Abstract
Accessing the nucleus through the surrounding membrane poses one of the major obstacles for therapeutic molecules large enough to be excluded due to nuclear pore size limits. In some therapeutic applications the large size of some nucleic acids, like plasmid DNA, hampers their access to the nuclear compartment. However, also for small oligonucleotides, achieving higher nuclear concentrations could be of great benefit. We report on the synthesis and possible applications of a natural RNA 5′-end nuclear localization signal composed of a 2,2,7-trimethylguanosine cap (m3G-CAP). The cap is found in the small nuclear RNAs that are constitutive part of the small nuclear ribonucleoprotein complexes involved in nuclear splicing. We demonstrate the use of the m3G signal as an adaptor that can be attached to different oligonucleotides, thereby conferring nuclear targeting capabilities with capacity to transport large-size cargo molecules. The synthetic capping of oligos interfering with splicing may have immediate clinical applications.
INTRODUCTION
Nucleocytoplasmic transport of endogenous molecules is a regulated process where small molecules can diffuse through the nuclear pore complex of the nuclear membrane, while molecules >40 KDa require the use of a signal and energy-mediated processes. Import of nuclear proteins requires nuclear localization signals (NLS) in the form of specific amino-acid sequences which mediate the interaction with carrier proteins (1). The best studied NLS sequence is the SV40 large-T antigen. It mediates the interaction between the cargo protein, bearing the NLS signal, and an import receptor consisting of an adaptor protein, importin α, which directly binds the NLS signal, and importin β which is the mediator of the actual import process through the nuclear pores (2).
An archetypical NLS, which is mediating the interaction with several different types of import receptors, is found on the ribosomal L23a protein. This NLS, which has a higher degree of complexity and harbours very basic regions, is thought to have evolved prior to the evolutionary divergence of import receptors (3).
In addition to the cargo and import receptor interactions, there are other factors needed in the process of nuclear import. It is the asymmetric distribution of the factors Ran, RCC1, RanGAP1 and RanBP1, creating a steep RanGTP gradient across the nuclear envelope that allows the directional movement of the cargo–importin receptor complex to the nucleus (4,5).
Apart from nuclear proteins some RNAs, in the form of small nuclear ribonucleoprotein complexes (snRNP), use signals for nuclear import. These RNAs are comprised of the major spliceosomal U snRNAs, such as U1, U2, U4 and U5 and are the major building units of the spliceosomal complex. U snRNAs are transcribed in the nucleus by RNA polymerase II after which they acquire a 7-methylguanosine (m7G) cap structure at their 5′-end. This cap structure acts as a nuclear export signal that is recognized by the cap-binding complex (CBC). The CBC complex is in turn recognized by the export receptor CRM1 with the help of the PHAX adapter leading ultimately to the nuclear export of the U snRNA (6).
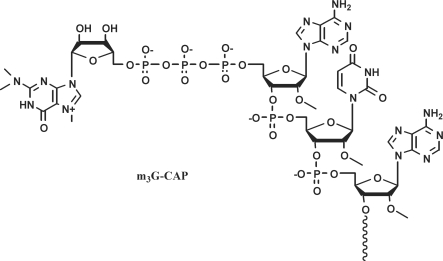
After release in the cytoplasm the U snRNA is recognized by the survival of motor neuron complex (SMN) that directs the proper assembly with a group of Sm proteins (7–10). Subsequently, the m7G cap is hypermethylated to a 5′ 2,2,7-trimethylguanosine (m3G) cap structure (Figure 1) by the small-nuclear RNA cap hypermethylase (11,12). The matured snRNP is then imported back into the nucleus. This nuclear transport involves two different pathways and two very distinct import signals, both of which, however, recruiting importin β (13–16). The first pathway uses a still ill-defined import signal present in the Sm core domain of the snRNP formed by the Sm proteins (17). The second pathway involves the use of the m3G-CAP structure (18). The m3G-CAP signal is recognized by the import adaptor protein snurportin (SPN1) (14,19,20), which in turn is recognized by importin β (14–16,21).
Figure 1.
Structure of the 2,2,7-trimethylguanosine cap.
Nuclear import of therapeutic molecules is of great importance for many applications. For example, specific targeting of exogenous proteins, such as antibodies, to the nucleus for the purpose of radioimmunotherapy, is seen as a way to increase cytotoxicity effects in cancer cells (22). In particular, one of the areas where nuclear targeting has been getting a lot of attention is the gene delivery/gene therapy field. This is especially true in non-viral or synthetic vector development, since these transport systems need to mimic most of the virus strategies used to overcome several cellular barriers of which the nuclear membrane is the ultimate one.
Some approaches to nuclear delivery of nucleic acids, like plasmid DNA, have relied on the DNA sequence itself (23). A plethora of other types of strategies for nucleic acid delivery have been based, however, on the direct or indirect attachment of NLS peptides to the nucleic acid molecules as a way to promote binding of the importin α/importin β heterodimer to the nucleic acid construct and its subsequent nuclear translocation (24). The SV40 large T antigen NLS (especially in its shortest form pkkkrkv) has been one of the most employed NLS peptides. It has been associated to DNA via ionic interactions (25); via chemical coupling (26); through the use of peptide nucleic acids (PNA) linked to the NLS and bound to DNA in a sequence-specific manner (27,28); through the use of a triple helix forming oligonucleotide system (29) and by the use of biotinylated DNA bound to NLS conjugated streptavidin (30). An example of another peptide NLS used is the non-classical NLS defined by the M9 sequence, coming from the heterogeneous nuclear ribonucleoprotein (hnRNP) A1, which has been used together with a DNA binding peptide for increased nuclear delivery after lipofection of non-dividing cells (31).
