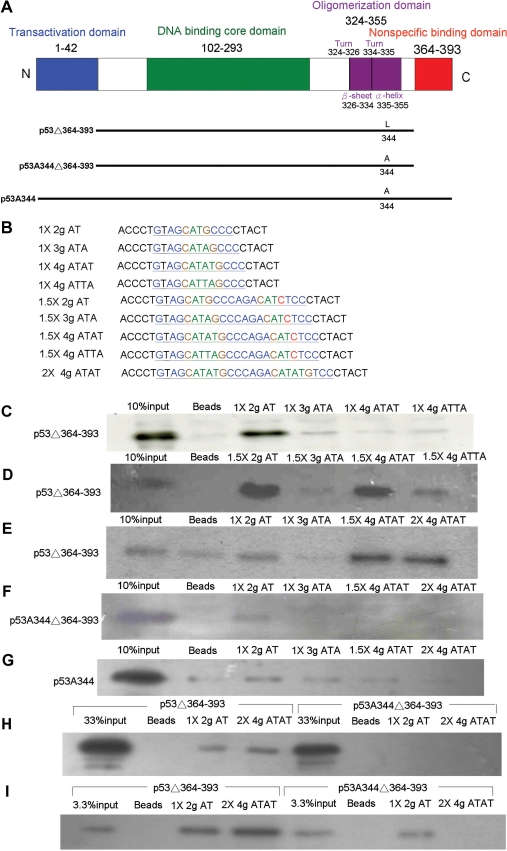
By using DAPA to investigate the p53-binding character to a two-base A/T core sequence and to a four-base A/T core sequence (Figure 6B), three p53 mutants, p53Δ364–393, p53A344Δ364–393 and p53A344, were constructed and expressed in H1299 cells (Figure 6A). The p53Δ364–393 mutant, consisting of the last 30 amino acids of a C-terminal deletion p53 mutant, avoided non-specific binding and sliding-off effects (5,11). First, 5′-labeled biotin probes with 2–4 gaps of A/T bases in a p53 half-site sequence were used to examine the binding character of p53Δ364–393. The results indicated that p53Δ364–393 can only bind to two-base-gap CATG core sequences but not to three-base-gap CATAG, four-base-gap CATATG, or CATTAG core sequences (Figure 6C). Surprisingly, with probes with two to four gaps of A/T bases in a 1.5-fold p53 half-site sequence, p53Δ364-393 not only can bind to two-base-gap CATG, but also can bind to four-base-gap CATATG or CATTAG core sequences (Figure 6D). However, p53Δ364–393 cannot bind to a three-base-gap CATAG core sequence. The binding affinity patterns in DAPA correlated to the function assay of the sequence activated by p53 (Figures 3B and 6D). The p53-binding affinity and activity of two four-base-gap CATATG and CATTAG core sequences were different; CATATG-binding affinity and activity was higher than for CATTAG (Figures 3B and 6D). p53Δ364–393 can bind two-base-gap CATG in both p53 half sites and 1.5-fold half sites as probes, but it can only bind to four-base-gap CATATG or CATTAG core sequences in 1.5-fold p53 half sites, not to p53 half sites (Figure 6C and D). p53Δ364–393 can also bind to a p53 whole site with four-base A/T gaps in each core sequence (Figure 6E). To evaluate the binding-pattern discrepancy between two-base-gap CATG to four-base-gap CATATG, perhaps due to dimeric or tetrameric-binding character differences, dimeric p53A344Δ364–393 and p53A344 were used (4,12). The DAPA results show that both dimeric p53 mutants, p53A344 and p53A344Δ364–393, can only bind to two-base-gap CATG core sequences in a p53 half site, but cannot bind to four-base-gap CATATG core sequences in a 1.5-fold or 2-fold half site (Figure 6F and G). Because of weak binding of dimeric p53A344Δ364–393 to the two-base-gap CATG core sequences in a p53 half site (Figure 6F), different dosages of nuclear extract were used in the DAPA reaction to compare the affinity of the dimeric and the tetrameric p53 binding with two- or four- base-gap CATG core sequences. In the low-dosage DAPA assay, dimeric p53A344Δ364–393 was not detected in two-base-gap CATG core sequences as a p53 half site, but tetrameric p53Δ364–393 bound both to two-base-gap CATG core sequences in a p53 half site and four-base-gap CATATG core sequences in a p53 whole site (Figure 6H). In the high-dosage DAPA assay, dimeric p53A344Δ364–393 bound to two-base-gap CATG core sequences in a p53 half site but could not bind to four-base-gap CATATG core sequences in 2-fold half site (Figure 6I). According to the results, we believed that the four-base A/T gap can be bound only by tetrameric p53, but not by dimeric p53.
