Abstract
DNA polymerase θ (Pol θ) is a low-fidelity DNA polymerase that belongs to the family A polymerases and has been proposed to play a role in somatic hypermutation. Pol θ has the ability to conduct translesion DNA synthesis opposite an AP site or thymine glycol, and it was recently proposed to be involved in base excision repair (BER) of DNA damage. Here, we show that Pol θ has intrinsic 5′-deoxyribose phosphate (5′-dRP) lyase activity that is involved in single-nucleotide base excision DNA repair (SN-BER). Full-length human Pol θ is a ∼300-kDa polypeptide, but we show here that the 98-kDa C-terminal region of Pol θ possesses both DNA polymerase activity and dRP lyase activity and is sufficient to carry out base excision repair in vitro. The 5′-dRP lyase activity is independent of the polymerase activity, in that a polymerase inactive mutant retained full 5′-dRP lyase activity. Domain mapping of the 98-kDa enzyme by limited proteolysis and NaBH4 cross-linking with a BER intermediate revealed that the dRP lyase active site resides in a 24-kDa domain of Pol θ. These results are consistent with a role of Pol θ in BER.
INTRODUCTION
DNA sequences homologous to the mus308 gene of Drosophila melanogaster were identified in human genome database, and the corresponding cDNA was cloned and mapped to chromosome 3q13.31. As the partial human cDNA encoded a 1762-amino-acid DNA polymerase with a molecular mass of 198 kDa, this new polymerase was designated DNA polymerase θ (Pol θ), and the corresponding vertebrate locus was named POLQ (1). Subsequently, a full-length cDNA for POLQ was cloned and shown to encode a 2592-amino-acid polypeptide with an amino-terminal helicase domain, a carboxy-terminal polymerase domain and an intervening spacer region (2). The mammalian POLQ and Drosophila mus308 gene products have similar domain arrangements. In addition to these orthologs, vertebrates have paralogs of the POLQ gene, HEL308 and POLN. The HEL308 gene product has homology to the helicase domain of POLQ but does not have a polymerase domain, whereas the POLN gene product has homology to the polymerase domain of POLQ, but does not contain a helicase domain (3,4).
DNA polymerase θ belongs to the family A polymerases (2,5) typified by Escherichia coli Pol I (6). Biochemical studies showed that purified full-length human Pol θ exhibited template-directed DNA polymerase activity on nicked double-stranded DNA and on a single-primed DNA template. Consistent with family A DNA polymerases, this activity was resistant to aphidicolin and inhibited by dideoxynucleotides. In addition, Pol θ exhibited a single-stranded DNA-dependent ATPase activity (2). Fidelity measurements of human Pol θ revealed that the polymerase generates single base pair substitutions at a rate 10- to 100-fold higher than other characterized family A DNA polymerases, making it one of the least faithful member of the family A DNA polymerases (7).
Pol θ is capable of conducting translesion DNA synthesis by inserting bases opposite an AP site or thymine glycol residue in the template strand. It also can extend an unpaired primer base opposite these lesions (8). Thus, Pol θ is sufficient for both the insertion and extension steps required for bypass of an abasic site. In contrast, Pol θ cannot insert bases opposite a cyclobutane pyrimidine dimer or a (6–4) photoproduct, but is able to extend primers after DNA polymerase ι had inserted a base opposite these UV-induced photoproducts (9). A role for Pol θ in somatic hypermutation (SHM) of immunoglobulin genes has been proposed where the excision of uracil by uracil–DNA glycosylase (UDG) during SHM generates an apurinic/apyrimidinic (AP) site that is repaired by either translesion bypass or a base excision repair (BER) process involving several DNA polymerases and accessory proteins (10–14). Recently, Takeda and associates (15) suggested that Pol θ and Pol β are involved in BER of DNA damage in DT40 cells. They reported that POLQ−/− cells exhibited no significant increase in sensitivity to methyl methanesulfonate (MMS) or HmdUrd, but deletion of the genes for both POLQ and Pol β resulted in cells more sensitive to these base-damaging agents. Furthermore, subcellular localization studies demonstrated that Pol θ is rapidly recruited to sites of base damage that are induced by laser-delivered irradiation (15). These data along with extract-based in vitro repair assays suggested that Pol θ participates in BER in mammalian cells under certain conditions.
To understand the potential involvement of Pol θ in BER in mammalian cells, we cloned, overexpressed and purified to near homogeneity the C-terminal 98-kDa region of Pol θ, corresponding to the core polymerase domain. We also prepared a polymerase-deficient variant of the enzyme by site-directed mutagenesis, and assessed activities associated with BER. Our results reveal that the Pol θ polymerase domain possesses both 5′-deoxyribose phosphate (dRP) lyase and DNA polymerase activities that function together during single-nucleotide BER in vitro. The 5′-dRP lyase activity was also fully active in the polymerase-deficient mutant. Our observation that Pol θ exhibits DNA polymerase and 5′-dRP lyase activities in vitro lends credence to the argument that Pol θ may have a role in BER in vivo.
MATERIALS AND METHODS
Materials
Synthetic oligodeoxyribonucleotides were from Oligos Etc, Inc. (Wilsonville, OR, USA) and The Midland Certified Reagent Co. (Midland, TX, USA). [α-32P]dCTP and [α-32P]ddATP (3000 Ci/mmol), and [γ-32P]ATP (7000 Ci/mmol) were from GE HealthCare (Piscataway, NJ, USA) and Biomedicals (Irvine, CA, USA), respectively. Optikinase and terminal deoxynucleotidyl transferase were from USB Corp. (Cleveland, OH, USA) and Fermentas Inc. (Hanover, MD, USA), respectively. Protease inhibitor complete (EDTA-free) was from Roche Molecular Diagnostics (Pleasanton, CA, USA). Leupeptin, aprotinin and phenylmethylsulfonyl fluoride were from Calbiochem (La Jolla, CA, USA). Recombinant human Pol β was overexpressed and purified as described previously (16). Human AP endonuclease (APE), uracil-DNA glycosylase (UDG) with 84 amino acids deleted from the amino terminus and DNA ligase I were purified as described previously (17–19).
