Abstract
Traditional chromatin analysis methods only test one locus at the time or use different templates for each locus, making a standardized analysis of large genomic regions or many co-regulated genes at different loci a difficult task. On the other hand, genome-wide high-resolution mapping of chromatin accessibility employing massive parallel sequencing platforms generates an extensive data set laborious to analyse and is a cost-intensive method, only applicable to the analysis of a limited set of biological samples. To close this gap between the traditional and the high-throughput procedures we have developed a method in which a condition-specific, genome-wide chromatin fragment library is produced and then used for locus-specific DNA fragment analysis. To validate the method, we used, as a test locus, the well-studied promoter of the divergently transcribed niiA and niaD genes coding for nitrate assimilation enzymes in Aspergillus. Additionally, we have used the condition-specific libraries to study nucleosomal positioning at two different loci, the promoters of the general nitrogen regulator areA and the regulator of secondary metabolism, aflR.
INTRODUCTION
The packaging of eukaryotic nuclear DNA in nucleosomes and higher order chromatin restricts the accessibility of DNA to regulatory factors (1). Local modifications of histones by acetylation, methylation, phosphorylation, sumoylation or ubiquitylation (2–8) and the function of ATP-driven nucleosome remodelling complexes (9,10) affect the positioning of nucleosomal cores and alter the structure of chromatin. It is a generally accepted view that strictly positioned nucleosomes and heterochromatic regions repress gene activities, whereas loss of tightly positioned nucleosomes is associated with active gene transcription. Regions where local modifications of chromatin structure occur can be detected by analysing the accessibility of DNA in this region to various nucleases. Traditionally, nucleases such as DNAse I, DNAse II or micrococcal nuclease (MNase) as well as sequence-specific restriction enzymes are used to test the accessibility of chromatin and link the digestibility of DNA to gene activity. DNAseI hypersensitive sites (DHSs) have been shown over the last two decades to be indicators for the activity of many different types of regulatory elements such as promoters, enhancers or locus-control-regions from simple eukaryotes to mammals (1,11–13). MNAse and site-specific restriction enzymes are used to map the accessibility of DNA in a series of phased nucleosomes or in one individual nucleosome, for example in a promoter region of interest (14,15). The positioning of nucleosomes in a region of interest is usually detected by electrophoretic separation of digestion products followed by Southern blotting and indirect end-labelling (16). This method tests one region per hybridization event and covers 300–800 bp with a moderate resolution of 20–50 bp. Ingram and colleagues (16–18) have improved this method by introducing a ligation-mediated PCR (LM-PCR) step after DNA digestion and separation of the amplification products by capillary sequencer. This method represents an important advance in locus-specific chromatin analysis as it allows a rapid and sensitive read-out with, at that time, unrecorded accuracy of product size determination. However, to test the chromatin structure of different loci in one specific condition, the primer extension, extension products capture, final PCR amplifications and labelling reactions must be repeated each time. Apart from being laborious, these steps potentially introduce a bias for certain digestion products depending on PCR primer efficiency, DNA target sequence and amplification conditions.
In recent years, genome-wide chromatin analysis methods have been considerably refined. DNaseI-hypersensitive sites have been studied by cloning and sequencing chromatin digestion fragments (19) and by hybridization of PCR-amplified chromatin fragments to tiled microarrays (20). The advent of massive parallel sequencing technologies developed by Illumina (Solexa) and Roche (454 Life Sciences) now allows chromatin accessibilities to be mapped on a genome-wide scale at single base pair resolution. For example, Boyle and co-workers (21) employed high-throughput sequencing to identify the locations of DHSs across the entire genome of human primary CD4+ T cells. While of great importance to the understanding of the role of chromatin in global gene expression patterns in different cell types or in response to external stimuli, these high-throughput methods are highly demanding in resources. Massive parallel sequencing is a costly task and requires extensive bioinformatic processing of the large data set for each chromatin fragment library. Therefore, whole genome-wide approaches have so far been limited to a small number of studies which usually employed these techniques for the analysis of one cell line at one single condition.
This situation leaves a technical gap when the specific scientific question suggests studying the chromatin at several loci of the genome under a variety of different conditions, and/or in different cell types or mutant strains. Typically, a scientific question requiring the analysis of multiple loci under different conditions would be studying genes involved in the same signalling pathway, co-regulated metabolic or developmental genes or the chromatin structure in large gene clusters. The traditional indirect end-labelling methods are not suitable for such multi-locus tasks. On the other hand, a whole genome-wide approach would in many cases be beyond the scope of the scientific question and hardly affordable.
To fill this technical gap, we have developed a cost-effective intermediate method that relies on the construction of condition-specific MNase digestion linker selection libraries, similar to the material used for high-throughput sequencing. These libraries then can be used in a traditional sequencer-based fragment analysis step to analyse nucleosomal pattern at any genomic region of interest. To validate the method, we used, as a test locus, the bidirectional promoter of the divergently transcribed niiA and niaD genes coding for nitrate assimilation enzymes in Aspergillus (22). The chromatin structure in this promoter is subject to changes depending on the function of two positive transcriptional regulators, the general nitrogen activator AreA, which belongs to the GATA factor family (23,24), and the nitrate-specific factor NirA, a typical fungal C6-Zn2 binuclear zinc cluster protein (25,26). During nitrate induction, NirA is activated by nuclear retention, cooperates with DNA-bound AreA and subsequently binds to cognate targets in the promoter resulting in loss of positioning of six nucleosomes (27–31). Loss of nucleosomal positioning is associated with AreA-mediated histone H3 acetylation and during nitrate induction, also requires the function of NirA (32). In addition to the niiA-niaD test locus, we used this method to map the chromatin structure of the areA promoter, and found that under AreA inactivating conditions, nucleosomes are positioned over consensus GATA sites in this positively auto-regulated regulatory gene.
