Abstract
JWA was recently demonstrated to be involved in cellular responses to environmental stress including oxidative stress. Although it was found that JWA protected cells from reactive oxygen species-induced DNA damage, upregulated base excision repair (BER) protein XRCC1 and downregulated PARP-1, the molecular mechanism of JWA in regulating the repair of DNA single-strand breaks (SSBs) is still unclear. Our present studies demonstrated that a reduction in JWA protein levels in cells resulted in a decrease of SSB repair capacity and hypersensitivity to DNA-damaging agents such as methyl methanesulfonate and hydrogen peroxide. JWA functioned as a repair protein by multi-interaction with XRCC1. On the one hand, JWA was translocated into the nucleus by the carrier protein XRCC1 and co-localized with XRCC1 foci after oxidative DNA damage. On the other hand, JWA via MAPK signaling pathway regulated nuclear factor E2F1, which further transcriptionally regulated XRCC1. In addition, JWA protected XRCC1 protein from ubiquitination and degradation by proteasome. These findings indicate that JWA may serve as a novel regulator of XRCC1 in the BER protein complex to facilitate the repair of DNA SSBs.
INTRODUCTION
Viable cells suffer spontaneous DNA damage or can be damaged as a result of environmental exposure to a variety of insults. DNA single-strand breaks (SSBs) are one of the commonest DNA lesions, arising either indirectly during DNA base excision repair (BER) through enzymatic incision of an abasic site (AP) by apurinic/apyridinic endonuclease 1 (APE1) or a dual-function DNA glycosylase, or directly from deoxyribose damage due to reactive oxygen species (ROS) or from abortive top1 activity (1–3). If SSBs are not properly repaired, they may result in genetic instability, higher frequency of chromosomal aberrations (4,5) and conversion into DNA double-strand breaks (DSBs) during DNA replication (6), eventually leading to subsequent tumorigenesis (7,8).
The pathways for SSB repair (SSBR) in mammalian cells involve four basic steps which include damage detection, end processing, gap filling and DNA ligation (2,9). During the first step, one of the earliest responses to DNA strand breakage is the induction of poly (ADP-ribose) (PAR) synthesis (10,11). When DNA SSBs are present, PARP-1 rapidly binds to the DNA strand breaks and is activated, covalently automodifying itself and, to a lesser extent, other acceptor proteins with long chains of PAR (10,12,13). This step is required for cellular SSBs to recruit, stabilize or accumulate the scaffold protein, XRCC1 (14,15), which then mediates multiple interactions with enzymatic components of the repair process. For example, XRCC1 appears to interact with APE1 (16)/DNA polymerase β (17) and DNA polynucleotide kinase (PNK) (18)/PCNA (19), which play important roles in end processing and gap filling, respectively (9). XRCC1 also interacts with DNA ligase IIIa (20), which seals single nucleotide nicks in the process of BER (21,22).
Substantial evidence indicates that XRCC1 plays a critical role in SSBR. XRCC1-deficient cells (EM9 or EM11) are hypersensitive to DNA damage induced by alkylating agents, ROS or ionizing radiation (23–25). Additionally, these cells display increased rates of spontaneous sister-chromatid exchange and chromosomal aberrations (4,5,23). Furthermore, downregulation of XRCC1 expression in human breast cancer cell lines by RNA interference (RNAi) also resulted in decreased SSBR capacity and hypersensitivity to methyl methanesulfonate (MMS) (26). LigIII, which is stabilized by XRCC1, is also required for SSBR (27). It was shown that LigIII mutant cells possess relatively normal XRCC1 levels, but have an elevated frequency of sister-chromatid exchange (SCE) (27). Deficiency in either XRCC1 or LigIII results in embryonic lethality in mice (27–29).
Recent studies have helped clarify how DNA damage leads to changes in XRCC1 transcription or translation. Yacoub et al. (30) demonstrated that XRCC1 mRNA and protein levels were increased after DNA damage through activation of MAPK signaling cascades. E2F1, an active transcriptional factor located downstream of the MAPK signaling pathway (31,32), can bind the XRCC1 promoter to regulate XRCC1 transcription, which facilitates the SSBR/BER (33). However, there is a striking check point mechanism present in mammalian cells to ensure that the XRCC1 protein is maintained at the necessary level (34,35). XRCC1 is ubiquitylated by the E3 ubiquitin ligase CHIP and subsequently degraded through the ubiquitin-proteasome pathway (34).
The JWA gene, also known as ARL6ip5, was initially cloned from human tracheal bronchial epithelial cells after treatment with all-trans retinoic acid (36). Subsequent studies indicated that JWA is a structurally novel microtubule-associated protein, which regulates cancer cell migration via MAPK cascades (37) and mediates differentiation of leukemic cells (38,39). JWA is also involved in the cellular responses to heat shock and chemical-mediated oxidative stresses (40,41). Moreover, JWA plays a key role in protecting cells from DNA damage induced by oxidative stress, as evidenced by the increase in the level of BER protein XRCC1 and the reduction in PARP-1 expression (42). However, the underlying mechanisms by which JWA regulates XRCC1 are unclear.
Herein, we provide additional evidence confirming that JWA is necessary for cell survival and efficient DNA repair after oxidative DNA damage. Further mechanistic studies have demonstrated that JWA translocates into the nucleus and co-localizes with XRCC1 foci after oxidative DNA damage. In addition, JWA regulates XRCC1 expression at both the transcriptional and post-translational levels. These observations identify a role for JWA in DNA SSBR/BER pathway and provide a novel mechanism of JWA-mediated regulation of XRCC1 expression.
MATERIALS AND METHODS
Cell culture
NIH-3T3, HELF cells were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum. The cells were grown at 37°C in the presence of 5% CO2 in a humidified incubator.
Plasmids and reagents
The EGFP-C1-antisense JWA and pEGFP-C1-JWA expression plasmids were described previously (37,42). The JWA siRNA Expression Cassette and scrambled short-hairpin RNA (shRNA) (39) were subcloned into the linearized vector pSEC (Ambion, Austin, TX, USA) to produce JWA shRNA and control shRNA plasmids, respectively. The identity of the plasmids was confirmed by DNA sequencing. The remaining plasmids were obtained from other investigators, including pGL3-XRCC1 (–881 to +158) and pGL3-XRCC1 (–766 to +158) luciferase-reporter plasmids (Prof. Charles D. Lopez, Oregon Health and Science University, USA), and RFP-XRCC1 (Prof. Heinrich Leonhardt, Ludwig Maximilians University, Germany). XRCC1 siRNA or scramble control siRNA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). MMS, cycloheximide (CHX), MG132 and 3-aminobenzamide (3-AB) were purchased from Sigma–Aldrich (St Louis, MO, USA). U0126 and PD98059 were purchased from Cell Signaling Technology (Beverly, MA, USA).
Stable or transient transfections
Transfections of the pEGFP-C1-antisense-JWA plasmid and the pEGFP-C1 vector control plasmid into the NIH-3T3 and HELF cells were carried out using Lipofectamine 2000 transfection reagent (Invitrogen, Shanghai, China) following the manufacturer's protocol. The cells were selected with G418 at a final concentration of 500 μg/μl or 400 μg/μl for NIH-3T3 cells or HELF cells, respectively. The resulting NIH-3T3 and HELF cells were verified to possess stable knockdown of JWA expression (KD-JWA) or to express the control vector. For transient transfections, plasmid DNA or siRNA were transfected into cells with Lipofectamine 2000 according to manufacturer's instruction.
Luciferase reporter gene assays and host-cell-reactivation (HCR) assay
Cells were seeded onto 24-well plates (1 × 105 cells per well), and co-transfected with 2.25 μg of pGL3-XRCC1 (–881 to +158) or pGL3-XRCC1 (–766 to + 158) luciferase-reporter plasmid, together with the JWA shRNA or control shRNA plasmid with Lipofectamine 2000. All the plasmids were co-transfected with 10 ng of pRL-SV40, which contains the Renilla luciferase gene. Forty-eight hours after transfection, the cells were treated with 100 μM H2O2 for 30 min, then washed once with PBS and harvested. Cell lysates were prepared according to Promega's instruction manual. Luciferase activity was measured with a dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and the activity was normalized against the Renilla luciferase gene.