Overproduction and purification of the Pol θ polymerase domain
Based on the cDNA sequence of human Pol θ (1,2), a 2.7-kb cDNA clone was amplified for overexpression and production of a 98-kDa C-terminal fragment of human Pol θ using a 5′-BamH1 primer: 5′-TGC CAA TCA TGA TGG ATC CTC ATC CCT CTT ACC-3′ and a 3′-Sal1 primer: 5′-CTC TGT TCT TTG CAG TCG ACT GCA TCT GCA C-3′. The amplified DNA was digested with BamH1 and Sal1 restriction enzymes, and the correct DNA fragment was gel-purified and ligated into BamH1–Sal1 digested pQE80L (Qiagen). The resulting plasmid pQE80L-θ2.7 was transformed into E. coli BL21-CodonPlus-RP cells (Stratagene) for protein production.
Pol θ was purified from 1 to 2 l of E. coli BL21-CodonPlus-RP cells transformed with pQE80L-θ2.7 following induction with 1.0 mM IPTG at a temperature of 30°C for 3.5 h. After harvesting by centrifugation and storage at –80°C, cells were resuspended in buffer (35 ml) containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM 2-mercaptoethanol, 5% glycerol and a cocktail of proteinase inhibitors. Cells were disrupted in a French Press chilled to 4°C with a constant pressure of 25 000 psi. The lysate was clarified by centrifugation at 12 000 g for 20 min, and the supernatant was saved for further purification. Imidazole was added to the cleared lysate to a final concentration of 20 mM before being applied to a 2-ml Ni–NTA column (Qiagen). The resin was washed with 50 column volumes of buffer containing 500 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl, pH 7.5 and 20 mM imidazole, followed by several column volumes of a second wash buffer containing 500 mM NaCl, 0.005% NP-40, 50 mM Tris–HCl, pH 7.5 and 20 mM imidazole. Bound proteins were step-eluted with the second wash buffer also containing 0.25 M imidazole. The buffer was exchanged on a Fast Desalt HR 10/10 column (GE Healthcare) equilibrated in 25 mM potassium phosphate, pH 7.5, 10% (v/v) glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.005% (v/v) NP-40 and 75 mM KCl. The desalted protein fraction was applied to a 1.0 ml HR 5/5 Mono S column equilibrated in 25 mM potassium phosphate, pH 7.5, 10% (v/v) glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol and 75 mM KCl. Protein was eluted with a linear KCl gradient (0.075–0.5 M) in this buffer, and fractions containing Pol θ were flash frozen in liquid nitrogen in small aliquots.
Generation of polymerase variant
The DNA polymerase activity of Pol θ was inactivated by disrupting the ability of two essential carboxylate side chains to bind Mg2+ within motif C of the polymerase active site. Polymerase domain residues Asp829 and Glu830, corresponding to residues 2540 and 2541 in full-length Pol θ, were changed to Asn and Gln, respectively, using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and the following oligonucleotides: forward Pol−: 5′-CAT CCT TCA ACT CCA TAA TCA ACT CCT ATA TGA AGT G-3′ and reverse Pol−: 5′-CAC TTC ATA TAG GAG TTG ATT ATG GAG TTG AAG GAT G-3′. The resulting plasmid bearing the polymerase activity mutations was sequenced in its entirety to confirm the D829N and E830Q substitutions. E. coli BL21-CodonPlus-RP cells were transformed with this plasmid, pQE80L-θPol−, followed by induction and purification of the protein as described above.
DNA polymerase assay
Pol θ activity was measured with a gel-based oligonucleotide extension assay as described previously (20–23). An 18-mer primer (5′-TGA CCA TGT AAC AGA GAG-3′) was gel purified, labeled at the 5′ end with [γ-32P]ATP and annealed in a 1:1.4 ratio to a 36-mer template (3′-ACT GGT ACA TTG TCT CTC GCA CTC ACT CTC TTC TCT-5′) in 10 mM Tris–HCl, pH 7.5 by heating to 90°C for 5 min followed by slow cooling to room temperature. One picomole primer/template (32P-end labeled primer) was incubated with 5 nM Pol θ in a reaction mixture (10 μl) containing 25 mM HEPES, pH 7.5, 2 mM 2-mercaptoethanol, 0.1 mM EDTA, 5 mM MgCl2, 50 µg/ml acetylated bovine serum albumin and 100 µM each dNTP, as indicated. Following incubation at 37°C for 10 min, reactions were stopped on ice by addition of 10 μl formamide gel-loading dye. Reaction products were separated by electrophoresis in a 15% polyacrylamide gel containing 8 M urea in 89 mM Tris–HCl, 89 mM boric acid, and 2 mM EDTA, pH 8.8. Imaging and data analysis were performed with a Typhoon PhosphorImager and the ImageQuant software (GE HealthCare).
Substrate preparation and dRP lyase activity assay
Preparation of the dRP lyase substrate and the dRP lyase assay conditions were essentially as described previously (24). Briefly, the reaction mixture (20 µl) contained 50 mM HEPES, pH 7.5, 20 mM KCl, 2 mM DTT, 1 mM EDTA and 50 nM preincised 32P-labeled AP site containing DNA. The reaction was initiated by adding the indicated amounts of Pol θ, mutant Pol θ or Pol β, followed by incubation at 37°C. Aliquots (9 µl each) were transferred at the indicated time intervals into the tubes that contained 1 µl of freshly prepared 200 nM NaBH4. Reaction mixtures were shifted to 0–1°C (on ice) and incubation was continued for 30 min. After heating to 75°C for 2 min, the reaction products were separated by electrophoresis in a 15% polyacrylamide gel containing 8 M urea as described above. Imaging and data analysis were performed by PhosphorImager and ImageQuant software.