MATERIALS AND METHODS
Culture conditions and DNAse treatment
Aspergillus nidulans wild-type (areA+ nirA +) strains were pre-grown in 1000 ml Erlenmeyer flasks containing 250 ml glucose minimal medium (33) inoculated with 106 conidia per millilitre for 14 h (over night) at 37°C and shaking at 180 r.p.m. and 5 mM di-ammonium D-(1)-tartrate as nitrogen source. Mycelia were harvested by filtering, washed with sterile deionized water and aliquots were transferred to 30 ml minimal medium (in 100 ml Erlenmeyer flasks) containing either 5 mM ammonium D-(1)-tartrate, for repressed conditions and 10 mM NaNO3 for induced conditions. The cultures were further incubated on a rotary shaker for 2 h and mycelia was harvested by filtering, grind to fine powder under liquid nitrogen and lyophilized overnight. Approximately 20 mg of these lyophilized samples were suspended in 1 ml of MNase buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 0.5 mM CaCl2, 3 mM MgCl2) and 300 µl of these suspensions were treated for 5 min with 0.5, 1 and 3 U of MNase (USB Corp., Cleveland, OH, USA) at 37°C. The reaction was stopped with stop buffer (2% SDS, 40 mM EDTA) and DNA was prepared by phenol/chloroform extraction, precipitated with isopropanol, washed with 70% ethanol and dissolved in a final volume of 100 µl H2O.
In vitro DNA digestion and preparation
DNA was extracted from overnight grown cultures with DNA extraction buffer (0.2 M Tris, 1% SDS, 1 mM EDTA) as described above, and dissolved to a final concentration of 1 µg/µl in H2O. DNA solution of 40 µl was diluted in 260 µl of MNase buffer and treated for 5 min with 0.1, 0.3 and 1 U of MNase at 37°C. Digested DNA was precipitated with isopropanol, washed with 70% ethanol and dissolved in final volume of 50 µl H2O.
Restriction enzyme accessibility assays
The restriction enzyme accessibility assays were carried out essentially following our published protocol (27). Briefly, a total of 20 mg (dry weight) of lyophilized mycelia was transferred into 1.2 ml buffer Y Tango+ (MBI Fermentas, Heidelberg, Germany). Two hundred microlitres aliquots of these crude fungal cell preparations were treated for half an hour with 0, 100 and 200 U of HaeIII restriction enzyme. The reactions were stopped with stop buffer (2% SDS, 40 mM EDTA) and the DNA was prepared by phenol/chloroform extraction, precipitated with isopropanol, washed with 70% ethanol and dissolved in a final volume of 100 µl H2O.
Enzymatic blunt-ending and phosphorylation
DNA digestion with MNase produces fragments which are not blunt-ended. To allow for ligation with the blunt-end of the double-stranded adaptors, 15 µl of purified MNase digested DNA fragments were blunt-ended in a final volume of 20 µl by Klenow fragment of Escherichia coli DNA polymerase (New England Biolabs, CA, USA) and phosphorylated by T4 polynucleotide kinase (New England Biolabs) in a final volume of 30 µl.
Adaptor ligation
The blunt-ended and phosphorylated DNA fragments were ligated to the double-stranded adaptors A and B as described in (34) with the following modifications. The partially complementary nucleotide fragments Adaptor Along and Ashort as well as Blong and Bshort are dissolved in H2O to a final concentration of 100 pmol/µl and annealed to double strands (dss) by heating to 95°C for 5 min, followed by immediate transfer to 65°C and incubation for 5 min. Finally, the reaction mixture is allowed to slowly cool down to room temperature. Annealed adaptors were kept at −20°C and thawed on ice for further use. For adaptor ligation, the total reaction mixture of blunt-ended and phosphorylated DNA (30 µl) was mixed with 7.5 µl (375 pmol) of ds-adaptor A, 7.5 µl (375 pmol) of ds-adaptor B, 1× Quick Ligase Reaction Buffer (New England Biolabs) and 50 U Quick Ligase (New England Biolabs) and filled up with H2O to a final volume of 120 µl. This mixture was incubated for 20 min at 25°C, purified twice over a Qiaquick PCR Purification column, and eluted in 30 µl of 55°C Buffer EB (Qiagen).
Nick repair
Nicks at the 3′-junctions between DNA fragments and adaptors were repaired by the strand-displacement activity of Bst DNA polymerase, Large Fragment (Fermentas). Fifteen microlitres of the adaptor ligated DNA were incubated for 30 min at 65°C in 1× ThermoPol Reaction Buffer (New England Biolabs), 8 µg Bovine Serum Albumin (BSA) (New England Biolabs), 20 nmol dNTPs and 16 U Bst DNA polymerase, Large Fragment (New England Biolabs) in a total volume of 20 µl.