The HCR assay was used to measure the DNA repair capacity (DRC) (43,44). The pGL3-control luciferase vector (LUCcon) (Promega, Madison, WI, USA) backbone containing the SV40 promoter and enhancer sequences was used. The LUCcon DNA was oxidatively damaged in vitro by dilution to 50 μg/ml and exposure to the indicated concentration of H2O2 (v/v) at room temperature for 1 h. Undamaged control plasmids were treated with the vehicle solutions without exposure to the damaging agents. After all treatments, the damaged or undamaged DNA was purified by ethanol precipitation, and resuspended in TE buffer (pH 7.8) at a final concentration of 500 μg/ml. NIH-3T3 cells were then transfected with the damaged or undamaged LUCcon plasmids. The methods for transfection and luciferase activity measurement were the same as used in the luciferase reporter assay described above. DRC (%) was calculated as the ratio of the damaged plasmid luciferase activity to the undamaged plasmid luciferase activity, multiplied by 100%.
Survival curves and determination of intracellular NAD(P)H
Cells were plated into 96-well plates (3000–5000 cells/well). The next day, the cells were exposed to H2O2 for 30 min or MMS for 1 h at the indicated concentrations in complete medium at 37°C. After washing with PBS, cells were further incubated for 3 days in drug-free medium. Cell viability was measured using the MTT assay as described (41). All measurements were done in triplicate and experiments were repeated at least twice.
Depletion of intracellular NAD(P)H was monitored as the described method (45) using a CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan). Briefly, cells were seeded into 96-well plates for 24 h, then treated with H2O2 or MMS at the indicated concentrations, and 1/10 vol of CCK-8 solution in the presence or absence of 10 mM 3-aminobenzamide (3-AB) was added to each well (Sigma). Visible absorbance at 450 and 600 nm as a reference filter was measured 4 h after the start of the treatment. Measurements were done in triplicate, and error bars represent the standard deviation of six parallel measurements of one typical experiment.
Indirect immunofluorescent microscopy
Cells were grown on coverslips, rinsed in PBS and treated with 10 mM H2O2 in PBS at room temperature for 20 min at 37°C, then incubated in drug-free medium for 10 min. Coverslips were rinsed in PBS and fixed with 4% formaldehyde in PBS for 20 min at 4°C. Cells were permeabilized with PBS containing 0.01% Triton X-100 and 0.05% SDS, pH 7.4 for 5 min at 4°C. Permeabilized cells were rinsed in PBS supplemented with 0.5% Tween-20 (PBST), and cells were blocked with blocking solution (0.1% saponin and 0.2% BSA in PBS, pH 7.4) for 1 h. Cells were rinsed in PBST three times and incubated with either a mixture of anti-JWA rabbit polyclonal antibody (1:200 dilution in PBST; Research Genetics Inc., S.W. Huntsville, AL) and anti-poly (ADP-ribose) mouse monoclonal antibody (Chemicon; 1:200 dilution in PBST) overnight at 4°C, or a mixture of anti-JWA rabbit polyclonal antibody (1:200 dilution in PBST) and anti-XRCC1 mouse monoclonal antibody (NeoMarkers; 1:200 dilution in PBST). After rinsing in PBST, coverslips were incubated in a mixture of Texas red-conjugated anti-rabbit IgG secondary antibody or fluorescein isocyanate (FITC)-conjugated anti-mouse IgG secondary antibody (KPL, Inc., Gaithersburg, MD, USA), at a 1:200 dilution in PBST for 1 h at 37°C. Cell nuclei were counterstained with 0.000025% 4′, 6′-diamidino-2-phenylindole (DAPI). The intracellular distributions of the target proteins were analyzed using confocal fluorescent microscopy. Confocal images were sequentially acquired with Zeiss AIM software on a Zeiss LSM 510 confocal microscope system (Carl Zeiss Inc, Thornwood, NY, USA) with excitation at 488 nm (for FITC), 543 nm (for Texas red) and 340 nm (for DAPI).
Quantitative real-time RT-PCR assay
NIH-3T3 cells were transiently transfected with either the JWA shRNA plasmid or control shRNA vector. Forty-eight hours later, the cells were cultured with or without 100 μM H2O2 for 30 min. Total RNA was extracted from the cells using the Trizol reagent (Gibco BRL, Gaithersburg MD, USA). Approximately 1 μg of RNA was used for the reverse transcription reaction with OligodT (18T) (Invitrogen). The cDNA was amplified with the following primers: 5′-GCCGGTGCTGAGTATGTC-3′ (forward) and 5′-CTTCTGGGTGGCAGTGAT-3′ (reverse) for GAPDH; 5′-CTTTGTGGAGGTGCTAGTGG-3′ (forward) and 5′- ATGGCGAGTCCTTGCTGT -3′ (reverse) for XRCC1; 5′-GGAGGAGTCATTGTGGTGC-3′ (forward) and 5′-GAAGTCTCAGGGATGCGTG-3′ (reverse) for JWA.
Quantitative real-time PCR was carried out on the Light Cycler System using the double-strand DNA-binding dye evaGreen (Biotium Hayward, CA, USA) for the detection of PCR products. The following thermal cycling conditions were used: denaturation, 94°C for 5 min followed by 44 cycles of denaturation at 94°C for 35 s, annealing at 56°C (for JWA and XRCC1) or 59°C (for GAPDH) for 30 s and extension at 72°C for 35 s. The cycles were followed by a final extension step at 72°C for 8 min, and melting curves from 70°C to 90°C were determined. The fluorescence intensity of the evaGreen was read on the Light Cycler System after the end of each extension step. Data were expressed as the number of cycle thresholds (Ct), the PCR cycle number at which the fluorescent signal in each reaction reached a preset threshold above the background. A melting curve was created using the built-in melting curve program to confirm the presence of a single PCR product. GAPDH mRNA was used as an internal control for each sample, and the Ct value for each sample was normalized to GAPDH mRNA.
Subcellular fractionation and western blotting analysis
Nuclear extracts were obtained using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Rockford, IL, USA). Total cell lysates were prepared with a detergent lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF]. Western blots were performed as previously reported (32). For each treatment group, three parallel samples were applied, and equal amounts of proteins from the parallel samples were mixed and used for blots.
The antibodies used were the monoclonal anti-XRCC1 (1:1000) (Abcam, Cambridge, UK); polyclonal anti-poly (ADP-ribose) polymerase-1 (PARP-1) (1:500), anti-aldolase (1:500), anti-histone H1 (1:500), monoclonal anti-ubiquitin (1:500), anti-E2F1 (1:500) (Santa Cruz Biotechnology); polyclonal anti-phospho-p44/42 MAPK (Thr202/Tyr204) (1:1000), anti-p44/42 MAPK (ERK1/2) (1:1000) (Cell Signaling Technology, Beverly, MA, USA); monoclonal anti-LigIII (1:1000) (BD Transduction Laboratories, San Diego, CA, USA); polyclonal goat anti-JWA (1:1000) (Imgenex, San Diego, CA, USA) and polyclonal anti-β-actin (1:1000) (Boster Biotechnology, Wuhan, China). Each blot was repeated three times. β-Actin was used for the protein loading control, whereas aldolase and histone H1 were used for the cytoplasmic and nuclear controls, respectively.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed with a Biotin Gel Shift Kit (Pierce, Rockford, IL, USA), as described previously (42). Briefly, binding reactions were performed by adding 0.5 or 1 μg of the nuclear extracts to a mixture containing 40 fmol of biotin 3′-end-labeled, double-stranded probes: (5′-TAA TTT TTT TCG CGC GTG CGC GCG CGC GTA-3′) (underlined sequences indicate putative E2F1-binding sites) in 20 μl of binding buffer [100 mM HEPES (pH 7.6), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol (DTT), 1% Tween-20, 150 mM MgCl2, 1 μg/ml poly(dI–dC), 0.1 μg/ml l-lysine]. For supershift experiments, 1 μg of monoclonal E2F1 antibody was added to aliquots of the extracts, and incubated for 20 min on ice before addition of the reaction mixture. Competition reaction mixtures contained a 100-fold molar excess of nonlabeled double-stranded oligoDNAs or mutated nonlabeled double-stranded oligoDNAs: (5′-TAA TTT TCG AGC TGC GTA GACTCA CGC GTA-3′) (mutated nucleotides in putative E2F1-binding sites in italic and bold). The DNA–protein complexes were separated on a native 4.5% polyacrylamide gel at 100 V and then transferred onto a nylon membrane. The positions of the biotin end-labeled oligonucleotides were detected by a chemiluminescent reaction according to the manufacturer's instructions (Pierce) and visualized by autoradiography.