5′-End labeling and substrate preparation for dRP lyase and NaBH4 cross-linking reactions
Dephosphorylated 17-mer oligodeoxyribonucleotide (5′-UGTS-SGGATCCCCGGGTACBiotin-3′) containing a uracil residue at the 5′-end, a disulfide bond (S–S) 3 nt from the 5′-end, and biotin at the 3′-end was phosphorylated with Optikinase and [γ-32P]ATP. A 34-mer (5′-GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG-3′) template was then annealed with 15-mer (5′-CTGCAGCTGATGCGC-3′) and 17-mer 32P-labeled oligonucleotides by heating the solution at 90°C for 3 min and allowing the solution to slowly cool to 25°C. The 32P-labeled duplex DNA was treated with human UDG to generate the 32P-labeled deoxyribose sugar phosphate containing single nucleotide gapped substrate. The S–S bond was included in the substrate molecule to enable future studies on cross-linking within the dRP lyase active site.
Covalent cross-linking of DNA to Pol θ
A NaBH4 trapping technique was utilized to covalently cross-link Pol θ to DNA (25). The reaction mixture (10 μl) contained 50 mM HEPES, pH 7.4, 20 mM KCl, 1 mM EDTA, 200 nM 32P-labeled UDG-treated duplex DNA, 150 nM wild-type Pol θ, mutant Pol θ or 20 nM Pol β and 1 mM NaBH4. Reactions were incubated first on ice for 60 min and then for 10 min at room temperature. After these incubations, 10 µl SDS–PAGE gel-loading dye was added to each reaction, boiled for 5 min and protein–DNA cross-linked complexes were separated by electrophoresis in a 10% NuPAGE Bis–Tris gel with a MOPS running buffer system. Radioactive bands were visualized with a Typhoon PhosphorImager.
Kinetic measurements of dRP lyase activity of Pol θ
Kinetic analysis of the dRP lyase activity of Pol θ was performed with a 32P-labeled 34-bp substrate that had been pretreated with UDG to generate a 32P-labeled dRP flap within a single-nucleotide gap in the duplex DNA. The reaction mixture contained 50 mM HEPES, pH 7.4, 20 mM KCl, 1 mM EDTA and 100 nM 32P-labeled UDG-treated duplex DNA. For the time course experiment, the reaction mixture (50 µl) was assembled at 0–1°C in the above buffer. Reactions were initiated by adding 400 nM Pol θ or the dilution buffer (control), as indicated in figure legends, and incubated at 37°C. Aliquots (9 µl each) were withdrawn at different time intervals and transferred to 0–1°C to stop the reaction. DNA products were stabilized by addition of 20 mM NaBH4 and incubated for 30 min on ice. Then, an equal volume of gel-loading buffer was added, and the reaction mixture was incubated at 75°C for 2 min. The reaction products were separated by electrophoresis in a 15% polyacrylamide TBE-Urea gel (Invitrogen, Pre-cast gel) for 30 min at constant voltage (200 V). To quantify the reaction products, gels were scanned on a PhosphorImager and the data were analyzed as above. Reaction rates were determined by plotting the amount of substrate released as a function of time, and data were fitted to the appropriate equation by nonlinear least squares methods. To examine the influence of protein concentrations on dRP removal, the reactions were assembled on ice, as above, and initiated by adding appropriate dilutions of Pol θ, as indicated in the figure legends. Reaction mixtures were incubated at 37°C for 10 min and processed as above.
In vitro SN-BER
Repair assay was performed in a final reaction volume of 30 μl. A 35-bp oligonucleotide duplex DNA (250 nM) containing uracil at position 15 was incubated in a BER reaction mixture that contained 50 mM HEPES, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 20 mM KCl, 4 mM ATP, 5 mM phosphocreatine, 100 μg/ml phosphocreatine kinase, 0.5 mM NAD, 15 nM UDG, 15 nM APE, 200 nM DNA ligase I and 400 nM or 800 nM purified Pol θ, as indicated. Repair reactions were initiated by the addition of 10 mM MgCl2 and 2.2 μM [α-32P]dCTP (specific activity, 1 × 106 dpm/pmol), followed by incubation at 37°C. Aliquots (9 µl) were withdrawn at the indicated time intervals. Reactions were terminated by the addition of an equal volume (9 µl) of DNA gel-loading buffer. After incubation at 75°C for 2 min, the reaction products were separated by electrophoresis in a 15% polyacrylamide gel containing 8 M urea, and the data were analyzed as above.
Limited proteolysis and amino-terminal sequencing of Pol θ
The purified human Pol θ 98-kDa polymerase domain (66 µg) was subjected to limited proteolysis by mixing with trypsin (0.66 µg) at a 1:100 weight ratio (trypsin:Pol θ) in 100 mM Tris–HCl, pH 8.0 and then incubating the solution at 25°C. The final reaction mixture volume was 90 µl. Aliquots (20 µl each) were withdrawn at 5-, 15-, 30- and 60-min time intervals. A portion of each sample (18 µl) was mixed immediately with 10 µl SDS–PAGE gel-loading buffer, boiled for 5 min and separated by electrophoresis in a 12% NuPAGE Bis–Tris gel with a MOPS running buffer system. Proteins were electrophoretically transferred onto an Immun-Blot PVDF membrane (7 × 8.4 cm) (Bio-Rad) using a transfer buffer that contained 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] and 10% methanol, pH 11.0 at 50 V (7 V/cm) for 1 h. The membrane was stained briefly with 0.2% (w/v) Coomassie blue R-250 in 45% methanol and 10% acetic acid and destained with 90% methanol and 10% acetic acid. The membrane was air-dried, and protein bands were cut with a scalpel. Amino-terminal sequencing was performed using a Model 492 Procise sequencing system (Applied Biosystems) at Wake Forest University, Winston-Salem, NC. The remaining portion of the trypsin digested sample (2 µl) was subjected to NaBH4 cross-linking to a 5′-end labeled dRP lyase DNA substrate, as described above. The resulting gel was subjected to phosphorimaging, and then the same gel was silver-stained for protein detection.