Figure 6.
p53-binding character to various A/T core sequences of p53 response element by DAPA assay (A) Diagrams of the p53 protein structure and oligomerization mutant constructs: There are four major functional domains within 393 amino-acid residues; N-terminal transactivation domain, middle DNA-binding core domain, C-terminal oligomerization and non-specific binding domains. p53Δ364–393: tetrameric form of p53 with non-specific binding domain deletion. p53A344Δ364–393: dimeric form of p53 with non-specific binding domain deletion. p53A344: dimeric form p53. (B) Oligomerization mutants of p53 expression vectors were transfected into H1299 cells. After 48 h incubation, the nuclear proteins were extracted and incubated with various probes in a DAPA assay. Various A/T core sequences with p53 response element designed primers as probes. One hundred micrograms of nuclear extracts were used in C, D, E, F and G; 30 μg of nuclear extracts were used in H; 300 μg of nuclear extracts were used in I. All of the input samples were 10 μg of nuclear extract as 10% loading control in C, D, E, F and G; 33% in H; 3.3% in I. (C) The tetrameric form of p53Δ364–393 can bind to two-base A/T gap CATG core sequences but cannot bind to three-base A/T gap CATAG, four-base A/T gap CATATG, or CATTAG core sequences when the probes as the half site. (D) The tetrameric form of p53Δ364–393 can bind to two-base A/T gap CATG, four-base A/T gap CATATG and CATTAG core sequences but cannot bind to three-base A/T gap CATAG core sequences when the probe was the 1.5-fold half site. Among four-base A/T gap core sequences, the p53-binding affinity in CATATG was higher than CATTAG. (E) The tetrameric form of p53Δ364–393 can bind to four-base A/T gap CATATG core sequences when the probes were the 1.5-fold half site or the 2-fold half site. (F) The dimeric form of p53A344Δ364–393 can bind to the two-base A/T gap CATG core sequence when the probe was the half site but cannot bind to four-base A/T gap CATATG core sequences even with probes as the 1.5-fold half site or 2-fold half site. (G) The other p53 mutant, dimeric form of p53A344, can bind to two-base A/T gap CATG core sequences when the probe as the half site. (H) Tetrameric p53Δ364–393 can bind to two-base A/T gap CATG in a half site or four-base A/T gap CATATG in a 2-fold half site, but dimeric p53A344Δ364–393 can not be detected in all reactions with low nuclear extract samples. (I) Both tetrameric p53Δ364–393 and dimeric p53A344Δ364–393 can bind to two-base A/T gap CATG when the probe was a half site. However, tetrameric p53Δ364–393 can bind to four-base A/T gap CATATG when the probe was a 2-fold half site, but dimeric p53A344Δ364–393 lost it binding affinity at all even reaction with high nuclear extract sample.
DISCUSSION
Based on the functional assay and DAPA results, except for the two-base A/T gap core sequence of the p53 response element, the four-base A/T gap core sequence also acts as a p53 response element. The p53 target gene, the 14-3-3σ core sequence of p53 response element GtAGCATTAGCCCAGACATGTCC, is thought to be a whole p53 response element. We tested two four-base A/T gap variant core sequences of p53 response elements, CATATG and CATTAG. Both p53-binding affinity and activity of CATATG were higher than for CATTAG (Figures 3B, 4B and 6D). It has been demonstrated that two-base A/T gap core sequences in different A/T arrays had different responses for p53 activation (13). The p53-induced core sequence activity with AT or TA was higher than for AA or TT. It may explain how the four-base A/T gap CATATG activity was higher than for CATTAG in our experiments. However, A/T arrays in the core sequence with four-base gaps may have 16 variants; different A/T arrays in the core sequence with four-base gaps may have different p53 responses. It needs further investigation.