Co-immunoprecipitation
NIH-3T3 cells were grown to confluence and processed for co-immunoprecipitation by standard procedures described previously (32). Briefly, the cells were harvested and lyzed in cold lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) NP-40, 10% (v/v) glycerol, 1 mM PMSF]. The cell extracts were centrifuged at 12 000 g at 4°C for 15 min, and the supernatant was then incubated with protein A/G agarose beads or control IgG (Santa Cruz) as a pretreatment. Precleared lysates were then incubated with anti-PARP-1 polyclonal antibody, anti-JWA polyclonal antibody, anti-LigIII monoclonal antibody, anti-XRCC1 monoclonal antibody or control IgG for 1 h, then incubated overnight with protein A/G agarose beads. The beads were collected by centrifugation, washed three times with the lysis buffer and resuspended in 1 × SDS loading buffer. The immunoprecipitates were eluted from the beads by incubation at 95°C for 5 min. The eluted proteins were separated by SDS–PAGE and western blotting was subsequently performed with indicated antibodies.
Statistical analysis
Data were expressed as the means ± SD. Two-factor analysis of variance procedures and the Dunnett's t-test were used to assess differences within treatment groups. Differences were considered significant when P < 0.05.
RESULTS
JWA is required for DNA repair following oxidative stress
Chen et al. (42) previously showed that the DNA damage in JWA knockdown cells was greater than that in vector control cells as determined using the comet assay. JWA knockdown also decreased the DNA repair frequency following damage induced by oxidative stress. The alkaline comet assay can thus be used to intuitively observe damaged DNA under fluorescent microscopy. However, alkylating agents and oxidants were also reported to induce either alkaline-labile base lesions or AP sites leading to SSBs under basic conditions (46).
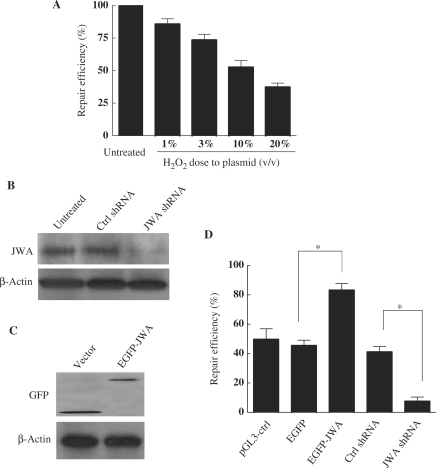
Studies were undertaken to determine if endogenous JWA plays a physiological role in the repair of H2O2-induced DNA lesions. The HCR assay, which was previously validated in a study of BER proteins and H2O2-induced DNA lesions (44), was conducted in this study. First, a dose-repair model for the LUCcon plasmid was performed, and then the DRC was determined for the plasmids damaged by H2O2. The DRC was reduced to ∼55% for 10% (v/v) H2O2-treated LUCcon plasmids compared to undamaged control plasmids (Figure 1A). In NIH-3T3 cells, the JWA protein level was significantly reduced by ∼97% after transient transfection with JWA shRNA (Figure 1B). Subsequently, the effect of JWA in NIH-3T3 cells was examined by HCR assay, and it was found that the DRC was reduced by more than 80% in JWA-knockdown cells compared to cells transfected with the control vector (Figure 1D). In contrast, overexpression of JWA in the NIH-3T3 cells markedly increased the DRC by up to 2-fold (Figure 1C and D).
Figure 1.
JWA is required for repairing H2O2-damaged DNA. (A) The repair efficiency of NIH-3T3 cells on damaged plasmid DNA was detected by HCR assay. NIH-3T3 cells were transfected with control LUC plasmid DNA (pGL3-control) or the same plasmid damaged by hydrogen peroxide at concentrations of 1, 3, 10 or 20% (v/v). DNA repair rates were measured as the ratio of LUC activity in extracts from cells transfected with a damaged plasmid to the LUC activity in extracts from cells transfected with an undamaged plasmid. The experiment was done in triplicate. (B) NIH-3T3 cells were transiently transfected with plasmids to knockdown JWA (JWA shRNA), or with the corresponding control vector (Ctrl shRNA), or were left untreated. After 48 h, whole-cell lysates were collected for detection of target proteins by immunoblotting. (C) NIH-3T3 cells were transiently transfected with a plasmid expressing human JWA protein (EGFP-JWA) or a control vector (EGFP-C1). Expression of target proteins 48 h after transfection was examined in whole-cell lysates by immunoblotting. (D) Knockdown of JWA inhibits DRC of damaged plasmids, while overexpression of JWA enhances DRC in NIH-3T3 cells. NIH-3T3 cells were transfected with either the EGFP-C1 control or EGFP-JWA plasmid, control shRNA or JWA shRNA together with undamaged or 10% (v/v) H2O2-damaged LUC plasmids. The pGL3 (undamaged or H2O2-damaged) plamids were transfected as a control for co-transfection efficiency. Twenty-four hours after transfection, the DRC of the damaged LUC reporter was assayed as indicated in (A). The renilla luciferase reporter (internal control, Promega) was used to normalize the activity of the LUC reporter. *P < 0.05.
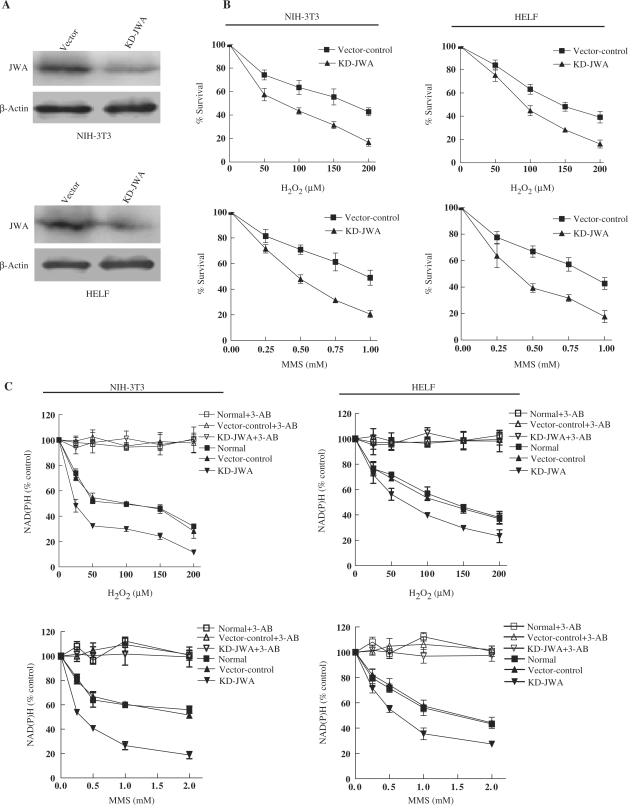
To confirm the requirement for JWA in DNA damage repair, additional SSBR endpoints were investigated. Cell survival for 72 h was determined by MTT assay after induction of DNA damage by H2O2 or MMS. Stable transfection to knockdown JWA expression was conducted in two cell lines (KD-JWA NIH-3T3 and KD-JWA HELF) and the loss of JWA was determined by western blot analysis. We observed that JWA protein levels were reduced by 85% or 90% in KD-JWA NIH-3T3 or HELF cells compared to the vector control cells (Figure 2A). Compared to the control cells, both KD-JWA NIH-3T3 and KD-JWA HELF cells showed significantly greater sensitivity to H2O2 and MMS, which causes SSBs in DNA (Figure 2B). These data indicate an important role for endogenous JWA in SSBR/BER of oxidative-stress-induced DNA lesions.
Figure 2.
JWA-knockdown cells are hypersensitive to DNA-damaging agents. (A) JWA expression was decreased in KD-JWA NIH-3T3 cells (top panel) or HELF cells (bottom panel) compared with vector control cells. (B) JWA knockdown enhances the cell death induced by DNA-damaging agents (MMS and H2O2). Vector control and KD-JWA NIH-3T3 or HELF cells were treated with the indicated doses of H2O2 for 30 min or MMS for 1 h, then the cells were incubated for further 72 h in drug-free medium. Cell survival was determined using the MTT assay. The relative% surviving cells are presented as the means ± SD of three independent samples. (C) Intracellular NAD(P)H levels in living cells (vector control and KD-JWA cells) were determined by CCK-8 assay. Both NIH-3T3 and HELF cells were exposed to H2O2 (25, 50, 100, 150 or 200 μM) for 1 h or MMS (0.25, 0.5, 1.0 or 2 mM) for 4 h in the absence or presence of the PARP inhibitor, 3-AB (10 mM). Mean ± SD were from triplicate experiments.
