Abstract
Recent technological advances have enabled us to visualize the organization and dynamics of local chromatin structures; however, the comprehensive mechanisms by which chromatin organization modulates gene regulation are poorly understood. We designed a human artificial chromosome vector that allowed manipulation of transgenes using a method for delivering chromatin architectures into different cell lines from human to fish. This methodology enabled analysis of de novo construction, epigenetic maintenance and changes in the chromatin architecture of specific genes. Expressive and repressive architectures of human STAT3 were established from naked DNA in mouse embryonic stem cells and CHO cells, respectively. Delivery of STAT3 within repressive architecture to embryonic stem cells resulted in STAT3 activation, accompanied by changes in DNA methylation. This technology for manipulating a single gene with a specific chromatin architecture could be utilized in applied biology, including stem cell science and regeneration medicine.
INTRODUCTION
Our ability to analyze human gene regulation and function has been advanced by the complete sequencing of the genome and our increased knowledge of the interactions between regulatory proteins and DNA elements. However, our ability to analyze chromatin dynamics and epigenetic regulation, including DNA methylation and histone modification, is limited, hindering our understanding of the comprehensive mechanisms of gene regulation. In particular, analyses using transgene have often been dependent on transient expression of exogenous genes and silenced or ectopic expression of integrated genes. Novel technologies that allow direct analysis of native gene expression based on chromatin structure are needed.
Chromosome technologies that utilize large-sized DNA packaged into chromatin structures have been developed. The main techniques involve the construction of chromosome fragments from human native chromosomes or human artificial chromosomes (HACs) from naked DNA (1–3) and the introduction of chromosomes using microcell-mediated chromosome transfer (MMCT) (4). HACs can be reproducibly generated de novo in human HT1080 cells by transfection of long alpha-satellite DNA containing CENP-B boxes (5–7). During their generation, alphoid precursors are built up to mega-base size (2). Previous studies have shown that chromosome fragments and HACs replicate and segregate once per cell division cycle to be stably maintained in mice (8,9), and can carry large human genes that contain native regulatory regions into human and animal cells (8–14). HACs provide several potential advantages over viral and integrating vectors for evaluating gene expression, including long-term stability, low toxicity and huge size of inducible DNA.
Here, we designed and constructed a chromosome vector that contained elements essential for chromosome maintenance and a platform suitable for gene expression. To assess the use of chromosome vectors as tools for analyzing gene regulation, we examined the construction, maintenance and epigenetic changes of the chromatin architecture using human STAT3 on the chromosome vector as a model.
MATERIALS AND METHODS
Cell lines
HT1080 cells were cultured in DMEM (Sigma) supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. CHO cells were cultured in Ham's F-12 nutrient mixture (Sigma) supplemented with 10% FBS at 37°C and 5% CO2. TT2F mouse embryonic stem (ES) cells were cultured in DMEM supplemented with 20% Knockout SR (Invitrogen), 1000 U/ml ESGRO (Chemicon) and 0.1 mM β-mercaptoethanol. NIH/3T3 cells were cultured in DMEM supplemented with 10% calf serum (CS). PK15 cells were cultured in MEM (Sigma) supplemented with 5% CS at 37°C and 5% CO2. DT40 cells were cultured in RPMI1640 (Sigma) supplemented with 10% FBS, 1% chick serum and 0.05 mM β-mercaptoethanol at 37°C and 5% CO2. A6 cells were cultured in DMEM (Sigma) supplemented with 10% FBS at 26°C and 5% CO2. AB.9 cells were cultured in DMEM (Sigma) supplemented with 15% FBS at 28°C and 10% CO2. Cell lines harboring a chromosome vector were selected with G418 (Sigma) at 300 μg/ml (TT2F), 400 μg/ml (HT1080, CHO), 600 μg/ml (NIH/3T3), 750 μg/ml (PK15), 1000 μg/ml (DT40), 1400 μg/ml (AB.9) or 2000 μg/ml (A6).