RESULTS
Production of Pol θ polymerase domain
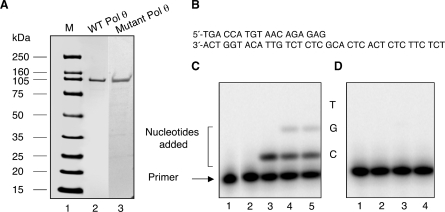
To facilitate the high-yield production of recombinant human Pol θ in E. coli, we expressed several nested POLθ cDNA clones and screened them to find the largest fragment that retained DNA polymerase activity. From the full-length human Pol θ cDNA (1,2, Copeland, unpublished data), we amplified 2.7-, 2.1-, 1.8- and 1.3-kbp cDNA fragments containing the DNA polymerase domain. Expression of the 2.7-kbp cDNA fragment, corresponding to the 98-kDa polymerase domain at the C-terminus of Pol θ (amino acids 1712–2590), produced soluble protein with the highest yield (data not shown). A (His)6 affinity tag was added to the N-terminus by transferring this fragment to the pQE80L expression plasmid, and protein was overproduced in the E. coli host BL21-CodonPlus-RP cells. As described in ‘Materials and Methods’ section, recombinant protein was purified to apparent homogeneity by sequential passage over a Ni–NTA affinity column, a buffer exchange column, and a Mono S cation exchange column (Figure 1A, lane 2). As a control for analysis of the polymerase activity, a mutant variant of the Pol θ polymerase domain was constructed that abolished DNA polymerase activity. This was achieved by altering critical carboxylate residues that bind active site Mg2+ ions. Similar alterations in other DNA polymerases are known to abolish DNA polymerase catalytic activity without dramatically affecting the overall structure of the protein (26). Asp829 and Glu830 (corresponding to residues 2540 and 2541 in the full-length Pol θ) in motif C were changed to Asn and Gln, respectively. This mutant of the Pol θ domain harboring D829N and E830Q substitutions was overexpressed and purified using the same protocol as for the wild-type 98-kDa Pol θ preparation (Figure 1A, lane 3).
Figure 1.
Purification of the 98-kDa Pol θ from E. coli. (A) The 98-kDa polymerase domain of Pol θ was purified as described in Materials and Methods section, resolved by SDS–PAGE, and visualized by staining with Coomassie blue. Lane 1, molecular weight markers; lanes 2 and 3, 0.4 μg of the wild-type and mutant Pol θ polymerase domains after Mono S chromatography, respectively. (B) Schematic representation of the primer/terminus sequence. (C and D) DNA polymerase activity of wild-type and mutant Pol θ was assayed on a synthetic oligonucleotide primer/template as described in the Materials and Methods section. (C) Polymerase activity of the wild-type 98-kDa Pol θ. Lane 1 is the 5′-32P-end-labeled primer alone; lane 2, primer and enzyme with no added nucleotides; lane 3, addition of 100 μM dCTP; lane 4, addition of 100 μM dCTP and 100 μM dGTP; lane 5, addition of all four dNTPs at 100 μM each. (D) Polymerase activity of the mutant 98-kDa Pol θ. Experimental conditions are the same as in lanes 2–5 in (C).
DNA polymerase activity of the purified wild-type and mutant Pol θ proteins was determined using an oligonucleotide primer/template as substrate (Figure 1B). The wild-type 98-kDa Pol θ readily incorporated a single nucleotide when only dCTP was included in the reaction mixture (Figure 1C, lane 3) and could incorporate 2 nt when the next correct nucleotide also was added (lane 4), but the enzyme was unable to extend the primer further when all four nucleotides were present (lane 5). Thus, the 98-kDa Pol θ C-terminal domain exhibited altered activity compared to the full-length 298-kDa Pol θ, which easily extends DNA primers to the end of an oligonucleotide template (2,9 and Copeland, unpublished data). Control incubations utilizing the 98-kDa Pol θ double mutant failed to display any DNA polymerase activity (Figure 1D), indicating complete inactivation of Pol θ polymerase function, as well as the absence of any significant contaminating nuclease or polymerase activities that may have been carried over from the E. coli host. A comparison of the single-nucleodide primer-extension DNA polymerase activity of full-length Pol θ and the 98-kDa Pol θ is shown in Table 1. The polymerase activities were similar and also were similar to the activity reported by Seki et al. (8) for the full-length enzyme.
Table 1.
Steady-state kinetic parameters for polymerase activities of full-length and 98-kDa fragment of Pol θ
| Enzyme | Km | kcat | kcat/Km | References |
|---|---|---|---|---|
| µM | min−1 | µM−1 min−1 | ||
| Full-length 290-kDa Pol θ | 7.0 | 0.67 | 0.096 | (8) |
| Full-length 290-kDa Pol θ | 9.0 | 0.42 | 0.047 | This study |
| 98-kDa fragment Pol θ | 9.7 | 0.7 | 0.072 | This study |
These measurements were made by a single nucleotide-primer extension assay.