Isolation of the single-stranded A-B adaptor fragment library
Twenty microlitres of stock M-270 Streptavidin beads (Dynal, Oslo, Norway) were washed twice in a 20 µl volume of 2× Beads buffer and washing buffer (B&W) (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 2 M NaCl), by precipitating, between the washes, with the magnetic particle concentrator (MPC, Dynal). After the second wash, the beads were resuspended in 20 µl of 2× B&W buffer and the entire 20 µl of the Bst polymerase-treated fragment library was added. The sample was kept at room temperature for 10 min for biotin streptavidin binding. After MPC collection of the beads, the supernatant containing non-ligated fragments and fragments ligated at both ends to adaptor A was discarded. The beads are resuspended in 40 µl of 2× B&W and incubated at 95°C for 5 min to melt the dss. By this procedure, hybrid single strands carrying the non-biotinylated adaptor oligonucleotides Bshort at the 3′-end and Along at the 5′-end are released from the beads and represent the single-stranded A-B adaptor fragment library. After the denaturation step, the supernatant was collected using MPC and the melting procedure was repeated once with 40 µl of 2× B&W buffer and both supernatants were pooled to result in a final volume of 80 µl of single stranded A-B adaptor fragment library.
Library amplification and analytical PCR conditions for locus-specific fragment generation
The single-stranded A-B adaptor fragment library was amplified by nested PCR as follows: 10 μl of template DNA (single-stranded A-B library), 20 pmol of forward and reverse primer (Amplifier-Aout and Amplifier-Bout), 1× Taq Polymerase mix REDTaq™ Ready-Mix™ (Sigma-Aldrich) in a total volume of 50 µl were amplified using following cycle times: 5 min at 94°C (hot start initiation) and 35 cycles (30 s at 94°C, 45 s at 64°C and 60 s at 72°C). The reaction was purified over a Qiaquick PCR Purification column and eluted with 30 µl of 55°C Buffer EB (Qiagen). The reaction product was diluted 100-fold for the following nested PCR amplification using primers Amplifier-Ain and Amplifier-Bin. The resulting A-B library represents chromatin accessibility fragments of the whole genome.
From this library, the specific regions of interest were amplified by a nested PCR approach to improve specificity. The first PCR always employed a locus-specific primer that was not labelled and primer Amplifier-Ain hybridizing to adaptor A. In the second, nested PCR, a 5′6-carboxyfluorescein (FAM) or Alexa 488 labelled locus-specific nested primer was employed along with primer Amplifier-Ain. In the case, where the primer Amplifier-Ain primer was FAM-labelled, the nested locus-specific primer was left unlabelled. Adaptor, amplifier and other oligonucleotides used throughout this work are specified in Table 1.
Table 1.
Oligonucleotides used in this study
| Oligonucleotides | Sequence |
|---|---|
| Adaptor oligonucleotides | |
| Adaptor-Ashort | 5′CTGAGACAGGGAGGGAACAGATGGGACACGCAGGGATGAG |
| Adaptor-Along | 5′CCATCTCATCCCTGCGTGTCCCATCTGTTCCCTCCCTGTCTCAG |
| Adaptor-Bshort | 5′CTGAGACACGCAACAGGGGATAGGCAAGGCACACAGGGGA |
| Adaptor-Blong Biotin labelled | 5′(Bio)CCTATCCCCTGTGTGCCTTGCCTATCCCCTGTTGCGTGTCTCAG |
| Library amplification PCR primers | |
| Amplifier-Aout | 5′TCCCTGCGTGTCCCATCTGT |
| Amplifier-Bout | 5′CCCTGTGTGCCTTGCCTATCC |
| Amplifier-Ain | 5′CCATCTGTTCCCTCCCTGTC |
| Amplifier-Bin | 5′CCTATCCCCTGTTGCGTGTC |
| Analytical PCR primers | |
| Orf-2out | 5′CTAGCTTGACGAGTTTCTCAC |
| Orf-2in | 5′(Alexa488)GACGAGTTTCTCACTAAGCCA |
| Nuc-2out | 5′GGAGCATGAGATAATTAAATGGG |
| Nuc-2in | 5′(Alexa488)GGGCCAATGACTATGTGGTAACAC |
| Nuc-1out | 5′CTGTCATTGTTTGGTGGATGT |
| Nuc-1in | 5′(Alexa488)CATCGTGGATGGCTTCGATG |
| Nuc+2out | 5′CCGTCTGGCTTGCCGTTGACCC |
| Nuc+2in | 5′(Alexa488)TTGCCGTTGACCCGGGCTGCAGTA |
| P442out | 5′GGGGGGTTGCAGGGATGGG |
| P442in | 5′(FAM)GGATGGGATCGAGCTCCAG |
| P689out | 5′GGTCGAGGATGCAGGGAAG |
| P689in | 5′(FAM)AAGAAGCGAAGAAGGTGCA |
| P900out | 5′ATAGGACAGGGAAGGGGATG |
| P900in | 5′(FAM)AGAGCGACGAGAGGAGTGA |
| Alfrout | 5′GATATTTGCATATGATACAGGCCCGCATTG |
| Aflrin | 5′(FAM)GGCCCGCATTGCGGATTGAACCG |
| areAclaIF | 5′ATCGATGGATCTATTAAGG |
| areAscaIR | 5′AGTACTAGCAGCTGGTTGCATTAC |
Sequencing and processing of fragment profile data
The fragments amplified in the analytical PCR were purified using the Qiaquick PCR Purification kit. The products were analysed with an ABI 3700 sequencer (Applied Biosystems) by denaturing capillary electrophoresis. ROX 1000 Ladder (Sigma-Aldrich) was used as internal standard for fragment lengths. Electrophoretic profiles were analysed using ABI gene scan software and fragment lengths were calculated from the second-order polynomial coefficients obtained from the software by using the following formula.
where A0, A1 and A2 are coefficient values provided by the ABI gene scan software for each of the analysed histogram, L is the fragment length in base pairs and x values are retention time units of the capillary electrophoresis. Chromatograms were captured as screen shot, analysed and processed using ImageJ (U. S. National Institutes of Health, Bethesda, MD, USA). To generate the overlapping profiles as shown in various figures of this work, absolute numbers of fragment lengths were related to primer positions in the corresponding region. The final graph was created by placing the individual chromatograms into a plot representing a base pair scaling of the region of interest.