Next, we measured the activity of PARP-1 by quantifying intracellular NAD(P)H, which is also a reliable index to monitor the imbalance in DNA-strand break repair in BER-deficient cells (45). We demonstrated that JWA knockdown caused an enhanced reduction of intracellular NAD(P)H in both NIH-3T3 and HELF cells in response to MMS or H2O2 (Figure 2C). The presence of 3-AB, a specific PARP-inhibitor, almost completely blocked the DNA-damage-induced decrease in NAD(P)H in all tested cells, suggesting that the observed NAD(P)H depletion is due to increased PARP-1 activation. The trypan blue exclusion assay demonstrated no significant differences in cell viability at the noted MMS or H2O2 concentrations, indicating that the depletion of NAD(P)H is not due to a reduction in the number of viable cells (data not shown). Additionally, our data indicate that there was a greater deficiency in SSB repair in the JWA-deficient cells than in the control cells.
JWA as a novel BER protein is involved in the SSBR pathway
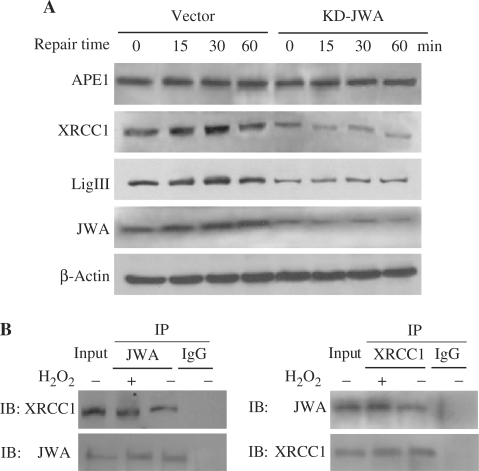
Our previous studies indicated that XRCC1 protein levels were regulated by JWA under oxidative stress conditions (42). To elucidate if JWA is involved in the XRCC1-associated signaling pathways, JWA knockdown NIH-3T3 cells and control cells were treated with 100 μM H2O2 for 30 min, and then cultured in H2O2-free medium to allow for DNA rejoining. In the cells transfected with the control vector, the levels of XRCC1, LigIII and JWA all gradually increased for the first 30 min during repair, then returned to almost normal by 1 h. The levels of XRCC1 and LigIII were downregulated in the JWA knockdown NIH-3T3 cells, while no change in APE1 protein (upstream of XRCC1) level was observed in JWA knockdown cells compared to the control cells (Figure 3A).
Figure 3.
JWA as a novel BER protein is involved in the SSBR pathway. (A) The expression levels of BER complex components (APE1, XRCC1, LigIII) during DNA repair after H2O2 treatment (100 μM, 30 min) in JWA stable knockdown cells and vector control cells were detected by immunoblotting. The indicated time points represent the amount of time after withdrawal of the DNA-damaging agent. Whole-cell extracts were prepared for immunoblotting, and equal protein loading was confirmed by comparison with β-actin. (B) The NIH-3T3 cells were pretreated with 100 μM H2O2 for 30 min and endogenous protein–protein interaction between JWA and XRCC1 was determined by immunoprecipitation (IP) with JWA or XRCC1 antibodies followed by immunoblotting. IgG was used as negative control for IP.
Chen et al. (42) demonstrated that JWA interacts with XRCC1 in intact NIH-3T3 cells. Moreover, XRCC1, as a scaffold protein, interacts with LigIII and PARP-1 to assemble a DNA repair complex for BER (47). In the present study, co-immunoprecipitation assays were employed to establish whether JWA is involved in the BER protein complex. As shown in Figure 3B and Supplementary Figure S1, JWA interacts with XRCC1, LigIII and PARP-1, not only in intact NIH-3T3 cells, but also in treated cells (100 μM H2O2 for 30 min). Moreover, the amount of the proteins (JWA-XRCC1 and JWA-LigIII) in the extracts of impaired cells was markedly enhanced compared with those in intact cells. However, no increased JWA-PARP-1 complex in the H2O2-treated cells was found. These data indicated that JWA may be a component of BER protein complex in response to DNA damage.
Oxidative damage induces JWA translocation into the nucleus
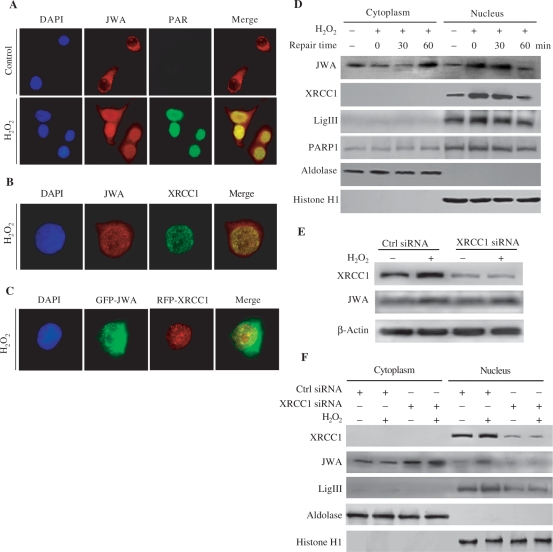
In response to DNA-strand breaks, activated PARP-1 catalyzes the transfer of the ADP-ribose moiety from NAD+ to a number of protein acceptors, resulting in PAR accumulation in the SSB sites (11). These steps are required for the recruitment of scaffold protein XRCC1 to DNA damage sites. In our study, immunofluorescent microscopy was used to determine the intracellular distribution of JWA in NIH-3T3 cells with or without H2O2 treatment. As shown in Figure 4, in addition to the expected uniform cytoplasmic distribution, JWA was also present in small intense regions of the nucleus in untreated cells; however, the PAR was not detected in untreated cells (Figure 4A). After exposure to 10 mM H2O2 for 15 min, most of the JWA in the cells were translocated into the nucleus. We also observed that JWA is rapidly assembled into discrete nuclear foci after oxidative DNA damage at sites of PAR synthesis (Figure 4A). Moreover, JWA foci were co-localized with XRCC1 foci after H2O2 treatment (Figure 4B). To exclude the nonspecificity of the JWA and XRCC1 antibodies, we observed the intracellular distribution JWA and XRCC1 in JWA knockdown cells. As shown in Supplementary Figure S2, knockdown of JWA decreased the XRCC1 staining in untreated cells (as indicated by the yellow arrow in Figure S2A) and in the impaired cells (10 mM H2O2 for 15 min) (as indicated by the single yellow arrow in Figure S2B; the double yellow arrows denote that the JWA was not decreased in the cell due to ineffective transfection). We also found that a large proportion of the GFP-JWA in the cells was translocated into the nucleus and co-localized with RFP-XRCC1 foci after H2O2 treatment (Figure 4C).
Figure 4.
H2O2 triggers JWA translocation into the nucleus and co-localization with the components of BER complex on DNA damage sites. (A) NIH-3T3 cells were treated with or without 10 mM H2O2 for 20 min and then incubated in H2O2-free medium for 10 min, fixed with 4% formaldehyde, permeabilized with 0.01% Triton X-100 and then immunostained with anti-JWA (red) or anti-PAR antibody (green). (B) Cells were treated with H2O2 as in (A) and stained with anti-JWA (red) or anti-XRCC1 antibody (Green). (C) NIH-3T3 cells were transiently transfected with a GFP-JWA or an RFP-XRCC1 plasmid. After 48 h, the transfected cells were split, grown on coverslips, exposed to H2O2 as in (A). Representative images were photographed and colored using a Zeiss LSM 510 confocal microscope system. (D) The intracellular distribution of JWA and the components of BER complex during DNA repairing after H2O2 exposure. NIH-3T3 cells were treated with 100 μM H2O2 for 30 min and further cultured in H2O2-free medium to allow for DNA repair. Cytoplasmic and nuclear extracts were prepared, and western blotting were employed to detect the expression of XRCC1, LigIII, PARP-1 and JWA. Aldolase and histone H1 were used as the cytoplasmic and nuclear loading controls, respectively. The experiments were repeated twice with similar results. (E) XRCC1 does not affect the expression of JWA. NIH-3T3 cells were transfected with a control siRNA pool or a XRCC1 siRNA pool to knockdown endogenous XRCC1. After 72 h, the transfected cells were treated with or without 100 μM H2O2 for 30 min. Whole-cell lysates were collected for detection of target proteins, including XRCC1 and JWA by immunblotting. β-Actin was used for the protein loading control. (F) XRCC1 induced subcellular redistribution of JWA and LigIII. The treatments were the same as in (E). Western blot analysis was performed to analyze XRCC1, LigIII and JWA in the cytoplasmic and nuclear extracts. Aldolase and histone H1 were used as the cytoplasmic and nuclear loading controls, respectively.