5′-dRP lyase activity of Pol θ
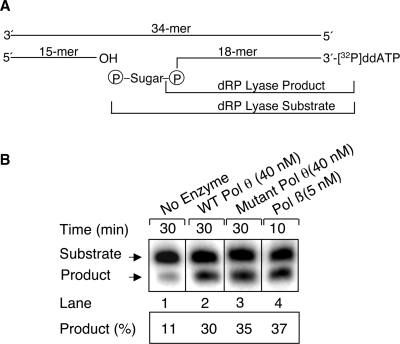
To examine a potential role of human Pol θ in BER and to further characterize its biochemical properties, we assessed the ability of the purified wild-type 98-kDa Pol θ and its polymerase-deficient mutant (D829N/E830Q) to process a BER intermediate substrate. Pol β was included in the experiments as a reference. A 34-bp oligonucleotide duplex that contained a uracil residue at position 16 (Figure 2A) was used to measure the release of the 5′-dRP group by dRP lyase activity. The uracil-containing DNA strand was 3′-end-labeled with [32P]ddAMP and annealed to its complementary strand; the duplex DNA then was pretreated with UDG and APE. The prepared DNA substrate, contained a 1-nt gap with a 5′-dRP group and 32P-label at the 3′-end of the dRP-containing strand (Figure 2A). The removal of the dRP group from the 32P-labeled substrate strand is visualized in a denaturing gel as a band migrating approximately one-half nucleotide faster than the substrate. The preincised substrate was incubated with wild-type Pol θ (98-kDa peptide), mutant Pol θ or Pol β. The results demonstrated that both wild-type Pol θ and the mutant Pol θ could release the dRP group from the substrate (Figure 2B), and the activities appeared to be similar. The apparent rates of dRP removal of Pol θ and Pol β were 0.016/min and 0.64/min, respectively. Under the same reaction conditions, the rate of dRP lyase activity of Pol θ appeared to be about 40-fold lower than that of Pol β (Figure 2B).
Figure 2.
dRP lyase activity of wild-type and mutant Pol θ. (A) A schematic representation of the dRP lyase substrate (18-mer + 32P-dRP) generated by pretreatment of 32P-labeled 34-bp DNA with UDG and APE and the expected products 32P-labeled 18-mer product formed as a result of Pol θ (sample) or Pol β (positive control) catalyzed excision of 5′-terminal dRP. (B) Preincised DNA substrate (50 nM) was incubated either with no enzyme (lane 1), 40 nM wild-type Pol θ (lane 2), 40 nM mutant Pol θ (lane 3), or 5 nM Pol β (lane 4). Aliquots were withdrawn at the indicated time intervals, and the DNA products were stabilized and analyzed as described under Materials and Methods section. A photograph of the phosphorimage, illustrating the reaction products, is shown. Reaction products were quantified using ImageQuant software, and the fraction of the substrate converted into product was indicated as a percentage under each lane. The apparent rates for the dRP lyase activity of Pol θ and Pol β were 0.016 and 0.64/min, respectively. The positions of substrate and the product are indicated.
Kinetics and mechanism of dRP lyase
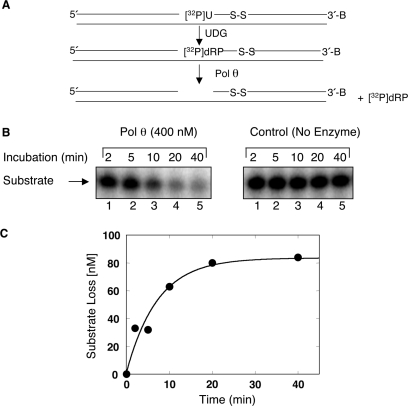
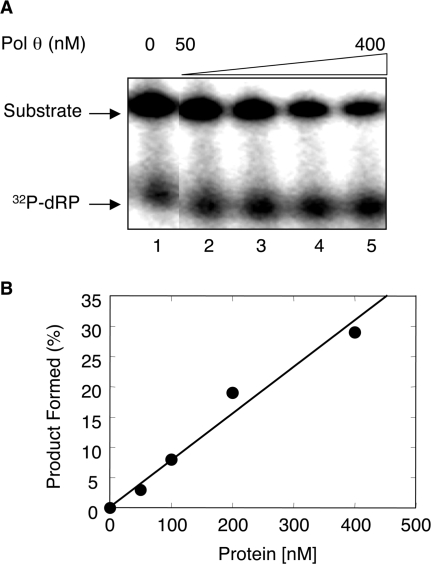
To determine kinetic parameters of 5′-dRP group removal by Pol θ, we prepared a 34-bp duplex DNA that contained uracil at position 16 and a nick between positions 15 and 16, by annealing both a 15-mer and a 5′-end 32P-labeled uracil-containing 17-mer to the 34-mer complementary DNA strand. The resulting 32P-labeled nicked DNA was pretreated with UDG to generate the 5′-dRP-containing single-nucleotide gapped substrate; the DNA substrate, thus prepared, contained a 32P-labeled dRP flap in a single-nucleotide gap (Figure 3A, 16-mer + 32P-dRP). The rate of dRP removal by Pol θ was determined under single turnover conditions (i.e. enzyme/DNA = 4). The results revealed that Pol θ cleaved 5′ dRP in a time-dependent manner as monitored by disappearance of the substrate (Figure 3B), with an observed rate of dRP removal of ∼0.14/min (Figure 3C). In other experiments where substrate was in excess, removal of the 5′-dRP group from the substrate was dependent on the enzyme concentration (Figure 4A and B).
Figure 3.
Kinetic measurements of dRP lyase activity of Pol θ. The loss of dRP from the nicked AP-site containing DNA substrate was examined as a function of incubation time, and the rate of dRP removal by Pol θ was determined under single turnover conditions (i.e. enzyme/DNA = 4). The reaction was performed with Pol θ as described under Materials and Methods section. (A) Schematic representation of the DNA substrate prepared by annealing a 15-mer oligonucleotide, a 17-mer oligonucleotide containing 32P-labeled uracil at the 5′-end, and a 34-mer oligonucleotide template. (B) The duplex DNA (100 nM) was pretreated with UDG and then reacted with 400 nM Pol θ. Aliquots were withdrawn at the indicated time intervals and stabilized by the addition of 20 mM NaBH4 and incubation on ice for 30 min. Then, an equal volume of gel-loading dye buffer was added, and the reaction products were separated by precast 15% TBE-Urea gels. A representative phosphorimage of the polyacrylamide gel illustrates product analysis at various times (lanes 1–5) with and without Pol θ. (C) Time course data were analyzed using ImageQuant software and fitted to an exponential equation by using nonlinear least squares methods to determine the reaction rate (kobs= 0.14/min).