RESULTS AND DISCUSSION
Rationale of method design
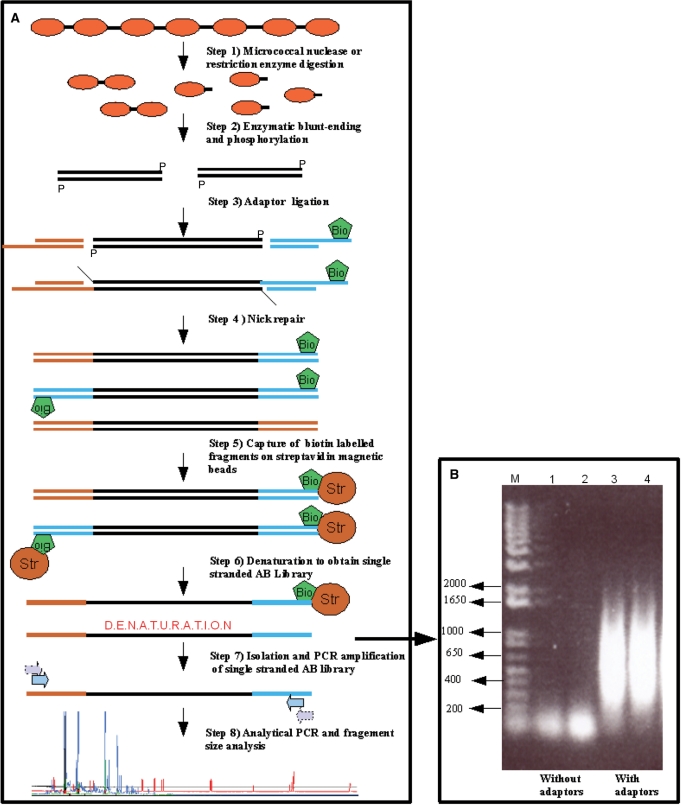
In order to avoid that for each analysed locus the linker annealing, primer extension and fragment capture steps have to be repeated as is the case in the LM-PCR method (17,18), we first created a linker-selection library of chromatin MNase or restriction enzyme digest fragments. The general set up of the library-based chromatin analysis method is presented in Figure 1. With some modifications, the method employs the strategy used to generate A-B linker genomic libraries for high-throughput sequencing. The two different adaptors are necessary for the amplification of the chromatin fragment library and to ensure directionality in the generation of the final analytical chromatin fragments (see ‘Materials and methods’ section for details and Figure 1). We have tested the robustness of this approach. First, we tested if amplification with adaptor primers occurs from a chromatin fragment library without adaptor ligation. Lack of amplification from the fragment library without adaptor ligation demonstrates that unspecific amplification with the adaptor primers does not occur. Additionally, this test revealed an average size of the chromatin digestion fragments present in the adaptor-ligated library (positive controls) between 200 bp and 1000 bp (Figure 1B). In a second test, we determined possible contaminations of the amplified A-B library by A-A and B-B adaptor fragments. Amplification of the eluted single-stranded A-B adaptor fragment library with amplifier primers A or B alone did not result in any detectable PCR amplification of fragments. In contrast, combined primers A and B resulted in a strong amplification, again showing an average size distribution of fragments between 200 bp and 1000 bp (data not shown).
Figure 1.
Overview of the experimental steps required to create and analyse a chromatin accessibility library. (A) Step 1: fungal mycelia pre-grown under specific conditions or isolated DNA (in vitro controls) are processed as described in Materials and methods section and digested with MNase or restriction enzymes of choice. Step 2: digested DNA is blunt-ended and phosphorylated by subsequent treatment of the chromatin with Klenow fragment polymerase, T4 polynucleotide kinase. This step produces blunt-ended DNA fragments for ligation with adaptors. Step 3: DNA fragments are ligated with double-stranded adaptors A and B, originating from oligonucleotides Adaptor-Ashort and Adaptor-Along or Adaptor-Bshort and Adaptor-Blong, where adaptor oligonucleotide Blong is biotinylated for later retention on the streptavidin beads. In this step, fragments containing all adaptor combinations (A-A, A-B and B-B) are generated. Step 4: the ligation step leaves nicks at the 3′-terminus that are repaired by Bst polymerase treatment. Step 5: all fragments containing biotinylated adaptor B are captured on streptavidin-coated magnetic beads. At this step, adaptor A-A fragments are lost. Step 6: after a washing step, the retained fragments (adaptors A-B and B-B fragments) are denatured at 95°C. The denaturation step results in the release of single strands which exclusively carry A-B adaptor fragments. Step 7: the single-stranded A-B adaptor fragment library is amplified by a nested PCR approach to give the final A-B fragment library. The input and output fragment libraries are quality controlled by amplification with single A and B, as well as mixed A-B primers. Only the A-B primer mix should result in the amplification of fragments in the range of 200–1000 bp (see Panel B). Step 8: the resulting A-B adaptor fragment library is diluted and aliquots are used for analytical PCR amplifications for fragment size analysis of specific loci of interest. In the final analytical PCR step, either gene-specific or adaptor-specific primers can be labelled for subsequent capillary sequencer analysis. The chromatograms are finally analysed by image analysis software. (B) Example of quality control of A-B adaptor fragment libraries. Two input chromatin fragment libraries without adaptor ligation (lanes 1 and 2) are compared to two output libraries with adaptor ligation as described in Materials and methods section (lanes 3 and 4). Libraries originating from nitrate-grown cells (lanes 1 and 3) as well as from ammonium-grown cells (lanes 2 and 4) are shown as an example. M, DNA size marker.