A precursor of the HAC vector, lox-BAC
Human beta-globin 5′ hyper sensitive (5′HS5) site (3.4-kb EcoRI fragment; 4818-8173 in GenBank NG000007) was cloned into the EcoRI site of pUC119, and 3′ hyper sensitive (3′HS1) site (5.6-kb SphI-SacI fragment; 8255-13891 in GenBank AC104389) was cloned into the SphI-SacI site of pTWV229 (TaKaRa) vector from YAC clone A201F4.3. CAG promoter/lox71/neo cassette (CAG-lox71-neo) was constructed from pCAGGS-bsr (15) by deletion of the HindIII-HindIII fragment containing bsr and insertion of neo from the PstI partial fragment of SV-neo and I-SceI recognition site. Then, 5′HS5, 3′HS1 and CAG-lox71-neo were inserted into HpaI-SphI, SalI and NotI sites of pBelo-BAC11 (New England BioLabs) by blunt-end ligation to construct lox-BAC that contains 5′HS5, CAG/lox71/neo and 3′HS1, in that order.
A precursor of the HAC vector, alphoid-BAC
A pBelo-BAC vector was modified by insertion of the kanamycin resistance gene in the BsmI-BsmI site, the ampicillin resistance gene in SexAI-AhdI and the MluI/SacII linker in the SalI-SalI site, followed by deletion of the loxP site (BAC-AK). Finally, 50-kb alphoid fragment was inserted into the MluI-SacI site of BAC-AK to construct alphoid-BAC.
Donor plasmid
The plox66-puro was constructed from a 1.2-kb HindIII-SalI fragment of pGK-puro (coding region of puro) and a 3.0-kb HindIII-XhoI fragment of lox66-Nlaczeo (15) (lox66). A 1.2-kb SpeI-KpnI fragment of plox66-puro (lox66-puro cassette) was blunt-ended and inserted into the HindIII site of pTWV229 (TWV-lox66-puro). The CMV promoter in pEGFP-C1 was replaced with the SV, EF1 and cyclin A promoters to construct SV-EGFP, EF1-EGFP and cyclin A-GFP. The cyclin A promoter was amplified by PCR from HT1080 genomic DNA promoters. AseI-MluI fragments of pEGFP-C1 (CMV-EGFP), SV-EGFP, EF1-EGFP and cyclin A-EGFP were inserted into the SalI site of TWV(lox66)puro by blunt-end ligation.
DNA transfection
To generate chromosome vectors, 1.0 μg of alphoid-BAC and 0.5 μg of lox-BAC were co-transfected into HT1080 cells (5 × 105) using lipofectamine2000 (Invitrogen) according to the manufacturer's instructions.
Red-ET recombination
Red-ET recombination in Escherichia coli was performed according to the manufacturer's instructions (Gene Bridges). To modify the loxP site in BAC clones with the lox66/puro cassette, a linear fragment with homologous arms was prepared. The 200-bp arms homologous to two sites of the BAC vector (Belo-BAC [6915-7114] and Belo-BAC [51-250]) were inserted into the HindIII site and EcoRI site in pTWV(lox66)puro to construct a circular vector. Then, the target linear fragments were prepared by digestion of the circular vector with HpaI and FspI, and purification of lox66/puro cassette with homologous arms.
Microcell-mediated chromosome transfer
Forty 10-cm dishes of HT-A9/25-4 cells were grown to 80% confluency and then cultured for 72 h in DMEM containing colcemid (0.05 μg/ml). Collected cells were incubated for 5 min at 37°C in serum-free DMEM containing cytochalasin B (20 μg/ml), then added to an equal volume of Percoll (GE Healthcare). The suspension was centrifuged in a Hitachi R20A2 rotor at 15 000 r.p.m. for 90 min at 37°C. The collected microcells were suspended in serum-free DMEM, then mixed with recipient cells and centrifuged at 2000 r.p.m. for 5 min. The resulting pellet was finally suspended in 1 ml 50% PEG1500 (Roche) and incubated at room temperature for 90 s. Five milliliters of serum-free DMEM was added and the mixture was centrifugated at 1000 r.p.m. for 5 min. The fused cells were washed twice in serum-free DMEM and were plated.
Pulsed-field gel electrophoresis
DNA in agarose blocks was digested for 4 h and size separated in a 1% GTG agarose gel (Cambrex) on the CHEF mapper system (Bio-Rad). The running conditions were dependent on the auto algorithm from 5 to 500 kb.
Southern analyses
DNA in the gel was transferred to a nylon membrane and hybridized with 32P-labeled DNA probes prepared from Belo-BAC vector DNA for detection of chromosome vectors or a DNA probe from the 1234-bp fragment (MscI-DraI) of the 5′ region and 1283-bp fragment (AflII-MscI) of the 3′ region of human STAT3 gene for detecting methylation status.