We are now exploring the possibility to use a different type of signal, the m3G-CAP, for the purpose of enhancing nuclear delivery of nucleic acids. We coupled the m3G pyrophosphate to 5′-phosphorylated 2′-O-methyloligoribonucleotides to achieve m3G-capped oligonucleotides resembling the 5′-end of endogenous U snRNAs. The presence of the m3G-CAP clearly showed an increase of the nuclear delivery of the oligonucleotide together with a large cargo protein (Streptavidin) after microinjections of Xenopus oocytes and when using a protein transfection method for cultured cells that delivers proteins to the cytosol. Furthermore we observe increased splice correction when the m3G-CAP is connected to an antisense oligonucleotide, which provides a functionally relevant example.
MATERIALS AND METHODS
Synthesis of m3G-capped oligonucleotides
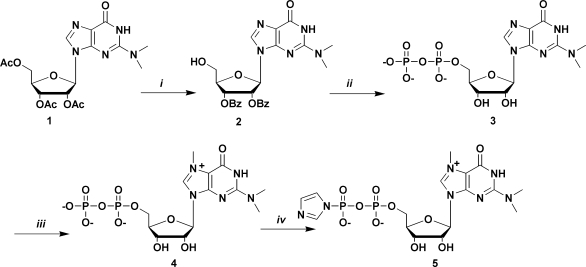
p-AS705 and p-PMO2 were converted to the m3G-capped derivatives by reaction with the m3G-5′-pyrophosphorylimidazolide 5 in a Mn(II) promoted (32) capping procedure. Synthesis of 5 (Scheme 1), was modified compared to previously published procedures (32,33) which includes a new one-pot, five-step 5′-Pyrophosphate 3 synthesis, based on known reactions (34–37). First 1 was converted to compound 2. To the purified compound 2 (171 mg, 0.33 mmol) in 2 ml of dry DMF was then added salicyl chlorophosphite (1.9 eq, 0.126 g, 0.627 mmol), whereupon the reaction mixture was stirred at r.t. [under Ar (g)]. After 15 min, a solution of tetra(tri-n-butylamine)pyrophosphate in DMF (2 eq, 0.66 mmol, 1.32 ml from a 0.5 mM stock solution, which was directly before reaction vortexed with 0.5 ml tri-n-butylamine) was added. After 20 min, a solution of iodine (0.091 g, 1.1 eq, 0.363 mmol) in 1.5 ml pyridine, containing water (0.33 mmol, 0.00 6ml, 1 eq) was added. After an additional 15 min ethylenediamine (0.11 ml, 1.65 mmol, 5 eq) was added and the reaction was stirred for 1 h. Solvents were evaporated and the crude product was dissolved in NH3(aq) and left to react overnight. After concentration, the crude product was dissolved in water and purified by preparative RP HPLC using a gradient of buffer B in buffer A (0–20% B over 30 min). Concentration of collected fractions gave N2, N2-dimethylguanosine 5′-pyrophosphate (3) as the tris(triethylamine)salt. RT = 23 min. Yield 55.8 mg, 22% (over five steps). 3 Was treated with sat. NH3 in methanol for two days at r.t. After concentration, the product was dissolved in water and washed with dichloromethane (3×), concentrated in vacuo, freeze-dried and redissolved in dry DMF (1 ml). Methyl iodide was then added (0.08 ml, 1.28 mmol, 20 eq) and the reaction was stirred at 40°C for 5 h. The solvent was evaporated and the product was dissolved in water and purified by preparative RP-HPLC using a gradient of buffer B in buffer A (0–10% B over 40 min). Evaporation of collected fractions gave N7, N2, N2-trimethylguanosine 5′-pyrophosphate 4 as the tris(triethylammine)salt (alternatively 4 was synthesized in an alternative fashion from 3, see Supplementary Material). Yield 52 mg, 20% (after 6 steps, from compound 2). 4 Was then converted to the imidazolide 5, which was used in reaction with the 5′-phosphorylated oligonucleotides to produce m3G-CAP-AS705 and m3G-CAP-PMO2 (Table 1).
Scheme 1.
Synthesis of the m3G 5′-pyrophosphateimidazolide. Reagents and conditions: (i) a: Py/NH3aq sat., 3 h, r.t.,; b: MMTrCl, Py/DMF, 24 h, r.t.; c: BzCl, Py, r.t., 24 h, D 80% AcOH, r.t., 5 h; (ii) a: salicyl chlorophosphite, r.t., 15 min; b: tri-n-buthylammine pyrophosphate, DMF, r.t, 20 min; c: I2, Py/H2O, r.t, 15 min; d: ethylenediamine, r.t., E NH3aq sat., 0°C 48 h; (iii) MeI, DMF, 40°C, 5 h; (iv) imidazole, triphenylphosphine, di-2-pyridyldisulphide, DMF 24 h. Py, pyridine; MMTrCl, monometoxy trityl chloride; DMF, dimethylformamide; r.t. = room temperature; BzCl, benzoyl chloride; AcOH, acetic acid; MeI, methyl iodide.
Table 1.
Oligonucleotides
 |
Oocyte microinjections, western blot analysis and fluorescence microscopy
Streptavidin-Alexa488 (Molecular Probes) + oligonucleotide constructs (Figure 2) were formed by incubating 3 µg STV (Streptavidin) reconstituted in 1× phosphate buffer saline (PBS, pH 7.4) with 2–4 times molar amount of biotinylated oligo constructs. The volume was adjusted with nuclease free water (Qiagen) to have a final concentration of 329 ng STV/µl and the incubation proceeded for 2 h at r.t. and if not used the same day the constructs were stored at 4°C.
Figure 2.