Figure 4.
Influence of protein concentrations on dRP release by Pol θ. The dRP lyase reactions were performed with different concentrations of Pol θ as described under Materials and Methods section. Reaction conditions and product analysis were as described in Figure 3. (A) UDG pretreated DNA substrate was mixed with the indicated amounts of Pol θ and incubated at 37°C. The migration positions of the substrate and the product (32P-dRP) are indicated. (B) Amounts of 32P-labeled dRP released from the substrate was quantified and plotted against Pol θ concentrations. Data were fitted to a straight-line equation.
To confirm that the 5′-dRP lyase activity of Pol θ was intrinsic to the 98-kDa peptide, we utilized the NaBH4 cross-linking technique (25). This cross-linking technique relies on the ability of the C1′-aldehyde group of the 5′-deoxyribose in the substrate to react with a lysine γ-amine of a protein to form a β-elimination intermediate (Schiff base) protein–DNA complex; this Schiff base is recovered as a covalent bond by NaBH4 trapping. We had previously shown that DNA polymerases, such as Pol β, Pol γ and Pol ι, catalyze dRP removal from the BER intermediate via β-elimination, and the Schiff base intermediate between these polymerases and DNA can be trapped by NaBH4 reduction (27). As demonstrated in Figure 5, both wild-type Pol θ and the polymerase-deficient mutant Pol θ could form covalent protein–DNA products with the BER intermediate substrate upon treatment with NaBH4. This indicates that the Pol θ polymerase domain possesses functional 5′-dRP lyase activity.
Figure 5.
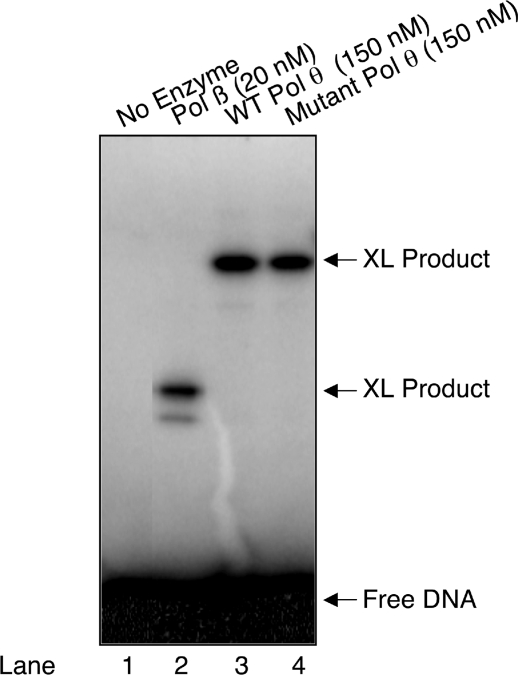
NaBH4 cross-linking of wild-type and mutant Pol θ. The reaction conditions and product analysis were as described under Materials and Methods section. UDG pretreated DNA substrate (200 nM) as described in Figure 3A was mixed with dilution buffer (lane 1), 20 nM Pol β (lane 2), 150 nM wild-type Pol θ (lane 3) or 150 nM mutant Pol θ (lane 4) and 1 mM NaBH4. The reaction mixtures were incubated on ice for 1 h followed by incubation at room temperature for 10 min. Covalently cross-linked DNA–protein products were separated by 10% NuPAGE Bis–Tris gel, and the gel was scanned on a PhosphorImager. The positions of cross-linked products and free DNA are indicated.
Domain mapping of the 98-kDa peptide of Pol θ
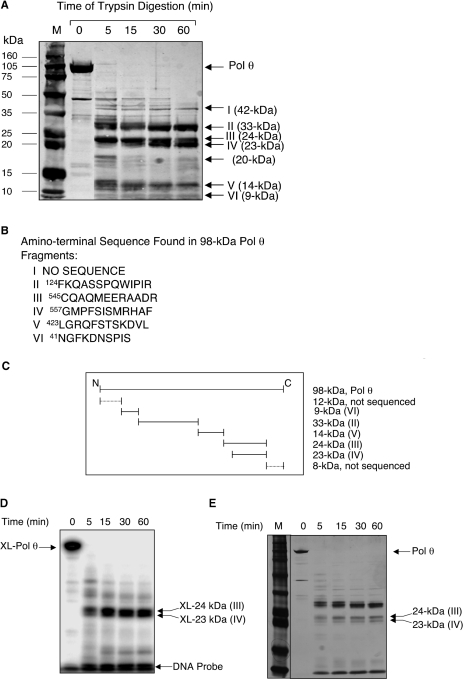
To probe domain organization of the Pol θ 98-kDa polymerase domain and to localize the 5′-dRP lyase active site of Pol θ, we subjected the purified protein to controlled proteolysis with trypsin, and then we identified the digestion products by amino-terminal sequencing. Samples from the incubation with trypsin were withdrawn at different time intervals. A portion of each sample was separated by SDS–PAGE, transferred onto a PVDF membrane and subjected to amino-terminal sequencing as described under ‘Materials and Methods’ section (Figure 6). Digestion for 5 min completely degraded the 98-kDa enzyme and produced two major fragments with molecular masses of about 33 and 24 kDa; several minor polypeptides of higher and lower molecular masses also were produced. With longer incubations of 30 and 60 min, four fragments persisted and were designated as fragments II, III, IV and V, respectively (Figure 6A). The 24-kDa fragment appeared to be further digested to a smaller molecular mass polypeptide resulting in a doublet in the gel, whereas the 20-kDa polypeptide and some high-molecular-mass polypeptides almost disappeared (Figure 6A). Amino-terminal sequence analysis of several of these protease-resistant fragments (denoted as I–VI in Figure 6A) revealed that the 33-, 24-, 23-, 14- and 9-kDA polypeptides (II to VI) begin with Phe124, Cys545, Gly557, Leu423 and Asn41, respectively (Figure 6B). These results indicated that the 23-kDa (IV) polypeptide resulted from removal of 12 amino acids from the amino terminus of the 24-kDa (III) polypeptide. Fragment I, with a molecular mass of 42 kDa, failed to exhibit reasonable sequence data due to low abundance. The domains of the 98-kDa Pol θ were aligned (Figure 6C) based on the molecular masses of these protease-resistant fragments and their amino-terminal sequences.