To confirm the observed translocation of JWA from the cytoplasm to the nucleus following H2O2 treatment, cells were subfractionated and immunoblotted with anti-JWA antibody. As expected, JWA underwent nuclear translocation under oxidative stress, with the amount of JWA in the nucleus increased during the first 30 min following withdrawal of the H2O2; at the same time, other DNA repair enzymes including XRCC1 and LigIII, were also elevated in the nucleus. Interestingly, JWA returned back to the cytoplasm by 60 min in the repair process. These changes in JWA distribution paralleled changes in XRCC1 and LigIII (Figure 4D).
It was unclear how JWA was being translocated into the nucleus without a classic nuclear translocation signal (NLS). We postulated that JWA is recruited into the nucleus either by an unknown target of XRCC1 or XRCC1 itself right after its synthesis in the cytoplasmic ribosomes as knocking down XRCC1 inhibited JWA translocation into the nucleus. Since XRCC1 is known to play a role as a scaffold in the BER complex, we initially determined whether XRCC1 could transport JWA into the nucleus after DNA damage. An siRNA was used to downregulate XRCC1, then the expression and distribution of JWA was measured. Seventy-two hours after transfection of NIH-3T3 cells with XRCC1 siRNA, we observed a 70% and 90% reduction in XRCC1 in the whole-cell lysates of untreated and H2O2-treated NIH-3T3 cells, respectively, whereas JWA level in these extracts was unaltered (Figure 4E). However, the nuclear JWA was markedly reduced in the XRCC1 knockdown cells (Figure 4F). Interestingly, cytoplasmic JWA tended to be increased by knockdown of XRCC1 (Figure 4F). Additionally, nuclear LigIII, which is stabilized by XRCC1, was decreased when XRCC1 was knocked down.
JWA via E2F1 regulates XRCC1 transcription
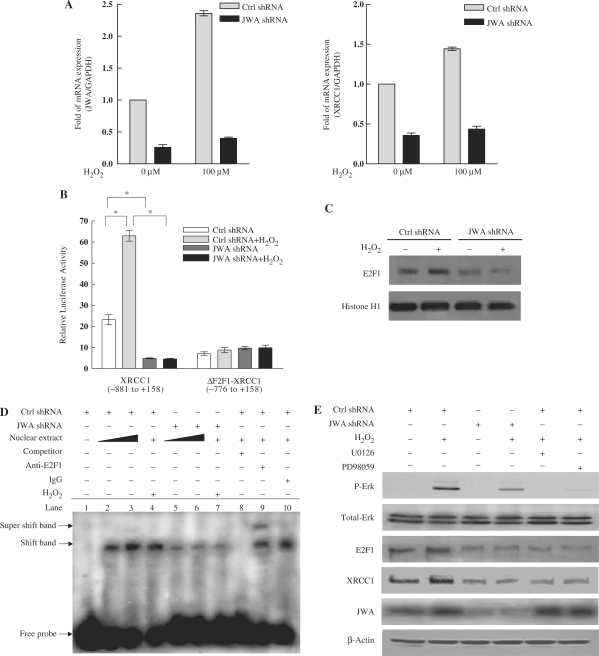
To address how JWA downregulates XRCC1, the level of XRCC1 mRNA in JWA knockdown and control NIH-3T3 cells was determined. Quantitative RT-PCR results showed that the levels of JWA and XRCC1 mRNA were increased 2.3-fold and 1.5-fold in control cells in response of H2O2 treatment, respectively. In contrast, XRCC1 mRNA expression was reduced in JWA knockdown cells, and there was no significant increase in either JWA or XRCC1 mRNA expression following H2O2 treatment (Figure 5A).
Figure 5.
JWA regulates XRCC1 transcription via the MAPK signaling pathway and E2F1. (A) JWA knockdown in NIH-3T3 cells significantly inhibits H2O2-induced transcription of XRCC1. NIH-3T3 cells were transfected with a control shRNA or a JWA shRNA plasmid, followed by treatment with or without 100 μM H2O2 for 30 min. Levels of JWA and XRCC1 transcription were detected by quantitative RT-PCR, and GAPDH was used as an endogenous control to normalize the differences in the amount of total RNA in each sample. (B) The E2F1-binding domain in the XRCC1 promoter is required for the JWA-mediated increase in XRCC1 expression after exposure to H2O2. NIH-3T3 cells were co-transfected with either control shRNA or JWA shRNA, together with the XRCC1 promoter-reporter (–881 to + 158, containing E2F1-binding domain) or an E2F1-binding site deleted XRCC1 promoter-reporter (ΔE2F1-XRCC1, –776 to + 158). After 24 h, the transfected cells were cultured with or without 100 μM H2O2 for 30 min, then the reporter activity was examined. The means ± SD of triplicate experiments are shown. *P < 0.05. (C) JWA is required for H2O2-induced E2F1 expression. NIH-3T3 cells were transfected with a control shRNA or JWA shRNA plasmid. Then 48 h after transfection, the cells were treated with or without 100 μM H2O2 for 30 min, and nuclear lysates were collected for detection of E2F1 by immunblotting. Histone H1 was used as the nuclear protein loading control. (D) JWA alters the affinity of E2F1 for the XRCC1 promoter, as detected by EMSA. The nuclear protein extracts of the NIH-3T3 cells (with or without treatment with 100 μM H2O2 for 30 min) were incubated with a biotin-labeled double-strand oligonucleotide probe of the XRCC1 promoter region, which contains an E2F1-binding domain (–826 to –797 bp). JWA shRNA transient transfection was used to knock down JWA expression in the NIH-3T3 cells. The DNA–protein complex (shift band) or DNA–protein–antibody complex (supershift band) is indicated by an arrow. Lane 1 contains no nuclear extracts. All other lanes contain 0.5-μg nuclear extracts except lanes 3 and 6 which contain 1-μg nuclear extracts. Lane 8 represents competition analysis using 100-fold unlabeled probes. The supershift band was observed when the E2F1 antibody was added (lane 9) and IgG was used as negative control for supershift (lane 10). (E) JWA regulates E2F1 expression via MAPK signaling cascades. JWA shRNA and control shRNA plasmids were transiently transfected into NIH-3T3 cells. After 46 h, the control shRNA vector transfected cells were incubated with 20 μM of PD98059 or 10 μM U0126 for another 2 h. All transfected cells were then cultured for another 30 min in the presence or absence of H2O2 (100 μM), and the whole-cell lysates were collected for western blotting.
It has been reported that transcription factor E2F1 binds to the XRCC1 promoter region and regulates XRCC1 transcription (33). To examine whether JWA regulates XRCC1 promoter transcriptional activity via E2F1, a JWA shRNA or control shRNA were cotransfected together with an XRCC1 promoter-luciferase reporter including (E2F1-XRCC1) or lacking the E2F1-binding site (▵E2F1-XRCC1). The luciferase activity of the XRCC1 promoter-luciferase reporter (E2F1-XRCC1) increased following exposure to H2O2, whereas the activity was significantly reduced both prior to and following H2O2 exposure in JWA konckdown cells. The luciferase activity of the ▵E2F1-XRCC1 promoter-luciferase reporter was significantly attenuated compared with that of the E2F1-XRCC1 promoter-luciferase reporter. In addition, the activity of the ▵E2F1-XRCC1 promoter did not change in response to either H2O2 treatment or knockdown of JWA (Figure 5B).
To confirm whether JWA regulates E2F1 expression at protein levels, we examined the E2F1 protein expression by western blotting. Our results showed that knockdown of JWA significantly decreased E2F1 protein level in both control and H2O2-treated NIH-3T3 cells (Figure 5C). In addition, EMSA was performed to determine whether JWA affects the affinity of E2F1 for binding to the XRCC1 promoter. As shown in Figure 5D, the proteins in the nuclear extracts of NIH-3T3 cells formed a complex with the labeled probe (XRCC1 –826 to –797-bp promoter region), which included the E2F1-binding sites. The shift band was eliminated by the addition of 100-fold unlabeled probe. When an antibody against E2F1 was added, a specific supershift band was observed, while no supershift complex was formed when a control IgG was substituted. The shift band and the supershift band were not affected by the addition of the same fold unlabeled mutated probe (Supplementary Figure S3). Furthermore, the DNA–protein complex or DNA–protein–E2F1 antibody complex was more abundant in the nuclear extracts from normal cells than from JWA knockdown cells (Supplementary Figure S3). We also found that the amount of DNA–protein binding increased further when more nuclear extracts from normal cells, but not from JWA knockdown cells, were added to the complex (Figure 5D). Following H2O2 stimulation, the affinity of E2F1 binding to the labeled probe was increased in the control cells (Figure 4D, lanes 2 and 4), but not in JWA knockdown cells (Figure 5D, lanes 5 and 7). These data provide further evidence that knockdown of JWA downregulates E2F1 levels and decreases the affinity of E2F1 binding to the XRCC1 promoter, resulting in decreased expression of XRCC1.