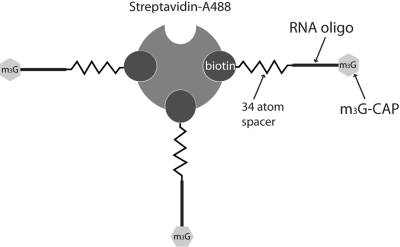
Schematic representation of the m3G-CAP oligo construct bound to the Streptavidin-Alexa488 (STV) used in the nuclear transport assays by cytoplasmic microinjections in Xenopus oocytes and by PULSin protein delivery to the cytosol of a mammalian cell line. Image proportions are exaggerated for a better understanding.
Prior to microinjections, the prepared STV-oligo constructs were spun down at 16 000g for 5 min. A volume of 41.4 nl of STV-oligo constructs were injected in the cytoplasm of the oocyte using the Nanoinjector 2000™ pump (World Precision Instruments, Inc.) followed by incubation at 19°C in OR2 buffer for 4 h. After incubation, oocyte nuclei were manually dissected in nuclear dissection buffer (140 mM KCl; 0.5 mM MgSO4; 20 mM Tris–HCl pH 7.2) and collected directly in 2× Laemelli sample buffer. Collected nuclei were then analysed by western blot (for details see Supplementary Material). Fluorescence microscopy imaging details are provided in the Supplementary Material.
Streptavidin-oligo complex transfections
U2OS cells were seeded on top of coverslips in a 24-well plate with DMEM+10% FCS the day before transfection so that they were confluent or close to confluency the next day. Streptavidin-Alexa488 (Molecular Probes) + oligonucleotide constructs (Figure 2) were formed by incubating 2 µg STV (Streptavidin) reconstituted in 1×PBS (pH 7.4) buffer with 2–4 times molar amount of biotinylated oligo constructs for 2 h at r.t. 20 mM HEPES buffer (pH 7.4) was then used to bring the total volume up to 100 µl and then 3.5 µl of PULSin transfection reagent (PolyPlus-Transfection, NY, USA) was added. The mixture was vortexed and left to incubate for 15 min at r.t. During this incubation time, the cells were washed twice with PBS and 900 µl of pre-warmed OPTI-mem I (Invitrogen) was added to the wells. The PULSin complexes were then added to the cells and left to incubate for 4 h. Subsequently, the medium was discarded and the cells washed twice with pre-warmed OPTI-MEM I. Finally, 400 µl of OPTI-MEM I was added to the wells after which the cells were further incubated for 2–3 h. Cells were then analysed by fluorescence microscopy and confocal microscopy (for details see Supplementary Material).
Oligonucletide transfections
HeLa/Luc705 cells were pre-seeded at a density of 35 000 cells per well in a 24-well plate the day before transfection, in order to have between 40% and 50% confluency the next day. Antisense oligonucleotide constructs were transfected by using Oligofectamine reagent (Invitrogen) and following the manufacturer's protocol. Cells were harvested in Luciferase lysis buffer (25 mM TAE, 10% glycerol, 1 mM EDTA, 1% Triton-X100) after 24 h and the level of luciferase expression was analysed based on a luciferase activity assay (Luciferase Assay Kit, Biothema AB, Handen, Sweden). Total protein was measured using a BCA protein assay (Micro BCATM protein assay kit, Thermo Scientific-Pierce Protein Research Products, Rockford, USA). RT–PCR was done by isolating total RNA from cells with RNeasy plus kit (QIAGEN) from which a total of 3ng of RNA was then used in each reaction (total volume per reaction was 20 µl) with the ONE STEP RT-PCR kit (QIAGEN) and following the manufacturers protocol. The primers used were: Fwd-TTGATATGTGGATTTCGAGTCGTC; Rev-TGTCAATCAGAGTGCTTTTGGCG. The program for the RT–PCR was as follows: (55°C, 35 min + 95°C, 15 min) × 1 cycle + (94°C, 30 s + 55°C, 30 s + 72°C, 30 s) × 29–30 cycles + 72°C, 10 min final extension. The PCR products were analysed in a 2% agarose gel in 1× TBE buffer and visualized by SYBR-Gold (Invitrogen, Molecular Probes) staining. Gel images were captured by a Fluor-S gel documentation system (BioRad) with the Quantity One software (BioRad).
Statistics
In Figure 3, a paired t-test with two-tailed P-value was used for analysis; In Figure 5, the 2 × 2 contingency table was analysed by the Fisher's exact test with two-tailed P-value.
Figure 3.
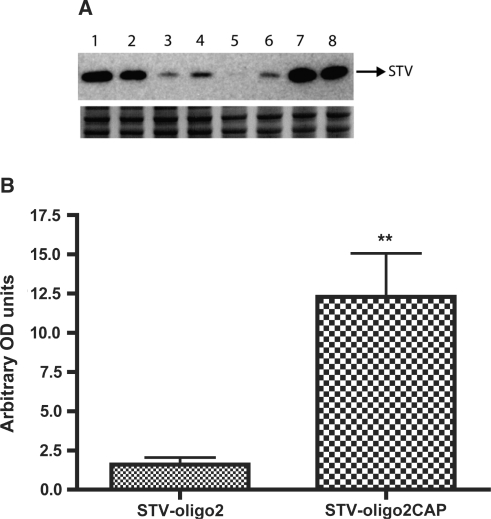
Accumulation of Streptavidin complexes in Xenopus oocyte nuclei after cytoplasmic injections. Oocytes were injected in the cytoplasm with either STV-oligo2 or STV-oligo2CAP complexes and after 4 h incubation the nuclei were dissected and collected for western blot analysis. (A) Upper panel shows western blot using anti-Streptavidin probing. Each lane corresponds to one group of four nuclei pooled together. Lanes 1–2–7–8: nuclei dissected from oocytes injected with STV-oligo2CAP (m3G-CAP-PMO2); lanes 3–4–5–6: nuclei dissected from oocytes injected with STV-oligo2 (p-PMO2). Lower panel shows a protein loading control by staining the PAGE gel with comassie blue after transfer. (B) Graph showing the quantification of western blot results by densitometry with normalization to the loading controls using Fluor-S MultiImager and Quantity One software® (BioRad) (SDs for n = 4 are shown). ** P < 0.005.