Figure 6.
Limited proteolysis of Pol θ with trypsin. (A) Purified 98-kDa Pol θ (66 µg) was digested at 25°C with trypsin (0.66 µg) at a weight ratio (trypsin:Pol θ) of 1:100 in 100 mM Tris–HCl, pH 8.0. Aliquots were withdrawn at 5-, 15-, 30- and 60-min time intervals, as indicated at the ‘top’ of the photograph. A portion of each digested sample was mixed with 10 µl SDS–PAGE gel-loading buffer and resolved on a 12% NuPAGE Bis–Tris gel. The proteins were transferred onto a PVDF membrane for amino-terminal sequencing, as described under Materials and Methods section. The positions of Pol θ, tryptic peptides and protein markers are indicated. (B) Amino-terminal sequencing was performed using the Procise sequencing system, Model 492 (Applied Biosystems). The amino-terminal sequences of peptides I–VI are shown. (C) The domain organization of 98-kDa Pol θ is depicted. (D) Portions of the trypsin digested samples from (A) were reacted with 5′-end-labeled dRP lyase DNA substrate and subjected to NaBH4 cross-linking as in Figure 5. The gel was scanned on a PhosphorImager. The positions of cross-linked Pol θ, 23- and 24-kDa peptides are indicated. (E) The gel in (D) was stained with silver to detect protein fragments. The positions of Pol θ, 23- and 24-kDA peptides are indicated.
Localization of the dRP lyase active site
To localize the dRP lyase active site within the 98-kDa Pol θ, portions of the tryptic digests of Pol θ (see above) were incubated with the 32P-labeled dRP lyase substrate and subjected to NaBH4 cross-linking as in Figure 5. This protocol was used to assess the capacity of the tryptic fragments for cross-linking and if successful to improve the yield of cross-linked material for sequencing. Reaction mixtures that were digested with trypsin for 5 min or 15 min produced major 33-kDa and 24-kDa fragments (Figure 6A), but only the 24-kDa fragment was radiolabeled following cross-linking (Figure 6D). Furthermore, longer incubations of 30 min and 60 min revealed that the radiolabeled 23- and 24-kDa peptides persisted and appeared as a doublet in the gel (Figure 6D), similar to the doublet observed upon staining with Coomassie blue or silver (Figure 6A and E, respectively). These observations suggested that even prolonged incubation with trypsin only partially converted the 24-kDa peptide to the 23-kDa peptide, and that both fragments contain the Schiff base dRP lyase nucleophile, i.e. dRP lyase active site. Taken together, these results indicate that the 24-kDa tryptic fragment is a domain of Pol θ that contains the dRP lyase active site and retains the ability to form a covalent DNA–protein complex with a BER intermediate (Figure 6D).
In vitro base excision repair activity
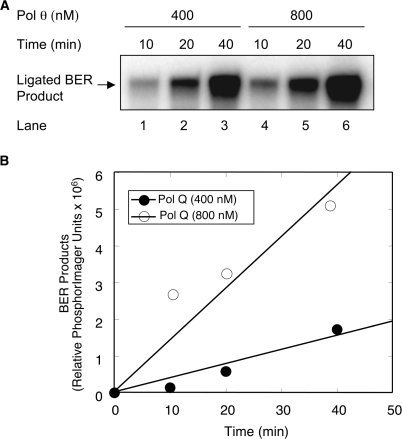
Since the 98-kDa Pol θ was able to cleave the 5′-dRP flap from a BER intermediate and also retained DNA polymerase activity, we asked whether the 98-kDa enzyme was capable of supporting BER in vitro. We assembled repair reaction mixtures containing a 35-bp DNA substrate with uracil opposite guanine and purified human enzymes, including UDG, AP endonuclease, DNA ligase I and the 98-kDa Pol θ. Reactions were initiated by the addition of Mg2+ and [α-32P]dCTP, and repair activity was monitored by the incorporation of [α-32P]dCMP in place of dUMP into a ligated 35-bp DNA product as a function of incubation time and concentration of Pol θ (Figure 7). In vitro repair activity was dependent on both enzyme concentration and incubation time (Figure 7A and B), confirming that human Pol θ catalyzed sufficient dRP removal and 1-nt gap filling to support single-nucleotide BER.
Figure 7.
In vitro BER with purified proteins. Gap-filling activity of Pol θ was evaluated on a uracil-containing BER substrate by measuring incorporation of [α-32P]dCMP as a function of incubation time and protein concentration. Reaction conditions and product analysis were as described under Materials and Methods section. A 35-bp oligonucleotide duplex DNA (250 nM) with a U:G mismatch was mixed with 15 nM UDG, 15 nM APE, 200 nM DNA ligase I, 400 or 800 nM purified 98-kDa Pol θ and incubated at 37°C. Aliquots were withdrawn at indicated time intervals, and an equal volume of gel-loading buffer was added to terminate the reaction. The reaction products were separated by 15% denatured PAGE, and a PhosphorImager was used to scan the gel. (A) Phosphorimage of denaturing PAGE demonstrating the in vitro BER activity of Pol θ is shown. The migration position of the ligated BER product is indicated. (B) BER products were quantified using ImageQuant software, and the relative phophorImager units were plotted against time of incubation.