RT–PCR
Total RNA was prepared using SV Total RNA Isolation System (Promega). Fifty nanograms of RNA were used for RT–PCR using the One Step RNA PCR Kit (AMV) (TaKaRa). For the full length human STAT3 gene (exons 2–24), the primers were hSTAT3F1: 5′-TCAGCTCTACAGTGACAGCTTCCCA-3′ and hSTAT3R1: 5′-GAGGTAGCGCACTCCGAGGTCAAC-3′. Samples were amplified at 50°C for 30 min, 94°C for 2 min, 30 cycles at 94°C for 15 s, 68°C for 15 s and 72°C for 2 min. For full length of mouse STAT3 gene (exons 2–24), the primers were mSTAT3F1: 5′-CCAGCTGTACAGCGACACGTTCCCC-3′ and mSTAT3R1: 5′-GAGGTAGCACACTCCGAGGTCAGA-3′. Samples were amplified at 50°C for 30 min, 94°C for 2 min, 30 cycles at 94°C for 15 s, 65°C for 15 s and 72°C for 2 min. To amplify human STAT3 exons 2–9, the primers were hSTAT3F1 and STATex9R: 5′-TTCATTAAGTTTCTAAACAGCTCC-3′. Samples were amplified at 50°C for 30 min, 94°C for 2 min, 30 cycles at 94°C for 15 s, 61°C for 15 s and 72°C for 1 min.
Fluorescence in situ hybridization
Metaphase chromosomes were prepared on glass slides after methanol/acetate (3:1) fixation. Fluorescence in situ hybridization (FISH) was performed using pBelo-BAC DNA and 11-4 alphoid DNA as probes for detecting HAC vectors according to a standard procedure. Digoxigenin-labeled pBelo-BAC was visualized with TRITC-conjugated anti-digoxigenin (Roche Diagnostics), and Biotin-labeled 11-4 alphoid DNA (16) was visualized with Alexa488-conjugated streptavidin (Invitrogen). Photographs were taken using a CCD camera mounted on a Zeiss microscope AxioPlan2. Images were processed using AxioVision.
Indirect immunofluorescence
Indirect immunofluorescence was performed using an anti-human STAT3 antibody (Epitomics) which does not cross-react with mouse Stat3. Mouse ES cells prepared on glass slides using cytospin (Shandon) were fixed in 2% paraformaldehyde for 30 min, reacted with an anti-human STAT3 antibody, then visualized with FITC-conjugated anti-rabbit IgG.
RESULTS
Constructing a circular chromosome vector in human cells
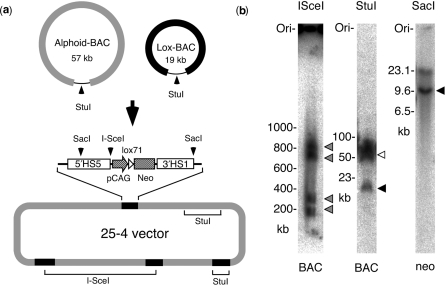
To construct a chromosome vector that contained the elements essential for both chromosome maintenance and gene expression, a 50-kb alphoid DNA fragment and a gene expression cassette cloned into bacterial artificial chromosomes (alphoid-BAC, Lox-BAC; Figure 1a) were co-transfected into HT1080 cells, transfectants were selected with G418 and analyzed by FISH. In vivo multimerization of the alphoid-BAC and Lox-BAC within the cells generated circular artificial chromosomes that harbored expression cassettes (Figure 1a). The expression cassette consisted of a CAG promoter-drive neomycin resistance gene (neo) with mutant loxP, lox71 and human beta-globin 5′HS5 and 3′HS1 sites, which possess chromatin boundary functions (17). The expression cassette was utilized during drug selection for the construction of the artificial chromosome and for gene insertion. While the number of expression cassettes in chromosome vectors varied, we used one of the vectors (25-4) in HT1080 cells (HT/25-4) that contained four gene expression cassettes for the remainder of the studies (Figure 1b).
Figure 1.