Figure 5.
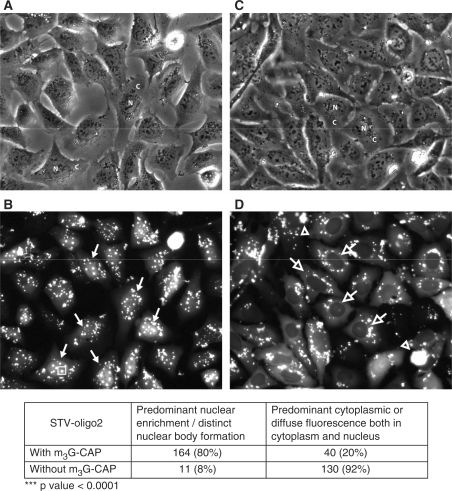
Localization of fluorescent Streptavidin bound to 3–4 biotinylated 2′-O-methyl RNA oligos with (pictures A and B) or without (pictures C and D) m3G-CAP (p-PMO2 and m3G-CAP-PMO2 oligos, respectively). Streptavidin-oligo complexes were transfected into U2OS cell lines by the use of PULSin reagent (protein transfection reagent). After 4 h incubation at 37°C, cells were washed and incubated for 2 h at 37°C before fluorescent microscopy (B and D) and phase contrast pictures (A and C) were taken. Fluorescent microscopy photographs were used to count the cells according to the described criteria and results are presented in the lower table. N and C mark some nuclei and cytoplasm of cells, respectively, in the phase contrast pictures. Solid white arrows indicate examples of cells counted as positive for nuclear enrichment; open white arrows point to cells counted as negative for nuclear enrichment; the open white arrowhead points to aggregates of fluorescent STV complexes occurring during the transfection procedure; open rectangular boxes were drawn around the ‘STV-m3G-oligo nuclear bodies’ formed by increased accumulation of STV complexes in the nucleus.
RESULTS
m3G capped RNA oligonucleotides are able to direct nuclear accumulation of a cargo protein in Xenopus oocytes
The use of Xenopus oocytes for the analysis of nuclear transport is a very well characterized system offering many advantages for this type of studies (38–41). First, they are very big cells with a large nucleus and easy to inject; second, it is quite straightforward to study the isolated compartments (cytoplasm and nuclei), since the nuclei are relatively easy to isolate by manual dissection. A further very important feature is that the oocyte is a non-dividing cell, which thereby maintains its nuclear membrane integrity throughout the experiments. Yet another reason to use the Xenopus oocyte for the initial analysis of m3G-capped oligos was that many studies dealing with import mechanisms of U snRNAs were initially done using these cells (12,18,38,42).
We decided to study the nuclear accumulation of a construct constituted of a fluorescent Streptavidin (STV) bound to 2–4 biotinylated 2′-O-methyl RNA oligos with (CAP-PM02) or without (p-PM02) m3G-CAP modification (Figure 2). The STV protein itself has a molecular weight of around 53 KDa and the addition of 2–4 biotinylated oligos equals a total MW for the construct of approximately 60–70 KDa. This should make it difficult for the STV-oligo construct to enter the nucleus by passive diffusion alone, since 45–50 KDa is the known cut-off of the nuclear pore. The cargo STV was linked through a long spacer to the carrier oligo, providing insight into the possibility of using the m3G-CAP signal distanced from its cargo.
Injections into the oocyte cytoplasm of STV-oligo constructs with or without added m3G-CAP were performed and after an incubation period of 4 h the nuclei were manually dissected to check for the relative amounts of STV protein that had been directed to the nuclear compartment. By analysing the amounts of STV recovered from the nuclear compartment with the use of western blot (Figure 3A), we verified that the addition of m3G-CAP oligos enhanced nuclear uptake at least 6-fold compared to the non-capped oligos.
There was still some STV recovered from the nucleus of oocytes injected with the non-capped oligo versions. This may be due to the fact that the outer nuclear membrane is in direct contact with the cytoplasm, hence there might be some residual unspecific binding of STV to the nuclear membrane. Due to this, the 6-fold higher nuclear uptake seen with the m3G-CAP may thus be regarded as the minimal difference. We can, however, not exclude that some STV-oligo complexes diffuse passively into the nucleus, since the total MW of the constructs is close to the nuclear pore cut-off size.
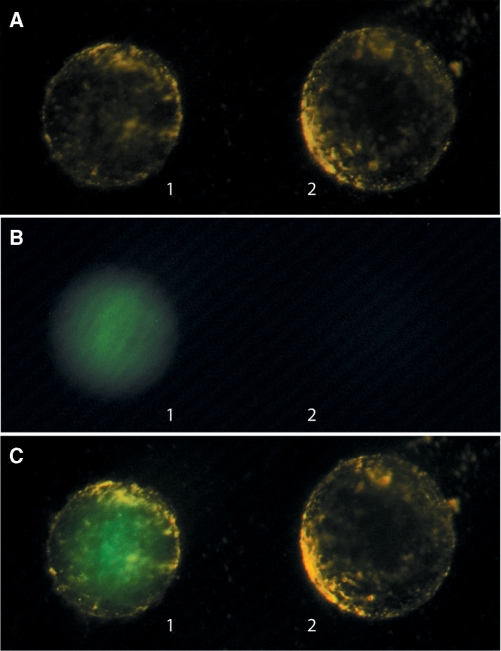
In addition to western blot analysis, we performed fluorescent microscopy of isolated nuclei after cytoplasmic injections of the STV complexes. As can be seen in Figure 4, the nuclei were isolated in pairs each representing a cytoplasmic injection of STV with (nucleus 1) or without (nucleus 2) capped oligonucleotide. By putting the nuclei side by side, it was easily seen that only the oocyte injected with STV-CAPoligo showed a fluorescent nucleus. The nucleus from oocytes injected with control oligo with no m3G-CAP lacked detectable fluorescence confirming the previous results of the western blot, where the capped oligo conferred enhanced nuclear transport capacity to the STV-oligo complex.