It was reported that E2F1 is located downstream of the MAPK signaling pathway and is activated by activation of the PKC/Raf/MEK/ERK pathway (31,32). Chen et al. (37) previously showed that phosphorylation of MEK-ERK, but not Raf, was completely blocked in JWA knockdown cells. Therefore, we postulated that JWA may activate E2F1 via the MAPK cascade. To test this hypothesis, western blot analysis for ERK and phosphorylated ERK expression following DNA damage in cell culture models was conducted. As shown in Figure 5E, phosphorylated ERK was detected following H2O2 treatment. As expected, both E2F1 and XRCC1 levels were enhanced following ERK activation. However, the activation of ERK was significantly blocked in JWA knockdown cells, and this was accompanied by decreased expression of E2F1 and XRCC1. In order to confirm the importance of ERK signaling in H2O2-induced upregulation of E2F1 and XRCC1, the MEK1/2 inhibitors PD98059 and U0126 were used. Both U0126 and PD98059 led to a substantial decrease in H2O2-induced phosphorylation of ERK, which subsequently reduced the H2O2-induced E2F1 and XRCC1 expression (Figure 5E). These data suggest that knockdown of JWA suppresses MAPK signaling and reduces E2F1 protein levels, leading to a decreased affinity of E2F1 for binding to the XRCC1 promoter, resulting in decreased XRCC1 transcription.
JWA is required for maintaining the stability of the XRCC1 protein
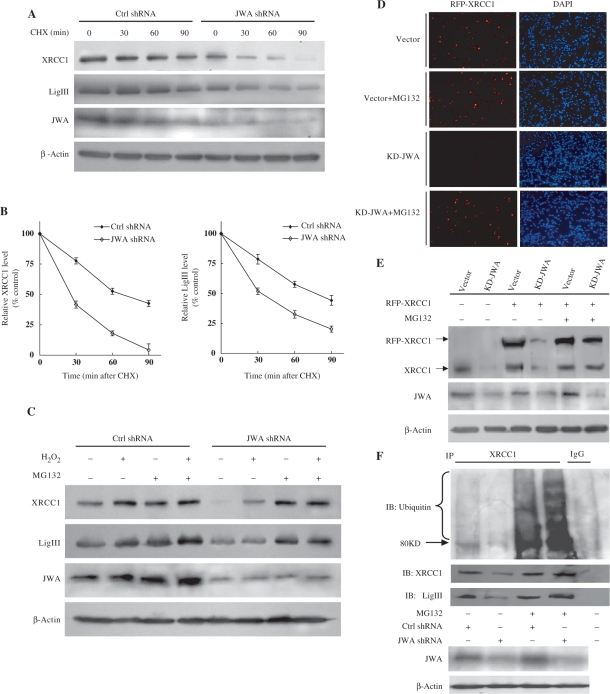
It was previously demonstrated that XRCC1 levels were reduced in stable JWA knockdown cells (42). We therefore postulated that JWA may be required for XRCC1 stability. When the cells were treated with CHX, an inhibitor of protein synthesis, we found that knockdown of JWA promoted the degradation of both endogenous XRCC1 and LigIII proteins in NIH-3T3 cells (Figure 6A and B). To determine whether the ubiquitin-proteasome pathway is responsible for XRCC1 degradation, cells were treated with the proteasome inhibitor MG132. We observed that loss of XRCC1 expression in JWA knockdown cells was inhibited by pretreatment with 10 μM MG132 for 4 h (Figure 6C). Similar effects were observed for LigIII (Figure 6C), suggesting that JWA inhibits the proteasomal degradation of these proteins. In order to confirm these data, an exogenous RFP-XRCC1 plasmid was transfected into stable selected vector control cells or KD-JWA cells with or without MG132 pretreatment. The transfected exogenous XRCC1 was also destabilized in JWA deficient cells (Figure 6D). The decreased expression of endogenous and exogenous XRCC1 in JWA knockdown cells was confirmed by western blotting (Figure 6E). However, the reduction in XRCC1 expression could be prevented by pretreatment with MG132 (Figure 6D and E).
Figure 6.
JWA is required for maintaining the stability of the XRCC1 protein. (A) JWA deficiency significantly enhanced the degradation of XRCC1 and LigIII. NIH-3T3 cells were transfected with JWA shRNA or the corresponding empty vector for 48 h, followed by exposure to cycloheximide (CHX) (50 μg/ml) for various time periods. Target proteins in whole-cell lysates were detected by immunoblotting using antibodies against XRCC1, LigIII and JWA. (B) The intensity of the XRCC1 and LigIII protein bands in (A) were analyzed by densitometry, after normalization to the corresponding β-actin level. The means ± SD are from three independent experiments. (C) The proteasome mediates the degradation of XRCC1 and LigIII. NIH-3T3 cells were transfected with JWA shRNA or the control vector. Forty-four hours later, cells were incubated with or without of MG132 (10 μM) for 4 h, then the cells were cultured for another 30 min with or without 100 μM H2O2. Cell lysates were used for immunoblotting with antibodies against XRCC1, LigIII and JWA. β-Actin was used as a loading control. (D) An RFP-XRCC1 plasmid was transiently transfected into stable selected EGFP-C1 vector control or KD-JWA NIH-3T3 cells. Forty-four hours later, the cells were incubated with or without of MG132 (10 μM) for 4 h. The cells were then fixed in methanol/acetone and counterstained with DAPI. The expression of RFP-XRCC1 in the cells (red) was observed under a fluorescent microscope. The nucleus of the cells was indicated by DAPI (blue). (E) NIH-3T3 or KD-JWA cells were transiently transfected with RFP-XRCC1 plasmid for 44 h, incubated with MG132 (10 μM) for 4 h, and western blotting was performed to confirm the levels of endogenous XRCC1 (lower molecule band) and exogenous XRCC1 (RFP-XRCC1). (F) Knocking down JWA results in the ubiquitylation and degradation of XRCC1. NIH-3T3 cells were transfected with JWA shRNA or the control vector. Forty-four hours later, the cells were incubated with or without of MG132 (10 μM) for another 4°h. Cell lysates were used for IP with the XRCC1 antibody and then blotted for XRCC1, LigIII and ubiquitin. Western blotting for JWA and β-actin in whole-cell lysates was utilized to check the JWA knockdown efficiency and to ensure equal protein loading.
To test whether XRCC1 degradation by the proteasome is initiated by XRCC1 ubiquitination, JWA shRNA- and control shRNA-transfected cells were cultured in the presence or absence of MG132. We found that XRCC1 was ubiquitinated in the presence of MG132, and that JWA depletion enhanced the ubiquitination of XRCC1 (Figure 6F). Furthermore, LigIII was immunoprecipitated by the anti-XRCC1 antibody in lysates from JWA-depleted cells, although the interaction was apparently decreased compared to control cells. However, the level of LigIII was preserved when the JWA knockdown cells were pretreated with MG132 (Figure 6F).
DISCUSSION
JWA was originally recognized as an all-trans-retinoic acid responsive and cytoskeleton-associated gene (36). Later Zhu et al. (40,41) noted that JWA was actively regulated by environmental stressors such as heat shock and oxidative stress, thereby suggesting that JWA might be a functionally active gene. JWA may play different biological functions under normal and different stress conditions due to its subcellular localization. Watabe et al. (48) demonstrated that the glutamate transport-associated protein for EAAC1 (GTRAP3-18), a homolog of JWA protein identified in rat, to interact with EAAC1 at the plasma membrane and thereby regulate neuronal glutathione levels. Our recent data showed that JWA was involved in cellular responses to chemically induced oxidative stress and also participated in the protection of cells from stress-induced DNA damage (42). The mammalian JWA proteins are highly conserved within their defined N- and C-terminal domains, and JWA-like proteins are also present in a wide range of eukaryotes from flies to human beings (http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene), indicating that JWA plays an important, evolutionarily conserved role. In this study, JWA, a novel component of the DNA damage response, was found to have a considerable impact in the repair of DNA SSBs.