Construction and configuration of chromosome vector 25-4. (a) The configuration of precursor BACs and the composition of 25-4. Alphoid-BAC contains 50 kb of alphoid DNA and Lox-BAC contains the CAG/lox71/neo cassette with insulator elements (5′HS5 and 3′HS1). The 25-4 is composed of alphoid DNA and four CAG/lox71/neo cassettes derived from Lox-BAC. (b) Southern analyses of 25-4. Genomic DNA of HT1080 cells containing 25-4 was digested with I-SceI, StuI or SacI, and hybridized with BAC vector sequences or neomycin resistant gene sequences (neo), respectively. I-SceI divided 25-4 into four fragments (800, 700, 300 and 200 kb) (gray), StuI cut 25-4 into precursor 57-kb fragments derived from alphoid-BACs (white) and 19-kb fragments derived from Lox-BACs (black) and SacI presented 9.5 kb of expression cassettes (black).
Transfer and availability of chromosome vectors into vertebrate cells
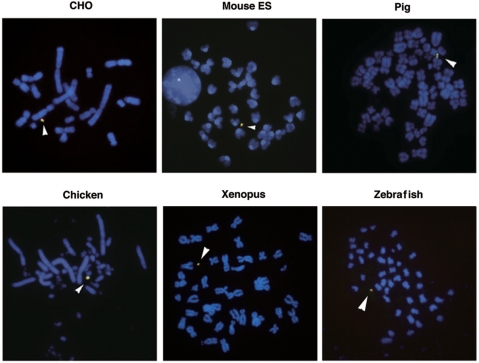
To utilize the chromosome vector as a tool in several studies, it must be transferable and be retained stably within the recipient cells. The useful method of transfer is MMCT, in which 25-4 was transferred via microcells generated from the A9/HT hybrid cell lines harboring 25-4 to the target cells. The 25-4 vector was successfully transferred and was maintained extrachromosomally in HeLa, NIH/3T3, pig (PK15), Indian Muntjac, rat (IAR20), quail (QNR/K2), chicken (DT40), Xenopus (A6), zebrafish (AB.9) and mouse ES cells (TT2F), with a chromosome loss rate of less than 0.5% per generation (Figure 2). These results indicated that chromosomal features and functions, including centromeres and replication origins, in 25-4 were maintained in several vertebrate cell lines and that 25-4 could be used as tool for stable gene expression.
Figure 2.
Stable retention of 25-4 vector in vertebrate cell lines. The vertebrate cell lines carrying 25-4 were established by MMCT. Transferred 25-4 was confirmed by FISH using metaphase cells: CHO, mouse ES (TT2F), pig (PK15), chicken B cells (DT40), Xenopus (A6) and zebrafish (AB.9). Green signals show alphoid DNA and red signals show BAC vector. Arrowheads show 25-4 vector.
Regulation of promoter-dependent gene expression from a chromosome vector
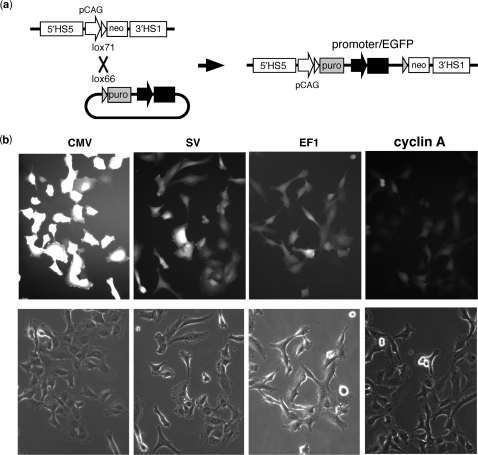
To investigate the ability of the 25-4 vector gene insertion and expression system, we inserted an enhanced green fluorescent protein (EGFP) gene driven by several different promoters (CMV, SV, EF1 and cyclin A) by Cre-lox recombination of the lox66 site at the promoterless cassette and lox71 site in the gene expression cassette of 25-4 in HT1080 cells (Figure 3a). The major event in puromycin-resistant cell lines was insertion of the EGFP gene in one expression cassette in four sites of 25-4. EGFP expression was evaluated by fluorescence intensity (Figure 3b). The relative level of fluorescence of CMV-EGFP was greater than that of EF1-EGFP or SV-EGFP, and two orders of magnitude greater than that of cyclin A-EGFP. The expression of EGFP was homogeneous in >90% of cells in each line, indicating little variation in gene expression between cells within a clonal HT/25-4 cell line (Figure 3b). This promoter-dependent expression of EGFP from the 25-4 vector was also observed in NIH/3T3 and CHO cells.