Figure 4.
Fluorescence imaging of the accumulation of Streptavidin complexes in Xenopus oocyte nuclei after cytoplasmic injections. Oocytes were injected in the cytoplasm with either STV-oligo2 or STV-oligo2CAP complexes and after 4 h incubation the nuclei were dissected. The nuclei were gently tapped with a glass rod, in order to release excess cytoplasm that might be still attached to the nuclear membrane after dissection. Photos were then taken with two nuclei side by side representing cytoplasmic injections of capped (nucleus 1) and non-capped (nucleus 2) STV-oligo constructs. (A) Photo taken with oblique visible light in order to get a phase contrast effect making it possible to see the almost transparent nuclear membrane; (B) same nuclei imaged with HBO light and fluorescence FITC filter; (C) same nuclei imaged both with low power visible light and with an HBO light with FITC filter. A number of 10 pairs of nuclei were analysed with all of them showing the same pattern as illustrated above.
m3G-capped RNA oligonucleotides are able to direct nuclear accumulation of a cargo protein in mammalian cells after cytosolic delivery by a transfection reagent
We went on to test if the previous constructs would behave similarly when delivered to a mammalian cell line and, importantly, by a transfection method that resembles the delivery routes of many gene transfer protocols. For this, we employed a method of protein and peptide delivery through the use of a reagent that promotes uptake of proteins into the cytosol of cells (PULSin) after endocytosis of protein–reagent complexes.
We first tested the feasibility of the method by using STV constructs bound to 2–3 biotinylated SV40-NLS peptides (pkkkrkv) or biotinylated SV40-mutatedNLS peptides (pktkrkv). When transfected to the cytosol of U2OS cells these constructs localized preferably either in the nucleus (SV40-NLS construct >90%) or in the cytoplasm (SV40-mutNLS >90%) as observed by the detection of fluorescence coming from the cargo STV-Alexa488 (data not shown).
We then tested the same constructs used in the Xenopus oocyte microinjections in the mammalian U2OS cell line. After transfection of the constructs we saw a completely different pattern between STV-oligo2CAP and STV-oligo2, both using 2–3 (see Supplementary Figure 5) or 3–4 oligos bound to STV (Figure 5). The cells did not only show a higher nuclear fluorescence but also a very specific pattern, with very prominent nuclear bodies containing most of the STV fluorescence, when the cargo was attached to the m3G-CAP oligos (Figure 5C). The size of these ‘STV-m3G-oligo bodies’ was easily correlated to the amount of STV found in the nuclear compartment and corresponded to most of the nuclear fluorescence in contrast to an overall diffuse pattern seen when using STV-NLS peptides.
By using fluorescence microscopy, cells were also enumerated using specific criteria. Cells counted as positive for enhanced nuclear transport were characterized as having a more overall nuclear fluorescence with distinct bodies. Cells assigned as negative were characterized as having diffuse, preferential staining in the cytoplasm or both in the cytoplasm and nucleus (which argues for that the STV was not being actively targeted to the nuclei, but instead could diffuse in and out of the nuclear compartment) (Figure 5D). Following these criteria, we found a very significant difference (P < 0.0001) between the STV constructs bound by 3–4 m3G-CAP-oligos (80% predominantly nuclear) and STV bound by 3–4 nonCAP-oligos (92% predominant cytoplasmic or overall diffuse staining). For the STV constructs carrying 2–3 oligos, the percentages were 92% nuclear and 81% cytoplasmic, respectively for m3G-CAP and nonCAP attached oligos (Supplementary Figure 5). It should be noted that when these STV-oligo constructs were created, together with the PULSin reagent, some aggregates were still remaining in the wells (these were visible as random bright spots of fluorescence of irregular shapes).
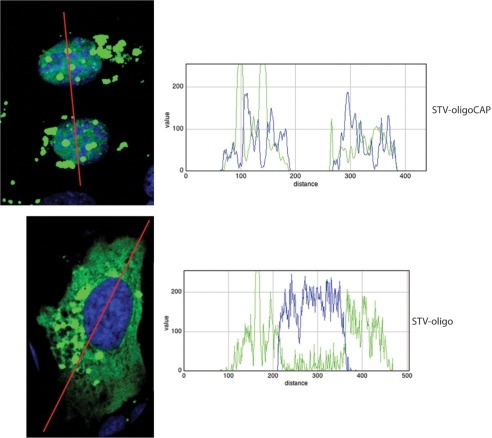
Confocal microscopy was used in order to further confirm the nuclear location of the STV-oligo-CAP (Figure 6). The line trace profile graphs clearly show the co-localization of the nuclear stain (DRAQ5 stain represented by blue colour; blue line) with the fluorescent STV-oligo-CAP constructs (ALEXA488 green fluorescence; green line). Inversely, when using the STV-oligo constructs with no m3G-CAP the line trace profile clearly shows an exclusive cytoplasmic localization. It is possible to see in the confocal images (Figure 6) as well as in the regular epifluorescence images (Figure 5) that some fluorescent STV–CAP complexes are outside the nucleus. We believe these are complexes still entrapped in vesicles in the cytoplasm or some form of irregular aggregates of the same complexes. This happened both for the non-CAP and CAP–STV transfected complexes.
Figure 6.