Fragment size analysis of locus-specific amplification products
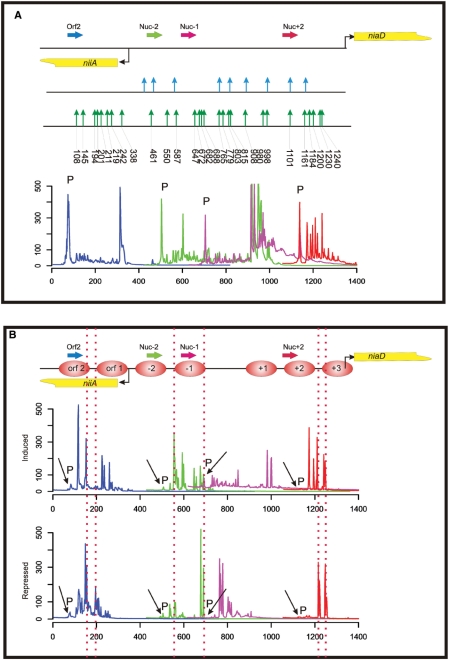
First, the amplified A-B adaptor fragment library from the in vitro digested DNA control (chromatin-free isolated DNA) was used in the fragment analysis step. We compared MNase sensitive sites in the region of the niiA-niaD promoter known from indirect end-labelling experiments (29) with sites detected by the present approach. To obtain optimal specificity for locus-specific amplification, it proved advantageous to use a nested PCR approach. Direct, single step amplification of the library resulted in a high background of unspecific fragments (data not shown). We used four different primers and four overlapping FAM-labelled nested primers to cover the majority of the regulatory niiA-niaD region (see overview of locus organization in Figure 2A). Primers have been selected on the basis of expected fragment sizes after PCR amplification. In average, fragments of up to 400 bp are obtained for one locus using a specific labelled primer and we thus used a distance of ∼300 bp on the locus map to produce overlapping chromatograms in the capillary sequencer analysis. Capillary sequencer analysis of the amplified PCR products (Figure 2A) shows an overall good correlation in naked DNA between previously determined MNase sensitive sites and sites detected by the library method. A major advantage of the library approach over gel electrophoresis methods is the possibility of precise fragment size and peak intensity determination of the sequencer chromatograms. These can be taken as an indication of MNase cutting frequency and hence of sensitivity of a target sequence to MNase digestion. The method is also more sensitive as additional MNase sensitive sites are revealed compared to the traditional gel analysis. The majority of these additional sites lie within the previously determined nucleosome-free region (nfr). Whereas the gel electrophoresis method only reveals a smear in this region, the sequencer-based analysis now allows a precise determination of MNase cutting positions at a single base pair resolution in this region.
Figure 2.
MNase accessibility assay of the A. nidulans niiA-niaD locus employing the library-based chromatin analysis method. (A) Overview of the bidirectional promoter driving gene expression of niiA and niaD (the transcriptional start points of the genes are indicated by bent arrows). A summary of MNase hypersensitive sites obtained from in vitro digested control DNA of the region is presented (indicated by vertical arrows). The MNase sites determined by indirect end-labelling and hybridization (29) are shown in the upper part of this summary by blue vertical arrows and sites determined with the library approach in this work are shown in the lower part by green arrows. Numbers below the green arrows indicate the exact nucleotide position of the MNase cut in the niiA-niaD region. The fluorescently labelled gene-specific primers used for analytical PCR and subsequent fragment analysis are shown as horizontal arrows. The colour code of the primers represents the colour code of the fragment size profiles. Below the locus overview, we show the overlapping fragment size profiles of this region composed by processing the original sequencer chromatograms as described in Materials and methods section. Signals originating from the labelled primers still present in the fragment analysis reaction mixture after the analytical PCR reaction (not incorporated primers), are indicated by a ‘P’ directly above the corresponding peak. (B) Overlapping fragment size profiles in the niiA-niaD region obtained by PCR amplification of MNase digest libraries with labelled locus-specific primers. Nucleosomes are numbered consecutively from the central nfr between nucleosome −1 and +1 [according to Muro-Pastor et al. (29)]. Nucleosomes positioned within the reading frame of niiA are depicted as orf 1 and orf 2. Two libraries are compared: (i) ‘induced’ indicates the profiles obtained from the library constructed from chromatin digestion of cells treated with nitrate as inducing agent; and (ii) ‘repressed’ indicates the profiles obtained from the library constructed from chromatin digestion of cells treated with ammonium as repressing agent. ‘P’ indicates signals originating from the labelled primers. Vertical dotted lines are drawn to highlight the highly accessible regions at the borders of positioned nucleosomes.