Our previous data provided direct proof that JWA is required for DNA-strand break resealing by comet assay and showed that reduced JWA levels partially inhibited DNA rejoining (42). Here, additional phenotypic evidence was provided confirming that JWA is essential for DNA damage repair. First, the HCR assay showed that endogenous JWA is required for the removal of H2O2-induced DNA lesions. Second, the intracellular NAD(P)H assay showed that DNA damage was greater in JWA-depleted cells than in JWA wild-type cells. Third, downregulation of JWA resulted in increased sensitivity of both mouse and human cell lines to H2O2 and MMS, which is in agreement with previous studies indicating that XRCC1−/− CHO cells or RNAi-mediated XRCC1 knockdown human cell lines are hypersensitive to DNA-damaging agents (23,26,49). These findings suggested that JWA mediates protective effects in cells in response to oxidative stresses.
BER is a ubiquitous mechanism for removing damaged DNA induced by spontaneous chemical reactions, ROS and a variety of environmental genotoxicants (1). Many repair enzymes are involved in this process, and a major characteristic of these repair proteins is the formation of distinct foci which persist for a period of time after DNA damage (50,51). At the earliest stage of SSBR, PARP-1 rapidly binds to SSBs and is thereby activated, leading to the rapid formation of poly (ADP-ribose) polymer (PAR) nuclear foci, followed by the appearance of XRCC1 nuclear foci at the same sites (14). In the present study, we revealed that a prominent feature of JWA is its ability to rapidly translocate into the nucleus and transiently assemble into discrete nuclear foci after oxidative DNA damage. The complete overlap of JWA localization with PAR and XRCC1 provides important clues for its function in DNA damage repair. It seems that these foci represent aggregates of proteins to facilitate DNA repair or assist in signaling that DNA damage is present (52). Moreover, the nuclear proteins in DNA repair foci have been immensely useful in the identification of new factors involved in DNA repair (53). These findings suggest that JWA is likely a novel DNA repair molecule.
A feature of mammalian SSBR appears to be the employment of protein–protein interactions to stimulate individual component steps and/or the overall repair reaction (16). Arguably, the most intriguing SSBR protein in this respect is XRCC1, a scaffold protein that interacts with PARP-1 and LigIII via its two BRCA1 carboxyl-terminal (BRCT) domains, denoted BRCT I and II, respectively (54). Our study revealed that JWA interacts with XRCC1, PARP-1 and LigIII in normal or stressed conditions, identifying JWA as a component of BER protein complexes. Furthermore, the interaction of JWA and XRCC1 may explain how JWA can translocate to the nucleus despite its lack of a classic NLS domain. It was presumed that JWA interacts with XRCC1, which contains a NLS at positions 239 to 266 in its amino acid sequence (54), and be subsequently transported into nucleus by XRCC1 following DNA damage. Notably, our studies showed that siRNA-mediated downregulation of XRCC1 inhibited JWA translocation into the nucleus. Despite the importance of JWA and XRCC1 in SSBR, it is possible that additional mechanisms may be involved in this process. For example, post-translational modifications, such as phosphorylation (55) and mono-ubiquitylation (56), may also influence a protein's localization. There are two potential phosphorylation sites (serine residues) and 10 ubiquitylation sites (lysine residues) in the JWA protein. Future studies will be necessary to determine which, if any, of these sites is important.
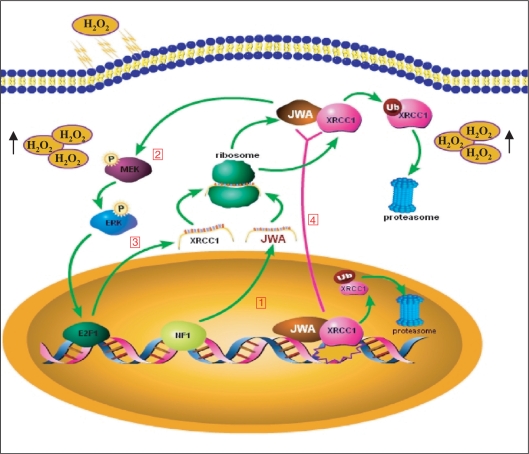
Current evidence suggests that DNA damage activates signaling pathways for damage recognition, DNA repair, cell-cycle progression and apoptosis (57,58). For example, generation of H2O2 is required for platelet-derived growth factor (PDGF) signal transduction (59); the EGFR (epidermal growth factor receptor)-MAPK signaling cascade appears to regulate the expression of XRCC1 in prostate carcinoma cells to repair DNA-strand breaks induced by radiation (30); and E2F1 is activated by the MAPK signaling cascade following DNA damage (31) and subsequently phosphorylated and stabilized via ATM/ATR (60) and Chk2 dependent pathways (61). Our present data confirmed that inhibition of ERK1/2 activity eliminated the H2O2-induced upregulation of the expression of E2F1 and XRCC1. We also found that knocking down JWA resulted in inactivation of the MEK-ERK signaling pathway and downregulation of E2F1. Since many repair genes have been identified as putative E2F1 targets, including TP53 (62,63) and XRCC1 (33), our study indicates that knockdown of JWA not only reduces E2F1 expression, but also decreases its affinity for binding to the XRCC1 promoter, leading to decreased transcription of XRCC1. Our previous and present studies also defined a novel signaling pathway wherein intracellular H2O2 triggers NF1 activation and binding to the JWA promoter region (42), resulting in increased JWA transcription, activating E2F1 and, subsequently, XRCC1 via the MAPK signaling cascades. This process facilitates DNA SSBR (Figure 7). However, recent publications have suggested that XRCC1 is phosphorylated by protein kinases DNA-PK (64) and CK2 (65) to enhance DNA repair and genetic stability. The role of JWA and a possible crosstalk between the MAPK and DNA-PK/CK2 pathways in XRCC1 activation needs to be examined in future studies.
Figure 7.
A model for the effects of JWA and XRCC1 on base excision repair. (1) Exposure to oxidative stress increases the generation of intracellular H2O2, which stimulates NF1 binding to the JWA promoter, enhancing JWA transcription and translation (42). (2) JWA regulates the expression of E2F1, leading to increased transcription of XRCC1. (3) Interactions between JWA and XRCC1 occur in both the cytoplasm and the nucleus when the cells are subjected to oxidative stress. XRCC1 transports JWA from the cytoplasm into the nucleus; however, JWA regulates and stabilizes nuclear XRCC1 by preventing its ubiquitination. (4) When cells are subjected to oxidative stress, both JWA and XRCC1 are recruited to DNA SSB sites and to exert a critical role in the SSBR/BER process.
XRCC1 not only interacts with enzymes during each step of BER, but also coordinates several repair proteins to enhance DNA-repair efficacy. It was reported that the formation of DNA-repair complexes on damaged DNA requires XRCC1 to act as a scaffold to stabilize the BER proteins, and that when the proteins are no longer needed for the repair complex, they are ubiquitylated by the E3 ubiquitin ligase CHIP and degraded by the proteasome (34). It was suggested that ubiquitination sites on the BER proteins within repair complexes may be masked by XRCC1, and that the absence of XRCC1 will immediately lead to their degradation (34). However, how the stability of XRCC1 itself is regulated is not clear. Our study demonstrated that JWA is a novel component of BER complex that can stabilize XRCC1. Interestingly, the half-life of both XRCC1 and its partner LigIII were shortened in JWA-depleted cells. Moreover, when JWA is absent, XRCC1 is rapidly degraded through the ubiquitin-proteasome pathway. Therefore, it is reasonable to presume that JWA might be the candidate protein that can mask the ubiquitlylation sites in XRCC1, thus stabilizing XRCC1 (Figure 7).