Confocal imaging of mammalian U2OS cells with STV-oligo complexes transfected by PULSin. Same conditions as for Figure 5 were used except that for confocal imaging there was further processing such as nuclear staining by DRAQ5 (blue) after cell fixation by 3.7% paraformaldeheyde in PBS. The line trace profile shows the intensity of the DRAQ5 (blue line in graph) and fluorescent STV-oligo complexes (green line in graphs) along the drawn line (in red). The confocal pictures confirm the co-localization of the green fluorescence coming from the STV-Alexa488 with m3G-CAP and the localization of the STV-Alexa488 without the CAP only in the cytoplasm of the cell as can be further analysed in the traced line profile graphs (graph 1 shows the line trace profile over cells transfected with STV-CAP-oligo; graph 2 shows the line trace profiles over a cell transfected with STV-oligo).
m3G-capped 2′-O-methyl RNA antisense oligonucleotides show increased efficiency in a splice correction assay
Intron splicing is a mechanism occurring only in the nuclei of cells and for this reason we used a splice correction assay to check for any increased activity of splice antisense oligos that could arise from their possible nuclear enrichment by the use of the m3G-CAP signal. We used HeLa/Luc705 cells stably expressing a luciferase gene, where the coding region was interrupted by a mutated β-globin intron at position 705. By targeting the region containing the mutation with antisense oligos, one can redirect the splicing so that the luciferase mRNA is successfully translated into protein (43).
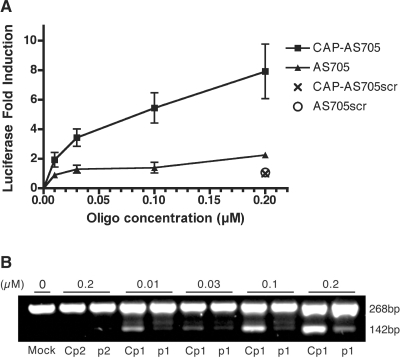
The oligonucleotide used for this study was a 2′-O-methyl oligoribonucleotide based on the same sequence as reported by Kang et al. (43) but with a 3-nt addition (AUA) to its 5′-end. The oligonucleotide included an overhanging phosphate group at this same 5′-end that was used later for the addition of the m3G-CAP (m3GpppAUA-). After transfection of both oligo versions, with (oligo1CAP) or without (oligo1) m3G-CAP addition, we found that the increase of luciferase activity relatively to untreated HeLa/Luc705 was dose-dependent and that the capped antisense oligo had an increasingly higher activity across the concentration range tested. At the highest concentration tested there was an ∼7.9-fold luciferase activity increase for the m3G-capped oligo version and only 2.3-fold for the uncapped oligo version (Figure 7A). Thus, we observed a maximum of 3.4-fold increase in efficiency when using the m3G-CAP signal.
Figure 7.
(A) Fold-induction of splice-correction 24 h after transfection of HeLa/Luc705mut with 2′-O-methyl antisense (AS) oligos at different concentrations. Correction was measured by testing for luciferase activity after AS oligo treatment relative to mock treatment. Both oligos have an additional tri-nucleotide extension (AUA) on their 5′-ends to which an m3G-CAP was added in one of them. Error bars show standard deviations for at least n = 3. (B) RT–PCR. Total cellular RNA was subjected to RT–PCR. The upper band (268 bp) and lower band (142 bp) correspond to the aberrant and correct luciferase mRNA, respectively. Cp1 and p1 correspond to the antisense oligo (AS705) with (oligo Cp1) or without (oligo p1) m3G-CAP added. Cp2 and p2 correspond to the scrambled (control) antisense oligo with (oligo Cp2) or without (oligo p2) m3G-CAP.
A control oligo with a scrambled sequence was also tested at the highest concentration with or without m3G-CAP. Both versions had no effect in correcting aberrant luciferase splicing attesting to that the m3G-CAP addition had no effect itself in restoring luciferase activity.
We confirmed the luciferase activity results with RT–PCR to directly show that the increase in luciferase activity was due to a correction of the aberrantly spliced luciferase by the antisense oligos. By subjecting total RNA from the treated cells to RT–PCR, we show that restoration of luciferase activity is correlated with an increase in the correctly spliced pre-mRNA and this was always more efficient when having capped-oligos compared to non-capped (Figure 7B).
In correlation to the previous results achieved with the cargo transport, this effect is most likely also due to a preferential accumulation of the antisense oligo in the nucleus. Despite the fact that the oligo itself is small enough to transverse through the nuclear pores by diffusion, by having an additional signal for nuclear targeting it is conceivable that this will shift the equilibrium in the direction of the nuclear compartment.
DISCUSSION
In this report, we describe a first approach to test the usability of the U snRNA m3G-CAP as an adaptor signal for the increase of nuclear transport of external cargoes such as proteins or nucleic acid molecules. To do this, we initially designed a construct composed of a fluorescent Streptavidin (STV-Alexa 488) binding a defined number of RNA oligonucleotides with or without the m3G-CAP at their 5′-ends. We chose initially the Xenopus oocyte to test our system, since the study of U snRNAs/snRNP particles has been well documented in these cells in previous experiments by others. Thus, using the Xenopus oocyte it was found that nuclear transport of the U1 snRNA species was very much dependent on the presence of the m3G-CAP in the RNA (18,44). Our experiments showed that the m3G-CAP was sufficient for enhancing the nuclear import of a cargo protein. More specifically, by using a defined construct with 2–4 m3G-capped RNA oligos, we achieved an at least 6-fold enhancement in nuclear import as compared to the uncapped counterpart. To our knowledge, there is the single study by Rollenhagen and Panté (45), in the Xenopus oocyte system, where, by pre-forming a complex composed of non-defined number of m3G-CAPs conjugated to gold particles together with recombinant snurportin, a significant change in nuclear import could be observed. Here, for the first time, we show that the m3G-CAP is working on its own in the context of a fully synthetic RNA oligo, which acquires nuclear import capacities efficient enough to drag an exogenous cargo protein. Moreover, this was shown to work in a mammalian cell system.