We next determined whether the sequencer-based analysis of library-generated fragments also mirrors the changes in nucleosome positioning of the nitrate promoter. Cultures have been grown under standard conditions for which a detailed nucleosome map of the niiA-niaD reference locus analysed by the traditional indirect end-labelling technique is available (29). Under nitrogen metabolite repressing conditions (ammonium as sole nitrogen (N) source, Figure 2B, chromatogram ‘repressed’) six nucleosomes are strictly positioned over this promoter and this positioning is lost upon nitrate induction (Figure 2B, chromatogram ‘induced’) in strains expressing functional variants of the two synergistic activators, AreA and NirA. When we tested MNase accessibilities in the region of nucleosomes −2 to +3, the pattern obtained from sequencer fragment analysis correlates with the positioning pattern determined by indirect end-labelling. In addition, we have tested here the positioning of two nucleosomes in the open reading frame (ORF) of the niiA gene previously not analysed. The pattern shows that under non-transcribing conditions (repressed), high accessibility to MNase is obtained only in the linker region between nucleosomes Orf2 and Orf1, consistent with the observations that MNase preferentially cleaves at the borders of strictly positioned nucleosomes (35). Under nitrate-inducing conditions, the positioning of nucleosome Orf1 is lost leading to high MNase accessibility to the DNA inside this nucleosome. This pattern is in agreement with high transcription rates of this gene under inducing conditions (data not shown).
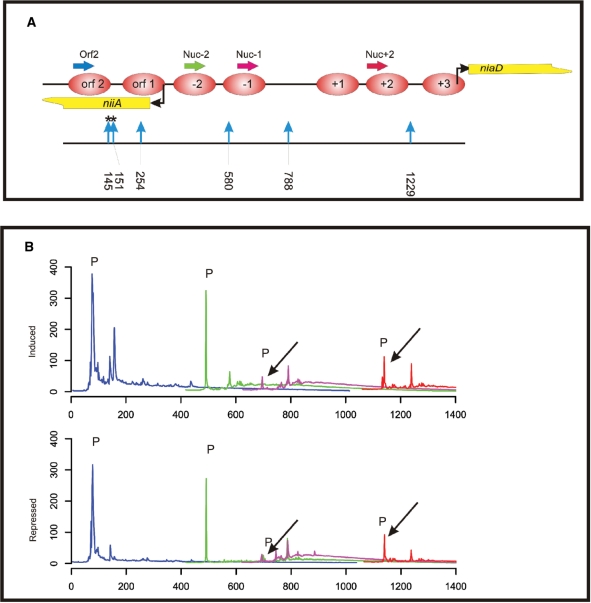
Library-based analysis of restriction enzyme accessibility
Nucleosome positioning in a region with known sequence can also be analysed by accessibility to specific restriction enzymes. Indirect end-labelling following electrophoretic separation (36) or quantitative PCR amplification after restriction enzyme digestion (27) has been used for this purpose. We have tested here if the library-based method can also be used for restriction enzyme accessibility tests and analysed the niiA-niaD Intergenic region (IGR) locus using the HaeIII restriction enzyme. The relative positions of all HaeIII sites in the IGR are shown in Figure 3A. The fragment analysis in Figure 3B shows that the majority of HaeIII sites are accessible under open chromatin, nitrate-induced conditions (chromatogram ‘induced’), but accessibility is restricted under ammonium-repressing conditions (chromatogram ‘repressed’).
Figure 3.
Chromatin restriction enzyme accessibility assay employing the library approach. (A) Overview of the positions of HaeIII restriction endonuclease sites in the niiA-niaD region. Restriction site positions are indicated by vertical blue arrows and numbers below the arrows indicate cutting site positions in the bidirectional promoter region. Asterisks above two sites (145 and 151) indicate cutting events obtained in the assay at positions which do not correspond to a HaeIII site and which probably represent sites of HaeIII star activity. Nucleosome positions, transcriptional start points and analytical PCR primer positions are as described in Figure 2. (B) Overlapping fragment size profiles in the niiA-niaD region obtained by fragment size analysis of libraries derived from HaeIII-treated chromatin. Primers and colour code are as in Figure 2. Two libraries are compared: (i) ‘induced’ indicates the profiles obtained from the library constructed from HaeIII chromatin digestion of cells treated with nitrate; and (ii) ‘repressed’ indicates the profiles obtained from the library constructed from HaeIII chromatin digestion of cells treated with ammonium. As in Figure 2, ‘P’ indicates signals originating from non-incorporated labelled primers.
Library-based analysis of nucleosome positioning in the areA promoter
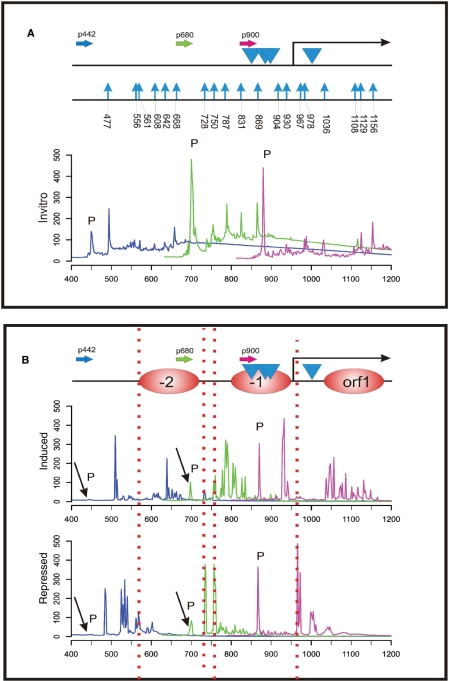
The GATA-factor AreA has multiple functions in the up-regulation of the nitrate assimilation genes (see ‘Introduction’ section). In addition to regulating many nitrogen metabolite repressible genes, AreA is positively autoregulated. AreA binds to cognate GATA sites in the niiA-niaD IGR, recruits histone H3 acetylating enzymes (32), interacts with the pathway-specific activator NirA and increases its in vivo binding site occupancy (22,27,29,30).