Figure 3.
Gene expression from the 25-4 vector. (a) Gene insertion into the 25-4 vector. The EGFP gene was inserted into 25-4 vector using Cre-lox recombination in HT1080 cells. Successful recombinants were selected by puromycin resistance (puro). (b) EGFP expression levels in 25-4. Fluorescence of CMV-, SV-, EF1- or cyclin A-driven expression of EGFP in HT1080 cells.
Construction of a mini-gene locus
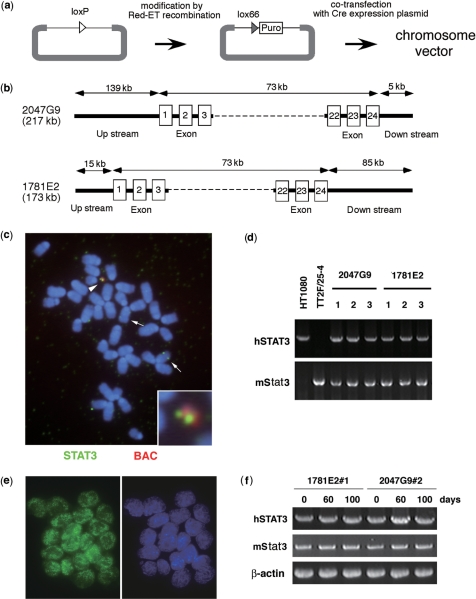
In order to develop a system for de novo construction of genes within the context of their native chromatin architecture under cellular conditions, naked genomic DNA was inserted into the gene expression cassette on the chromosome vector. The genomic DNA cloned into the BAC had been modified by addition of the lox/puromycin resistance (puro) cassette in place of the loxP by Red/ET recombination in E. coli. Subsequently, purified BAC DNA was inserted into the gene expression cassette of 25-4 in cells by co-transfection with a Cre expression plasmid (Figure 4a). The human STAT3 gene, composed of up-stream sequences, 24 exons and downstream sequences, was used as a representative genomic sequence. STAT3 is a signal transducer and transcriptional activator, and is essential for maintenance of undifferentiated mouse ES cells (18) (Figure 4b). More than 80% of transformants contained one copy of the intact human STAT3 gene at the correct position by Cre-lox recombination (Figure 4c). In ES cells, human STAT3 was actively expressed from the 25-4 vector and the complete length of the human STAT3 transcript was detected in 3 of 3 cell lines derived from BAC 2047G9 and in 10 of 11 cell lines from BAC 1781E2 at the same level as native human STAT3 expression in HT1080 (Figure 4d). Immunoanalysis using anti-human STAT3 antibody indicated that almost all cells in the cell lines expressed human STAT3 (Figure 4e), and transcription was maintained in mouse ES cells for 100 days in culture without noticeable silencing (Figure 4f). These results indicated that following insertion of naked DNA into chromosome vector in mouse ES cells in which the endogenous Stat3 was active, expression-permissive chromosomal architecture was constructed around the transfected human STAT3 gene.
Figure 4.
De novo construction of a human STAT3 mini-locus on 25-4 vector. (a) Strategy for modifying and inserting genomic DNA from BAC into chromosome vector. TT2F/25-4 cells that contained the human STAT3 genomic gene were selected with puromycin and drug-resistant cell lines were confirmed by PCR with CAG promoter and puro sequences, and by FISH using STAT3 and BAC sequences as probes. (b) The genomic composition of human STAT3 genes. Clone 2047G9 contains 139-kb of the upstream and 5-kb of the downstream regions of human STAT3. Clone 1781E2 contains 15-kb of upstream and 85-kb of downstream regions of human STAT3. The numbers in the boxes delineate exons. (c) FISH analysis of mouse ES cells containing human STAT3 mini-locus. Metaphase chromosomes were hybridized with human STAT3 cDNA (green) and BAC vector (red) sequences. Arrowhead shows 25-4 containing human STAT3; arrows show endogenous mouse Stat3. (d) Expression of human STAT3 from 25-4. The 2.2-kb fragments from human STAT3 transcripts were detected by RT–PCR in all ES cell lines that contained human STAT3. (e) Detection of human STAT3 products in TT2F/25-4 containing human STAT3. The STAT3 products were detected by anti-human STAT3 antibody in methanol-fixed cells. Green signals show human STAT3 products. (f) Expression of human STAT3 after long-term culturing. The TT2F/25-4 cells containing human STAT3 were cultured for 60 and 100 days and subsequently analyzed by RT–PCR. Human STAT3 transcripts were detected as before culturing.