It is important to point out that, in our study, to emphasise the possibility that the m3G-CAP can be used as a specific adaptor signal, we constructed our cargo–CAP complexes to have a defined number of m3G-CAP structures. With this, we showed that a small number (2–4) of m3G-CAP structures were sufficient for providing nuclear import. Another point worth noting is that the m3G-CAP is sitting on a short oligo bound to an STV through the use of a relatively long spacer (34 atoms long) with biotin at its end (see Supplementary 3. Structure of the m3G-CAP-PMO2 oligonucleotide and Figure 2). This shows that the cargo can be spaced away from the import complex formed by the m3G and the import receptors, which could be beneficial.
To further test the concept, we used a novel system where we performed a nuclear import assay in live mammalian cells by using a protein delivery reagent that is able to release the cargo into the cytosol of cells. This system resembles the pathways taken by molecules in gene delivery protocols, so we think it should be relevant in assessing the properties of the m3G-CAP for future uses in the gene delivery field. Both complexes with 2–3 or 3–4 (achieving saturation of the streptavidin binding sites) capped-oligos bound to STV were able to direct nuclear translocation of cargo efficiently. There was some minor variance in terms of percentage of cells showing nuclear uptake when using 2–3 or 3–4 oligos bound to STV, 2–3 being slightly more efficient. This slight variance could be explained simply by small differences in transfection efficiency due to the reagent itself. Alternatively, it could be due to the fact that by using 3–4 oligo bound per STV we are reaching the STV binding site saturation which then allows free capped-oligos present inside the cell to compete with the STV bound CAP-oligos for the import machinery.
As a proof of concept for the application of this nuclear import strategy in a therapeutic protocol we used a splice correction assay, where antisense oligonucleotides are employed to correct a malfunctioning splice occurring due to a mutation in the β-globin intron. In this method, the mutated β-globin intron is inserted in the middle of the luciferase gene whereby targeting the mutated site with an antisense oligo will restore the luciferase expression by correcting the aberrant splicing (43). Using antisense oligos with an incorporated m3G-CAP structure we achieved an enhanced splice correction, measured by increased activity of a luciferase reporter construct. These results were also confirmed by performing RT–PCR from the total RNA of the treated cells, which showed correction of the aberrant splicing at the mRNA level.
It is worth noting that the splice oligos only carry one m3G-CAP. In this way, we show that a single cap is sufficient in the context of an oligonucleotide. Natural snRNAs, always acquire an m3G-CAP after first being recognized and bound by the SMN–assembled protein complex (7–10), through an enzymatic reaction catalysed by a hypermethylase (11,12). Importantly, this protein complex already carries a signal for nuclear import (17). Thus, our capped-oligonucleotide based system allowed us to verify that in this context there is no need for the support of an extra NLS, further supporting the view of the m3G-CAP as an independent and efficient import signal.
Since small, single-stranded oligos should have easy access to the nuclear compartment in both dividing and non-dividing cells, we believe that in suboptimal conditions, such as low transfection efficiencies or less efficient oligos, higher intranuclear concentrations should be beneficial. It is also important to note that, in vitro, very high intracellular concentrations can be easily achieved using transfection reagents, whereas in vivo, in a clinical context, intracellular oligonucleotides levels may be a limiting factor. In this case having a nuclear targeting moiety in the oligo can help to increase local nuclear concentrations. Enhancing this process may have immediate clinical applications, since highly promising oligonucleotide-based treatments affecting splicing have been recently reported for Duchenne muscular dystrophy (46). An additional feature may be the possible localization of m3G-CAP cargoes into specific splice regions of the nucleus contributing to the biological effect. In preliminary experiments, we have studied the nuclear regions where the STV–CAP-oligo is concentrated, tentatively designated STV-m3G bodies. Interestingly, these regions differ from Cajal bodies as well as from the nuclear speckle pattern, both representing compartments where snRNPs and other splice machinery components have been found (unpublished data).