We used this method to investigate the currently non-resolved nucleosomal pattern of the promoter region of areA (Figure 4). We used the same MNase-derived A-B adaptor fragment libraries (repressed and induced) as in previous experiments (Figures 2 and 3) and amplified fragments with three different FAM-labelled primers binding to the areA promoter region. Fragment size analysis of the amplification products allowed us to resolve three positioned nucleosomes under repressed conditions (Figure 4B, chromatogram ‘repressed’). Interestingly, positioned nucleosome −1 covers a region which contains three putative AreA-binding GATA sites. Platt and co-workers (23) have shown that AreA is positively autoregulated. These GATA sites might therefore be excluded by the nucleosome for binding AreA under repressive conditions, leading to disruption of the positive autoregulation loop under nitrogen metabolite repressing conditions. In contrast, nitrate induction leads to chromatin remodelling as seen from the elevated MNase accessibility in the promoter region and in the first nucleosome of the areA ORF. Comparison of the induced MNase patterns obtained by the traditional end-labelling method and the fragment chromatograms again showed a good correlation of digestion profiles (Supplementary Figure S1).
Figure 4.
Nucleosome positioning analysis at the areA gene promoter. (A) Overview of the 700 bp promoter region of areA. The FAM-labelled gene-specific primers used for analytical PCR and subsequent fragment analysis are shown as horizontal arrows and named according to their relative position within the promoter sequence. The colours of the primers represent the colour code of the fragment size profiles. Blue arrowheads indicate the position of predicted AreA binding GATA sites in the promoter of this positively autoregulated gene. The start of the areA coding region (ORF) is indicated by the bent arrow. A summary of MNase hypersensitive sites obtained from fragment size analysis of the in vitro control DNA is presented (indicated by vertical blue arrows). Numbers below the blue arrows indicate the exact nucleotide position of the MNase cut in the areA promoter region. Overlapping fragment size profiles in the areA promoter obtained by fragment size analysis of the in vitro MNase digest library using the primers indicted above are shown below the locus overview. As in the other figures, ‘P’ indicates signals originating from non-incorporated labelled primers. (B) Overlapping fragment size profiles in the areA promoter obtained by fragment size analysis of the same MNase digest libraries as used for the analysis of the niiA-niaD region (Figure 2). Analytical PCRs were carried out using labelled primers indicated in panel A. The areA promoter profiles of the two libraries (induced, repressed) are shown here. The nucleosome positioned within the reading frame of areA is depicted as orf 1 and nucleosomes in the promoter region are designated −1 and −2. Under conditions in which AreA is active on its own promoter (induced) additional MNase cutting sites are revealed in nucleosome −2 at position ∼650 bp, at the end of nucleosome −1 at position ∼930 bp as well as in the orf1 nucleosome. ‘P’ indicates non-incorporated primer signals.
Fluorescently labelled linker primer is equally suitable for fragment amplification
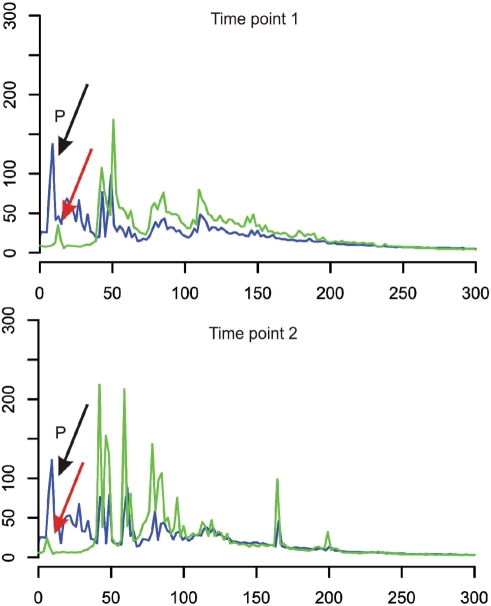
The approach used throughout this study requires one labelled primer for each locus amplification step. When multiple loci need to be analysed in one condition-specific chromatin fragment library, this strategy becomes costly due to the increased costs of primer labelling. We therefore have tested if also the opposite primer binding to the adaptor (primer Amplifyer-Ain) can be labelled and non-labelled locus-specific primers can be used for amplification. This strategy would considerably reduce consumables costs for multi-locus analysis. To this end, we generated two additional A-B adaptor fragment libraries and compared chromatin accessibility profiles in both libraries between the different primer labelling strategies. Figure 5 shows the results of both libraries comparing profiles obtained with gene-specific primer labelling (blue line profiles) with profiles obtained with adaptor primer labelling (green line profiles). Labelled and non-labelled gene-specific primers are amplifying an A. nidulans locus involved in secondary metabolism regulation (aflR promoter, gene number AN7802.3). Both, the green line (adaptor-specific labelled primer) and the blue line (gene-specific labelled primer) profiles match very well in both libraries indicating that labelling the adaptor-binding primer for fragment amplification is a suitable strategy to economize multi-locus chromatin accessibility analyses with the library-based method. The difference in peak intensities reflects the differences in PCR efficiencies in the individual reactions. Both reactions start with the same amount of FAM-labelled primers, in the case of the adaptor-specific primer the higher peak intensities of the PCR products correlate with the lower intensity of the primer peak.
Figure 5.