DISCUSSION
Abasic sites in DNA are common intermediates in several important DNA transactions within eukaryotic cells, including SHM, translesion DNA synthesis and BER. For example, SHM of immunoglobulin variable region genes is triggered by the action of activation-induced cytidine deaminase (28), which deaminates deoxycytosine to deoxyuracil. The abasic sites could be processed by AP endonuclease and Pol β in an error-free pathway, but such a repair system would not be beneficial for SHM. Several lines of evidence support the idea that faithful repair mediated by Pol β is suppressed during SHM. First, using a mouse model Rajeswky and his associates (29) demonstrated that Pol β is not required in SHM. Second, we recently showed that the expression of Pol β in the SHM-proficient human BL2 cell line was strongly down regulated (30). Third, Maizels and associates (31,32) reported that during immunoglobulin gene diversification in B cells, AP sites were preferentially processed by the MRE11-RAD50-NSB1 nuclease complex rather than by AP endonuclease.
Although Pol θ is a family A DNA polymerase, it exhibits unusual properties that permit efficient bypass of abasic sites. In fact, Pol θ is able to synthesize through these damaged sites with an efficiency that is comparable to that displayed on a corresponding undamaged template (8). The unique ability of Pol θ to insert and extend mispaired nucleotides opposite AP sites suggests an important role for Pol θ in SHM consistent with the observation that deletion of the Pol θ gene in mice decreased mutations in the Ig H chain by more than 80% (14). While these studies suggested involvement of Pol θ in translesion bypass during SHM, recent observations with DT40 cells also support some role for Pol θ in BER (15).
In this report we established that Pol θ, primarily known as a lesion bypass DNA polymerase, also possesses 5′-dRP lyase and single-nucleotide gap-filling activities that are usually associated with BER. The dRP lyase activity is independent of polymerase activity, since a polymerase inactive mutant retained full dRP lyase activity. Using limited proteolysis and sodium borohydride cross-linking techniques, we demonstrated that the dRP lyase activity resides in a 24-kDa domain in the 98-kDa carboxy-terminal region of Pol θ. This 24-kDa domain contains the three conserved motifs (A, B and C) found in all family A and B polymerases (1). In the 24-kDa domain Lys2383, near motif B, is conserved in Pol θ from vertebrate species as well as in other family A polymerases, and this residue could represent a candidate nucleophile for the dRP lyase activity. In investigating roles for the N-terminal 2/3 of the enzyme, it will be of interest to determine how the full-length enzyme influences overall BER efficiency in vivo. Similarly, Pol θ is highly expressed in germinal center B cells, where SHM and class switch recombination occur (33). Identification of protein partners or cofactors that interact with full-length Pol θ may help to assign roles for Pol θ in these processes.
FUNDING
Research Project Numbers Z01-ES050159-12 (to S.H.W.) and Z01-ES065078 (to W.C.C.), in the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences. Funding for open access charge: the Intramural Research Program, National Institutes of Health, National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Drs Bill Beard, Michelle Heacock and Mike Carrozza for critical reading of the manuscript; Dr Mark Lively, Wake Forest University, for protein sequencing; and Dr Bob Petrovich and Mrs. Lori Edwards, NIEHS Protein Expression Core Facility, for protein expression. We also thank Jennifer Myers for editorial assistance.
REFERENCES
- 1.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase θ (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 2.Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 4.Marini F, Wood RD. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2002;277:8716–8723. doi: 10.1074/jbc.M110271200. [DOI] [PubMed] [Google Scholar]
- 5.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seki M, Wood RD. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, O-Wang J. DNA polymerases η and θ function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 11.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J. Absence of DNA polymerase θ results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl Acad. Sci. USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, et al. Vertebrate POLQ and POLβ cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase β. Meth. Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 17.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 18.Strauss PR, Beard WA, Patterson TA, Wilson SH. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 19.Wang YC, Burkhart WA, Mackey ZB, Moyer MB, Ramos W, Husain I, Chen J, Besterman JM, Tomkinson AE. Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J. Biol. Chem. 1994;269:31923–31928. [PubMed] [Google Scholar]
- 20.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 21.Mendelman LV, Boosalis MS, Petruska J, Goodman MF. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 22.Copeland WC, Chen MS, Wang TS. Human DNA polymerases α and β are able to incorporate anti-HIV deoxynucleotides into DNA. J. Biol. Chem. 1992;267:21459–21464. [PubMed] [Google Scholar]
- 23.Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase γ. J. Biol. Chem. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- 24.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase τ by controlled proteolysis. J. Biol. Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 25.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 26.Copeland WC, Wang TS. Mutational analysis of the human DNA polymerase α. The most conserved region in α-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- 27.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase β deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 28.Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A⊕T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 29.Esposito G, Texido G, Betz UA, Gu H, Muller W, Klein U, Rajewsky K. Mice reconstituted with DNA polymerase β-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl Acad. Sci. USA. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poltoratsky V, Prasad R, Horton JK, Wilson SH. Down-regulation of DNA polymerase β accompanies somatic hypermutation in human BL2 cell lines. DNA Repair (Amst) 2007;6:244–253. doi: 10.1016/j.dnarep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- 32.Vallur AC, Yabuki M, Larson ED, Maizels N. AID in antibody perfection. Cell. Mol. Life Sci. 2007;64:555–565. doi: 10.1007/s00018-007-6434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]