In terms of development of new vector-based systems for nuclear targeting, we believe that the use of a signal that is naturally present in endogenous RNA molecules can have potential advantages over methods based on association of very positively charged NLS peptides with nucleic acids. These peptides can bind unspecifically by electrostatic interactions to the nucleic acids potentially hampering recognition of the NLS signal by the import receptors. This could make it difficult to control the properties of the nucleic acid–NLS complex and to assess their actual mechanism in transfection protocols. With the use of m3G-CAP this hurdle no longer poses any problem.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union grants FP6-018716 and FP6-037283 (to E.S.); the Swedish Foundation for Strategic Research grant Bio-X (to E.S., R.S.); Swedish Cancer Foundation (to Ö.W.); Swedish Research Council (to Ö.W., E.S., R.S.); Knut and Alice Wallenberg's Foundation (to Ö.W.). PhD fellowship from the Portuguese Foundation for Science and Technology (SFRH/BD/16757/2004 to P.M.D.M.). Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 2.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2006;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jäkel S, Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 5.Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitao S, Segref A, Kast J, Wilm M, Mattaj IW, Ohno M. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol. Cell Biol. 2008;28:487–497. doi: 10.1128/MCB.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J. Biol. Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 11.Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, Bordonne R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003;4:616–622. doi: 10.1038/sj.embor.embor863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plessel G, Fischer U, Luhrmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum. Mol. Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 16.Wohlwend D, Strasser A, Dickmanns A, Ficner R. Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J. Mol. Biol. 2007;374:1129–1138. doi: 10.1016/j.jmb.2007.09.065. [DOI] [PubMed] [Google Scholar]
- 17.Girard C, Mouaikel J, Neel H, Bertrand E, Bordonné R. Nuclear localization properties of a conserved protuberance in the Sm core complex. Exp. Cell Res. 2004;299:199–208. doi: 10.1016/j.yexcr.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 19.Strasser A, Dickmanns A, Schmidt U, Penka E, Urlaub H, Sekine M, Lührmann R, Ficner R. Purification, crystallization and preliminary crystallographic data of the m3G cap-binding domain of human snRNP import factor snurportin 1. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1628–1631. doi: 10.1107/S0907444904015380. [DOI] [PubMed] [Google Scholar]
- 20.Strasser A, Dickmanns A, Lührmann R, Ficner R. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 2005;24:2235–2243. doi: 10.1038/sj.emboj.7600701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber J, Dickmanns A, Lührmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Wang J, Hope K, Jin L, Dick J, Cameron R, Brandwein J, Minden M, Reilly RM. Nuclear localizing sequences promote nuclear translocation and enhance the radiotoxicity of the anti-CD33 monoclonal antibody HuM195 labeled with 111In in human myeloid leukemia cells. J. Nucleic Med. 2006;47:827–836. [PubMed] [Google Scholar]
- 23.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear import. Exp. Cell Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagstaff KM, Jans DA. Nucleocytoplasmic transport of DNA: enhancing non-viral gene transfer. Biochem. J. 2007;406:185–202. doi: 10.1042/BJ20070505. [DOI] [PubMed] [Google Scholar]
- 25.Collas P, Husebye H, Alestrom P. The nuclear localization sequence of the SV40 T antigen promotes transgene uptake and expression in zebrafish embryo nuclei. Transgenic Res. 1996;5:451–458. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- 26.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl Acad. Sci. USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandén LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999;17:784–787. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 28.Brandén LJ, Christensson B, Smith CI. In vivo nuclear delivery of oligonucleotides via hybridizing bifunctional peptides. Gene Ther. 2001;8:84–87. doi: 10.1038/sj.gt.3301345. [DOI] [PubMed] [Google Scholar]
- 29.Neves C, Byk G, Scherman D, Wils P. Coupling of a targeting peptide to plasmid DNA by covalent triple helix formation. FEBS Lett. 1999;453:41–45. doi: 10.1016/s0014-5793(99)00674-2. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke JJ, Zhang G, Sebestyén MG, Wolff JA. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell. Sci. 1999;112(Pt 12):2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, Ranganathan P, Diamond SL. Nuclear targeting peptide scaffolds for lipofection of nondividing mammalian cells. Nat. Biotechnol. 1999;17:873–877. doi: 10.1038/12860. [DOI] [PubMed] [Google Scholar]
- 32.Sawai H, Wakai H, Nakamura-Ozaki A. Synthesis and reactions of nucleoside 5'-diphosphate imidazolide. A nonenzymatic capping agent for 5'-monophosphorylated oligoribonucleotides in aqueous solution. J. Org. Chem. 1999;64:5836–5840. [Google Scholar]
- 33.Sekine M, Kadokura M, Satoh T, Seio K, Wada T, Fischer U, Sumpter V, Luhrmann R. Chemical synthesis of a 5'-terminal TMG-capped triribonucleotide m(3)(2,2,7)G(5)(')pppAmpUmpA of U1 RNA. J. Org. Chem. 1996;61:4412–4422. doi: 10.1021/jo952263v. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Xu Z, Liu H, Wennefors CK, Dobrikov MI, Ludwig J, Shaw BR. Synthesis of alpha-P-modified nucleoside diphosphates with ethylenediamine. J. Am. Chem. Soc. 2005;127:16782–16783. doi: 10.1021/ja055179y. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5'-0-(1-thiotriphosphates), 5'-triphosphates and 2',3'-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 36.Li P, Shaw BR. Convenient synthesis of nucleoside borane diphosphate analogues: deoxy- and ribonucleoside 5'-P(alpha)-boranodiphosphates. J. Org. Chem. 2004;69:7051–7057. doi: 10.1021/jo049094b. [DOI] [PubMed] [Google Scholar]
- 37.Han Q, Gaffney BL, Jones RA. One-flask synthesis of dinucleoside tetra- and pentaphosphates. Org. Lett. 2006;8:2075–2077. doi: 10.1021/ol060491d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 39.Pante N. Use of intact Xenopus oocytes in nucleocytoplasmic transport studies. Methods Mol. Biol. 2006;322:301–314. doi: 10.1007/978-1-59745-000-3_21. [DOI] [PubMed] [Google Scholar]
- 40.Bonner WM. Protein migration into nuclei. II. Frog oocyte nuclei accumulate a class of microinjected oocyte nuclear proteins and exclude a class of microinjected oocyte cytoplasmic proteins. J. Cell Biol. 1975;64:431–437. doi: 10.1083/jcb.64.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dabauvalle MC, Franke WW. Karyophilic proteins: polypeptides synthesized in vitro accumulate in the nucleus on microinjection into the cytoplasm of amphibian oocytes. Proc. Natl Acad. Sci. USA. 1982;79:5302–5306. doi: 10.1073/pnas.79.17.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stefanovic B, Hackl W, Lührmann R, Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang SH, Cho MJ, Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37:6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- 44.Rollenhagen C, Muhlhausser P, Kutay U, Pante N. Importin beta-depending nuclear import pathways: role of the adapter proteins in the docking and releasing steps. Mol. Biol. Cell. 2003;14:2104–2115. doi: 10.1091/mbc.E02-06-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rollenhagen C, Pante N. Nuclear import of spliceosomal snRNPs. Can. J. Physiol. Pharmacol. 2006;84:367–376. doi: 10.1139/y05-101. [DOI] [PubMed] [Google Scholar]
- 46.van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.