Either locus-specific or adaptor-binding primers can be labelled for analytical PCRs and fragment size analysis. The fragment size profiles of analytical PCRs using either the labelled locus-specific primer (green line profiles) or the labelled adaptor-binding primer (blue line profiles) are compared. Two independent A-B adaptor fragment libraries have been tested for this comparison and both profiles represent the MNase accessibility of the locus AN7802.3 at different time points 1 and 2. ‘P’ indicates non-incorporated primer signals.
CONCLUSIONS
The presented method allows the rapid and accurate analysis of chromatin accessibility and nucleosome positioning at many genomic loci in parallel using the same starting material, i.e. strain or condition-specific chromatin fragment libraries. This cost-effective method fills a technical gap between traditional chromatin analysis methods (end-labelling or LM-PCR analysis) and the resource-intensive genome-wide analysis methods such as high-throughput sequencing. Fragment size determination of chromatin digests results in a one base pair resolution and the capillary sequencer analysis set up allows the processing of hundreds of samples in parallel. This method is specifically designed for applications in which the specific scientific question requires analysis of many loci in different strains under a variety of experimental conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Science Fund FWF (P 19731-B11 to J.S.). Funding for open access charge: Austrian Science Fund, FWF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank an anonymous reviewer for many valuable suggestions how to improve this article.
REFERENCES
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 5.Emre NC, Berger SL. Histone post-translational modifications regulate transcription and silent chromatin in Saccharomyces cerevisiae. Ernst Schering Res. Found. Workshop. 2006;57:127–153. doi: 10.1007/3-540-37633-x_8. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 10.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 11.Bresnick EH, Johnson KD, Kim SI, Im H. Establishment and regulation of chromatin domains: mechanistic insights from studies of hemoglobin synthesis. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:435–471. doi: 10.1016/S0079-6603(06)81011-1. [DOI] [PubMed] [Google Scholar]
- 12.Keene MA, Corces V, Lowenhaupt K, Elgin SC. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl Acad. Sci. USA. 1981;78:143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinke H, Horz W. Anatomy of a hypersensitive site. Biochim. Biophys. Acta. 2004;1677:24–29. doi: 10.1016/j.bbaexp.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Cockell M, Rhodes D, Klug A. Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J. Mol. Biol. 1983;170:423–446. doi: 10.1016/s0022-2836(83)80156-9. [DOI] [PubMed] [Google Scholar]
- 15.Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 17.Ingram R, Gao C, Lebon J, Liu Q, Mayoral RJ, Sommer SS, Hoogenkamp M, Riggs AD, Bonifer C. PAP-LMPCR for improved, allele-specific footprinting and automated chromatin fine structure analysis. Nucleic Acids Res. 2008;36:e19. doi: 10.1093/nar/gkm1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram R, Tagoh H, Riggs AD, Bonifer C. Rapid, solid-phase based automated analysis of chromatin structure and transcription factor occupancy in living eukaryotic cells. Nucleic Acids Res. 2005;33:e1. doi: 10.1093/nar/gni001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl Acad. Sci. USA. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punt PJ, Strauss J, Smit R, Kinghorn JR, van den Hondel CA, Scazzocchio C. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell Biol. 1995;15:5688–5699. doi: 10.1128/mcb.15.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt A, Langdon T, Arst H, Kirk D, Tollervey D, Sanchez JM, Caddick MX. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 1996;15:2791–2801. [PMC free article] [PubMed] [Google Scholar]
- 24.Scazzocchio C. The fungal GATA factors. Curr. Opin. Microbiol. 2000;3:126–131. doi: 10.1016/s1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- 25.Strauss J, Muro-Pastor MI, Scazzocchio C. The regulator of nitrate assimilation in ascomycetes is a dimer which binds a nonrepeated, asymmetrical sequence. Mol. Cell Biol. 1998;18:1339–1348. doi: 10.1128/mcb.18.3.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burger G, Strauss J, Scazzocchio C, Lang BF. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol. Cell Biol. 1991;11:5746–5755. doi: 10.1128/mcb.11.11.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger H, Pachlinger R, Morozov I, Goller S, Narendja F, Caddick M, Strauss J. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol. Microbiol. 2006;59:433–446. doi: 10.1111/j.1365-2958.2005.04957.x. [DOI] [PubMed] [Google Scholar]
- 28.Bernreiter A, Ramon A, Fernandez-Martinez J, Berger H, Araujo-Bazan L, Espeso EA, Pachlinger R, Gallmetzer A, Anderl I, Scazzocchio C, et al. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell Biol. 2007;27:791–802. doi: 10.1128/MCB.00761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muro-Pastor MI, Gonzalez R, Strauss J, Narendja F, Scazzocchio C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter [published erratum appears in EMBO J. 1999;18:2670] EMBO J. 1999;18:1584–1597. doi: 10.1093/emboj/18.6.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muro-Pastor MI, Strauss J, Ramon A, Scazzocchio C. A paradoxical mutant GATA factor. Eukaryot. Cell. 2004;3:393–405. doi: 10.1128/EC.3.2.393-405.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendja F, Goller SP, Wolschek M, Strauss J. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 2002;44:573–583. doi: 10.1046/j.1365-2958.2002.02911.x. [DOI] [PubMed] [Google Scholar]
- 32.Berger H, Basheer A, Bock S, Reyes-Dominguez Y, Dalik T, Altmann F, Strauss J. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol. Microbiol. 2008;69:1385–1398. doi: 10.1111/j.1365-2958.2008.06359.x. [DOI] [PubMed] [Google Scholar]
- 33.Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 34.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 36.Barbaric S, Reinke H, Horz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.