Sauer et al. (11) reported p53 binding to p53 response elements with the electrophoretic mobility shift assay (EMSA). An oligonucleotide probe less than 66 bp may cause non-specific binding and sliding-off to reduce specific binding. Once the C-terminal 30 amino acids of p53 were deleted, the specific binding affinity was recovered (11). For DAPA, from our designed probes of 20–34 bp (Figure 5A) that were used to examine the binding character of 2–4-base A/T gap core sequences of p53 response elements, the 30 amino-acid C-terminal deletion mutant, p53Δ364–393, was chosen. Tetrameric p53 can bind to two-base A/T gap core sequences of p53 at half sites and 1.5-fold half sites, but it can only bind to four-base A/T gap core sequences of p53 1.5-fold half sites and whole sites, and not to half sites alone (Figure 6C, D and E). The wild-type p53 normally bound to two-base A/T core sequences of p53 2× half-site response elements in a tetrameric manner (4,5). p53 can bind to two-base A/T core p53 half-site sequences by one of its dimers and can leave the other dimer free (4), but this binding pattern was not observed in four-base A/T core sequence p53 half sites. Three tetramerization-domain-deleted monomeric p53 mutants can bind to two-base A/T core sequence 1.5-fold p53 half sites (5). We proposed that tetrameric wild-type p53 can bind to 1.5-fold p53 half sites by three of its monomers and can leave the other monomer free in both two-base and four-base A/T core sequence p53 1.5× half sites. For further characterization of the four-base A/T gap core sequence of p53 response elements at 1.5× half sites or whole sites that can only bind tetrameric p53 (Figure 6D and E), dimeric p53 mutant p53A344 and p53A344Δ364–393 were used in DAPA. Neither dimeric p53 mutant can bind to four-base A/T gap p53 response elements at 1.5-fold half sites and whole sites (Figure 6F and G), its different from two-base A/T gap core sequence p53 half sites can bind dimeric p53 mutants (Figure 6F and G). Therefore, dimeric p53 can only bind to two-base but not to four-base A/T gap p53 response elements.
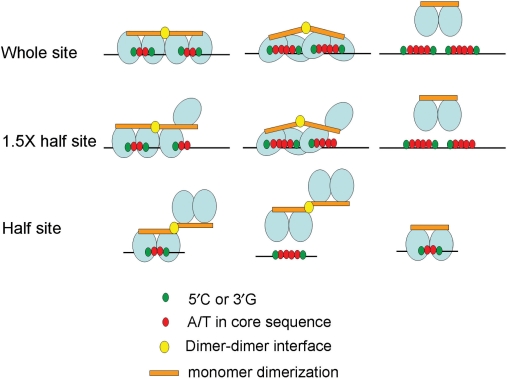
p53 is composed of an N-terminal transactivation domain, a DNA-binding core domain and a C-terminal oligomerization domain (Figure 6A) (14). The oligomerization domain is composed of a turn followed by a β-sheet, a turn and an α-helix (15,16). The β-sheet has a monomer dimerization motif, and the α-helix has a dimer–dimer interface motif. The oligomerization domain of p53 is also involved in DNA binding to the supercoil DNA and enhances core domain DNA-binding affinity (17,18). In our experiments, the dimeric p53A344Δ364–393 mutant bound to two-base gap core sequences of the p53 response element half site, but the affinity was much lower than for tetrameric p53Δ364–393 (Figure 6I; 1 × 2 g AT compares to input). It is indicated that the α-helix dimer–dimer interface of the p53 oligomerization domain enhanced core domain DNA binding. With an extended core sequence from two-base A/T gaps to four-base A/T gaps, not dimeric p53, but only tetrameric p53 can bind to both 1.5-fold p53 half sites and p53 whole sites (Figure 6E, F and G). Therefore, the α-helix dimer–dimer interface of the p53 oligomerization domain seems necessary for binding to four-base A/T core sequences of p53 response elements in both 1.5-fold p53 half sites and p53 whole sites (Figure 7).
Figure 7.
Model for p53 protein molecule-binding pattern to various A/T gap core sequences of the p53 response element. Wild-type p53 binds to a canonical p53 response element at the two-base A/T gap core sequence p53 whole site in a tetrameric manner, and each of the dimers binds to each p53 half site. Tetrameric wild-type p53 binds to two-base A/T gap core sequence at 1.5-fold p53 half sites by three of its monomers and leaves the other monomer free. p53 binds to two-base A/T gap core sequence p53 half-site core sequences by one of its dimers and leaves the other dimer free. Other binding patterns of p53 to a non-canonical p53 response element at four-base A/T gap core sequence of p53 response elements are similar to those of the two-base A/T gap core sequence in both the p53 whole site and 1.5-fold half site, but p53 cannot bind to the four-base A/T gap core sequence p53 half site. However, dimeric p53, p53A344,disrupting the dimer–dimer interface, can bind to two-base A/T gap core sequence p53 half sites, but not to four-base A/T gap core sequence p53 whole sites or 1.5-fold half sites.