Epigenetic maintenance and changes in gene architecture in the STAT3 locus
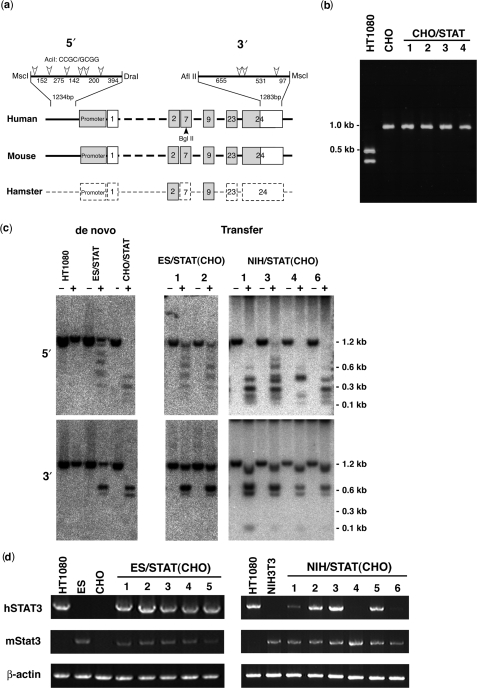
In order to examine the property of the de novo constructed gene locus, human STAT3 DNA was inserted into the 25-4 vector in CHO cells (CHO/STAT), which are often used for protein production. While the sequence of the Chinese hamster endogenous STAT3 gene was unknown, RT–PCR using primers for exons 2 and 9 of human STAT3, which are highly conserved between humans and mice, produced 0.9-kb fragments from both human and CHO cells (Figure 5a, b). Restriction analysis with BglII, a human STAT3-specific restriction site, indicated that human STAT3 was not expressed in any CHO/STAT cell line, despite active expression of hamster STAT3 (Figure 5a, b).
Figure 5.
Transfer of human STAT3 chromatin architecture. (a) Comparison of human, mouse and Chinese hamster STAT3 genes. Gray boxes show conserved regions, broken boxes show unknown regions. The numbers signify exons. AciI restriction enzyme sites in upstream of the promoter (5′) and in the last half of exon 24 (3′) are shown by open arrowheads. The BglII site is shown by a closed arrowhead. (b) RT–PCR of human STAT3 from CHO/STAT. STAT3 transcripts (about 1 kb) were detected in HT1080, CHO and four CHO lines that contained human STAT3 (CHO/STAT) by RT–PCR using primers corresponding to exons 2 and 9. Digestion of the transcripts with BglII produced 0.5- and 0.4-kb fragments of human STAT3 transcripts from HT1080 and nondigested 1.0-kb fragments from endogenous STAT3 in CHO cells and all CHO/STAT cell lines. (c) DNA methylation analysis in the 5′ and 3′ regions of human STAT3. Genomic DNA from HT1080, ES/STAT, CHO/STAT, two ES/STAT(CHO) and four NIH/STAT(CHO) cell lines was digested with MscI and DraI, or AflII and MscI, with (+) or without (−) AciI, and subsequently hybridized with 5′ or 3′ probes. The 1.2-kb fragments from digestion with AciI indicate a completely methylated state. (d) Detection of human STAT3 transcripts. Human STAT3 transcripts were detected in HT1080, mouse ES, CHO, NIH/3T3, five ES/STAT(CHO) and six NIH/STAT(CHO) cell lines by RT–PCR using human-specific primers corresponding to exons 2 and 24. A 2.2-kb fragment of the human STAT3 transcript was detected in all five ES/STAT(CHO) cell lines, while transcript detection varied in the six NIH/STAT(CHO) cell lines.