In summary, our present results indicate that JWA, a novel component of the BER complex which is recruited by XRCC1 to the DNA damage sites, mediates the repair of DNA damage induced by oxidative stress via activating the expression of XRCC1 through MAPK-E2F1 signaling pathway and protecting XRCC1 from ubiquitin-proteasome degradation. Our study not only provides novel mechanistic insights into the role of JWA in the regulation of BER, but further defines the involvement of XRCC1 in the SSBR/BER process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Natural Science Foundation of China (30771829); Jiangsu Provincial Graduates Innovative Project (to S.Y.W.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank professors R.W. Zhang and Donald L. Hill (the University of Alabama at Birmingham, AL, USA), and S. Kacew (University of Ottawa, Canada) for their critical review, helpful comments and suggestions for the manuscript.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 3.Xu YJ, Kim EY, Demple B. Excision of C-4'-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 4.Carrano AV, Minkler JL, Dillehay LE, Thompson LH. Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat. Res. 1986;162:233–239. doi: 10.1016/0027-5107(86)90090-4. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez I, Daza P, Natarajan AT, Cortes F. A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res. 1998;398:67–73. doi: 10.1016/s0027-5107(97)00241-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Caldecott KW. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 9.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Benjamin RC, Gill DM. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 1980;255:10502–10508. [PubMed] [Google Scholar]
- 11.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 12.Ohgushi H, Yoshihara K, Kamiya T. Bovine thymus poly(adenosine diphosphate ribose) polymerase. Physical properties and binding to DNA. J. Biol. Chem. 1980;255:6205–6211. [PubMed] [Google Scholar]
- 13.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 14.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 19.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor RM, Wickstead B, Cronin S, Caldecott KW. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 21.Petermann E, Keil C, Oei SL. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair (Amst) 2006;5:544–555. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zdzienicka MZ, van der Schans GP, Natarajan AT, Thompson LH, Neuteboom I, Simons JW. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 1992;7:265–269. doi: 10.1093/mutage/7.4.265. [DOI] [PubMed] [Google Scholar]
- 24.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green A, Prager A, Stoudt PM, Murray D. Relationships between DNA damage and the survival of radiosensitive mutant Chinese hamster cell lines exposed to gamma-radiation. Part 1: intrinsic radiosensitivity. Int. J. Radiat. Biol. 1992;61:465–472. doi: 10.1080/09553009214551221. [DOI] [PubMed] [Google Scholar]
- 26.Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol. Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tebbs RS, Thompson LH, Cleaver JE. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair (Amst) 2003;2:1405–1417. doi: 10.1016/j.dnarep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 30.Yacoub A, McKinstry R, Hinman D, Chung T, Dent P, Hagan MP. Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. Radiat. Res. 2003;159:439–452. doi: 10.1667/0033-7587(2003)159[0439:egfair]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Berkovich E, Ginsberg D. Ras induces elevation of E2F-1 mRNA levels. J. Biol. Chem. 2001;276:42851–42856. doi: 10.1074/jbc.M103596200. [DOI] [PubMed] [Google Scholar]
- 32.Pintus G, Tadolini B, Posadino AM, Sanna B, Debidda M, Carru C, Deiana L, Ventura C. PKC/Raf/MEK/ERK signaling pathway modulates native-LDL-induced E2F-1 gene expression and endothelial cell proliferation. Cardiovasc. Res. 2003;59:934–944. doi: 10.1016/s0008-6363(03)00526-1. [DOI] [PubMed] [Google Scholar]
- 33.Chen D, Yu Z, Zhu Z, Lopez CD. E2F1 regulates the base excision repair gene XRCC1 and promotes DNA repair. J. Biol. Chem. 2008;283:15381–15389. doi: 10.1074/jbc.M710296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 35.Sobol RW. CHIPping away at base excision repair. Mol. Cell. 2008;29:413–415. doi: 10.1016/j.molcel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Zhou JW, Di YP, Zhao YH, Wu R. In: Investigation on Cell Modulation: Signal Transduction, Apoptosis and Gene Expression. Ye XS, Shen BF, Tang, editors. Beijing: Military Medical Sciences Press; 1999. pp. 110–119. [Google Scholar]
- 37.Chen H, Bai J, Ye J, Liu Z, Chen R, Mao W, Li A, Zhou J. JWA as a functional molecule to regulate cancer cells migration via MAPK cascades and F-actin cytoskeleton. Cell Signal. 2007;19:1315–1327. doi: 10.1016/j.cellsig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Huang S, Shen Q, Mao WG, Li AP, Ye J, Liu QZ, Zou CP, Zhou JW. JWA, a novel signaling molecule, involved in all-trans retinoic acid induced differentiation of HL-60 cells. J. Biomed. Sci. 2006;13:357–371. doi: 10.1007/s11373-005-9068-0. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Shen Q, Mao WG, Li AP, Ye J, Liu QZ, Zou CP, Zhou JW. JWA, a novel signaling molecule, involved in the induction of differentiation of human myeloid leukemia cells. Biochem. Biophys. Res. Commun. 2006;341:440–450. doi: 10.1016/j.bbrc.2005.12.197. [DOI] [PubMed] [Google Scholar]
- 40.Zhu T, Chen R, Li A, Liu J, Gu D, Liu Q, H CC, Zhou J. JWA as a novel molecule involved in oxidative stress-associated signal pathway in myelogenous leukemia cells. J. Toxicol. Environ. Health A. 2006;69:1399–1411. doi: 10.1080/15287390500360612. [DOI] [PubMed] [Google Scholar]
- 41.Zhu T, Chen R, Li AP, Liu J, Liu QZ, Chang HC, Zhou JW. Regulation of a novel cell differentiation-associated gene, JWA during oxidative damage in K562 and MCF-7 cells. J. Biomed. Sci. 2005;12:219–227. doi: 10.1007/s11373-004-8186-4. [DOI] [PubMed] [Google Scholar]
- 42.Chen R, Qiu W, Liu Z, Cao X, Zhu T, Li A, Wei Q, Zhou J. Identification of JWA as a novel functional gene responsive to environmental oxidative stress induced by benzo[a]pyrene and hydrogen peroxide. Free Radic. Biol. Med. 2007;42:1704–1714. doi: 10.1016/j.freeradbiomed.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat. Res. 2002;509:165–174. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 44.Philpott SM, Buehring GC. Defective DNA repair in cells with human T-cell leukemia/bovine leukemia viruses: role of tax gene. J. Natl. Cancer Inst. 1999;91:933–942. doi: 10.1093/jnci/91.11.933. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura J, Asakura S, Hester SD, de Murcia G, Caldecott KW, Swenberg JA. Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res. 2003;31:e104. doi: 10.1093/nar/gng105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamae Y, Iwasaki K, Kinae N, Tsuda S, Murakami M, Tanaka M, Sasaki YF. Detection of DNA lesions induced by chemical mutagens using the single-cell gel electrophoresis (comet) assay. 2. Relationship between DNA migration and alkaline condition. Mutat. Res. 1997;393:107–113. doi: 10.1016/s1383-5718(97)00091-0. [DOI] [PubMed] [Google Scholar]
- 47.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 48.Watabe M, Aoyama K, Nakaki T. A dominant role of GTRAP3-18 in neuronal glutathione synthesis. J. Neurosci. 2008;28:9404–9413. doi: 10.1523/JNEUROSCI.3351-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan J, Wilson PF, Wong HK, Urbin SS, Thompson LH, Wilson DM., III XRCC1 down-regulation in human cells leads to DNA-damaging agent hypersensitivity, elevated sister chromatid exchange, and reduced survival of BRCA2 mutant cells. Environ. Mol. Mutagen. 2007;48:491–500. doi: 10.1002/em.20312. [DOI] [PubMed] [Google Scholar]
- 50.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Au WW, Henderson BR. Identification of sequences that target BRCA1 to nuclear foci following alkylative DNA damage. Cell Signal. 2007;19:1879–1892. doi: 10.1016/j.cellsig.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 53.Dellaire G, Bazett-Jones DP. Beyond repair foci: subnuclear domains and the cellular response to DNA damage. Cell Cycle. 2007;6:1864–1872. doi: 10.4161/cc.6.15.4560. [DOI] [PubMed] [Google Scholar]
- 54.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon SO, Shin S, Liu Y, Ballif BA, Woo MS, Gygi SP, Blenis J. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol. Cell. 2008;29:362–375. doi: 10.1016/j.molcel.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 57.Holbrook NJ, Liu Y, Fornace AJ., Jr. Signaling events controlling the molecular response to genotoxic stress. EXS. 1996;77:273–288. doi: 10.1007/978-3-0348-9088-5_18. [DOI] [PubMed] [Google Scholar]
- 58.Kharbanda S, Pandey P, Yamauchi T, Kumar S, Kaneki M, Kumar V, Bharti A, Yuan ZM, Ghanem L, Rana A, et al. Activation of MEK kinase 1 by the c-Abl protein tyrosine kinase in response to DNA damage. Mol. Cell Biol. 2000;20:4979–4989. doi: 10.1128/mcb.20.14.4979-4989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 60.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 61.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 62.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 63.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 64.Levy N, Martz A, Bresson A, Spenlehauer C, de Murcia G, Menissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34:32–41. doi: 10.1093/nar/gkj409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.