According to McLure and Lee (4), the dissociation constant (Kd) of tetrameric or dimeric p53 binding to the p53 whole site or half site varied. The binding Kd pattern was: tetrameric to whole site < dimeric to whole site < tetrameric to half site < dimeric to half site (4). Therefore, the highest p53-binding affinity was tetrameric binding to the whole site, followed by dimeric binding to the whole site, tetrameric binding to the half site, and the lowest p53-binding affinity is dimeric binding to the half site. There were similar results in our DAPA assay; the binding affinity of tetrameric p53Δ364–393 was higher than dimeric p53A344Δ364–393 bound to two-base A/T gap core sequence of the p53 half site (Figure 6I). Once the four-base A/T gap core sequence was substituted for the two-base A/T gap core sequence in the whole site, it could be bound only by tetrameric p53Δ364–393, but not by dimeric p53A344Δ364–393 (Figure 6I). Therefore, the integrity of the tetrameric state of p53 is necessary for p53 binding to the four-base A/T gap core sequence. It has been indicated that several other factors in cells can associate with the p53 oligomerization domain to affect the tetramerization state of p53. S100 family proteins and apoptosis repressors with caspase recruitment domains (ARC) can associate with the p53 oligomerization domain to inhibit p53 tetramerization and to repress the trans-activation function of p53 (19–23). The 14-3-3 family proteins can also associate with the p53 oligomerization domain to enhance p53 tetramerization (24). Therefore, the 14-3-3 family, S100 family and ARC may be important regulators for influencing the affinity and activity of p53 through the core sequence with a four-base A/T gap.
We identified a non-canonical p53 response element and found that a four-base A/T gap core sequence can be activated by p53. Also, the different A/T arrays in a four-base A/T gap core sequence may affect the p53-binding affinity and activation function. We also clarified that the p53-binding character for four-base A/T gap core sequence is different from the canonical p53 response element.
FUNDING
This work was supported by the Shin Kong Wu Ho-Su Memorial Hospital [SKH-8302-97-DR-11 to C. C.-F. and L. H.-C.]. This work was also supported in part by the C.Y. Foundation for Advancement of Education, Sciences and Medicine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We greatly appreciate Dr Li-Jung Juan and Dr Sheau-Yann Shieh for providing technical assistance and supplying plasmids.
REFERENCES
- 1.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 2.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3σ Is a p53-Regulated Inhibitor of G2/M Progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.McLure KG, Lee PW. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Schwedes JF, Parks D, Mann K, Tegtmeyer P. Interaction of p53 with its consensus DNA-binding site. Mol. Cell Biol. 1995;15:2157–2165. doi: 10.1128/mcb.15.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez D, Inga A, Snipe J, Krysiak O, Schönfelder G, Resnick MA. single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol. Cell Biol. 2007;27:2590–2600. doi: 10.1128/MCB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan JJ, Menendez D, Inga A, Nourredine M, Bell D, Resnick MA. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura Y, Futamura M, Kamino H, Yoshida K, Nakamura Y, Arakawa H. Identification of p53–46F as a super p53 with an enhanced ability to induce p53-dependent apoptosis. Cancer Sci. 2006;97:633–641. doi: 10.1111/j.1349-7006.2006.00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer M, Bretz AC, Beinoraviciute-Kellner R, Beitzinger M, Burek C, Rosenwald A, Harms GS, Stiewe T. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008;36:1900–1912. doi: 10.1093/nar/gkn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterman JL, Shenk JL, Halazonetis TD. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor–DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 15.Clore GM, Omichinski JG, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn AM. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994;265:386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 16.Clubb RT, Omichinski JG, Sakaguchi K, Appella E, Gronenborn AM, Clore GM. Backbone dynamics of the oligomerization domain of p53 determined from 15N NMR relaxation measurements. Protein Sci. 1995;4:855–862. doi: 10.1002/pro.5560040505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazdova M, Palecek J, Cherny DI, Billova S, Fojta M, Pecinka P, Vojtesek B, Jovin TM, Palecek E. Role of tumor suppressor p53 domains in selective binding to supercoiled DNA. Nucleic Acids Res. 2002;30:4966–4974. doi: 10.1093/nar/gkf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rippin TM, Freund SM, Veprintsev DB, Fersht AR. Recognition of DNA by p53 core domain and location of intermolecular contacts of cooperative binding. J. Mol. Biol. 2002;319:351–358. doi: 10.1016/S0022-2836(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 19.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc. Natl Acad. Sci. USA. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Blake M, Tang C, Zimmer D, Rustandi RR, Weber DJ, Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl Acad. Sci. USA. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Fernandez MR, Rutherford TJ, Fersht AR. Members of the S100 family bind p53 in two distinct ways. Protein Sci. 2008;17:1663–1670. doi: 10.1110/ps.035527.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, Ashton AW, Fu W, Mani K, Chin SF, Provenzano E, Ellis I, Figg N, Pinder S, Bennett MR, Caldas C, Kitsis RN. Regulation of p53 tetramerization and nuclear export by ARC. Proc. Natl Acad. Sci. USA. 2007;104:20826–20831. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36:5983–5991. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]