To investigate reasons for the differences in transcriptional status of human STAT3 in ES cells (expression) and CHO cells (repression), we analyzed the methylation status of human STAT3 in mouse ES (ES/STAT) and CHO (CHO/STAT) cells by restriction enzyme digestion (Figure 5c). A1234-bp sequence of 5′-upstream DNA and 1283 bp of 3′-untranslational region in exon 24 of human STAT3 were distinguishable from those of mouse ES and CHO cells (Figure 5a). The methylation status of human STAT3 was determined by the digestion pattern of AciI, which is sensitive to CpG methylation. The digestion pattern indicated that both the 5′ and 3′ regions of human STAT3 were completely methylated under the natural and expression-active conditions of the HT1080 cells. The majority of the human STAT3 genes in mouse ES cells (ES/STAT) were fully methylated and some were partially methylated, whereas human STAT3 genes in CHO cells (CHO/STAT) were unmethylated as compared to the native region in HT1080 cells (Figure 5c). These results indicated that methylation of the 5′ region of human STAT3 was important for attaining the epigenetic status necessary for active expression.
To assess whether the gene silencing was maintained or activated in different cellular conditions, human STAT3 genes within the context of repressed chromatin in CHO cells were transferred from CHO cells to mouse ES or NIH/3T3 cells using MMCT. The expression of human STAT3 in each mouse recipient cell line was detected by RT–PCR (Figure 5d) and the methylation status of the 5′ and 3′ regions was analyzed by AciI digestion (Figure 5c). RT–PCR indicated that human STAT3 transferred from CHO to ES cells [ES/STAT(CHO)] was activated in all of the independent cell lines, while human STAT3 transferred from CHO to NIH/3T3 cells [NIH/STAT(CHO)] was activated to varying degrees or remained repressed (Figure 5d). The DNA methylation pattern of human STAT3 transferred from CHO to ES cells was changed to match that of the de novo constructed STAT3 loci in ES cells, while STAT3 transferred from CHO to NIH/3T3 cells showed increased methylation in expression-active cell lines or remained unmethylated in expression-negative cell lines (Figure 5c). The correlation between gene expression and DNA methylation was indicated in the 5′ region of STAT3, where full methylation was required for gene expression.
These results demonstrate that the human STAT3 gene adopted different chromatin architectures in cell lines derived from different tissues or species, and once established, the chromatin architecture was maintained or changed according to the cellular conditions by mechanisms involving DNA methylation.
DISCUSSION
We created functionally open chromatin sites in HAC vectors that contained insulators (19), which protect gene expression from gene silencing, and a CAG promoter, which activates chromatin for gene expression (Figure 1). Recent insights suggested that insulators perform dual functions as enhancer blockers and as barriers against the spread of heterochromatin. Here, we showed that the human globin HS sequences could functionally divide the domains of HAC by providing a barrier structure or could influence the balance of activation and silencing functions by participating in transcription-specific chromatin architectures. The expression of EGFP from the HAC vector showed that these chromatin sites regulated gene expression according to the promoter rather than by simple ‘all or none’ activation (Figure 3). In the presence of the boundary elements, the inserted gene responded appropriately to the ambient cellular conditions and this may be related to chromatin structural and epigenetic (in particular, DNA methylation) status that is generated in the different cell types.
The de novo-constructed human STAT3 chromatin in mouse ES cell lines was actively expressed and the level of DNA methylation in the 5′ and 3′ regions of the gene was as high as that in native STAT3 genes. Therefore, genomic DNA inserted in an HAC vector can be packaged into chromatin with a natural DNA methylation pattern. Most human genes regulated by native promoters are thought to function in other mammalian cells, including murine cells. However, human STAT3 on chromosome vector was repressed in all CHO cells, while the endogenous CHO STAT3 was actively expressed (Figure 5b). Human STAT3 repression in CHO cells was probably a product of the chromatin architecture, including the DNA methylation pattern, rather than a lack of recognition of human sequences by the CHO transcription machinery, because methylation in the 5′ and 3′ regions of human STAT3 in CHO cells was lower than in human and mouse cells. In many genes, a low level of DNA methylation in the promoter and up-stream region is requisite for active expression, however, the highly methylated status that correlates with expression-active architecture might not have been appropriately positioned in the 5′ region of the human STAT3 gene in CHO cells.
In this study, the transcriptionally inactive chromatin architecture of the human STAT3 gene constructed in CHO cells was remodeled into transcriptionally active chromatin in ES cells by a mechanism involving DNA methylation. The methylation pattern underwent inter- and intra-cell line changes when human STAT3 architecture was transferred from CHO cells into mouse NIH/3T3 cells. In some cases, the architectural status was intermediate between ES and CHO types, while in others the status was unchanged from CHO type (Figure 5c). These results indicated that the cells have the potential to remodel or maintain chromatin structure through DNA methylation. Each epigenetic state was established by the balance of the two potentials. ES cells demonstrated a powerful force for remodeling as opposed to maintaining the human STAT3 chromatin status, corresponding with previous reports of their ability to reprogram somatic nuclei (20). The HAC vector technology described herein allows us to conduct experiments involving the establishment, modification and maintenance of chromatin epigenetics in various cell types. These types of technologies could be utilized in basic biological research as well as applied to stem cell science and regeneration medicine.
ACKNOWLEDGEMENTS
We thank S. Asakawa and N. Shimizu for the gift of the BAC clones containing human STAT3, D. Engel of Northwestern University for the YAC clone A201F4.3 and K. Yamamura for the plasmid pCAGGS-bsr and lox66-Nlaczeo.
FUNDING
Grant-in-Aid for Scientific Research from The New Energy and Industrial Technology Development Organization (NEDO); The Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. Funding for open access charge: The Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflict of interest statement. None declared.
REFERENCES
- 1.Larin Z, Mejia JE. Advances in human artificial chromosome technology. Trends Genet. 2002;18:313–319. doi: 10.1016/S0168-9525(02)02679-3. [DOI] [PubMed] [Google Scholar]
- 2.Grimes BR, Monaco ZL. Artificial and engineered chromosomes: developments and prospects for gene therapy. Chromosoma. 2005;114:230–241. doi: 10.1007/s00412-005-0017-5. [DOI] [PubMed] [Google Scholar]
- 3.Kuroiwa Y, Tomizuka K, Shinohara T, Kazuki Y, Yoshida H, Ohguma A, Yamamoto T, Tanaka S, Oshimura M, Ishida I. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat. Biotech. 2000;18:1086–1090. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- 4.Doherty AMO, Fisher EMC. Microcell-mediated chromosome transfer (MMCT): small cells with huge potential. Mamm. Genome. 2003;14:583–592. doi: 10.1007/s00335-003-4002-0. [DOI] [PubMed] [Google Scholar]
- 5.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromere and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997;4:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 6.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotech. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 7.Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomizuka K, Yoshida H, Uejima H, Kugoh H, Sato K, Ohguma A, Hayasaka M, Hanaoka K, Oshimura M, Ishida I. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat. Genet. 1997;16:133–143. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki N, Nishii K, Okazaki T, Ikeno M. Human artificial chromosomes constructed using the bottom-up strategy are stably maintained in mitosis and efficiently transmissible to progeny mice. J. Biol. Chem. 2006;281:26615–26623. doi: 10.1074/jbc.M603053200. [DOI] [PubMed] [Google Scholar]
- 10.Henning KA, Novotny EA, Compton ST, Guan XY, Liu PP, Ashlock MA. Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl Acad. Sci. USA. 1999;96:592–597. doi: 10.1073/pnas.96.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimes BR, Schindelhauer D, McGill NI, Ross A, Ebersole TA, Cooke HJ. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2001;2:910–914. doi: 10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mejia JE, Willmott A, Levy E, Earnshaw WC, Larin Z. Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeno M, Inagaki H, Nagata K, Morita M, Ichinose H, Okazaki T. Generation of human artificial chromosomes expressing naturally controlled guanosine triphosphate cyclohydrolase I gene. Genes Cells. 2002;7:1021–1032. doi: 10.1046/j.1365-2443.2002.00580.x. [DOI] [PubMed] [Google Scholar]
- 14.Kuroiwa Y, Kasinathan P, Choi YJ, Naeem R, Tomizuka K, Sullivan EJ, Knott JG, Duteau A, Goldsby RA, Osborne BA, et al. Cloned transchromosomic calves producing human immunoglobulin. Nat. Biotech. 2002;20:889–894. doi: 10.1038/nbt727. [DOI] [PubMed] [Google Scholar]
- 15.Araki K, Araki M, Yamamura K. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet. 1994;3:1245–1257. doi: 10.1093/hmg/3.8.1245. [DOI] [PubMed] [Google Scholar]
- 17.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raz R, Lee CK, Cannizzaro LA, d’Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl Acad. Sci. USA. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaszner M, Felsenfeld G. Insulator: exploiting transcriptional and epigenetic mechanism. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]