Abstract
RIG-I and MDA5 sense cytoplasmic viral RNA and set-off a signal transduction cascade, leading to antiviral innate immune response. The third RIG-I-like receptor, LGP2, differentially regulates RIG-I- and MDA5-dependent RNA sensing in an unknown manner. All three receptors possess a C-terminal regulatory domain (RD), which in the case of RIG-I senses the viral pattern 5′-triphosphate RNA and activates ATP-dependent signaling by RIG-I. Here we report the 2.6 Å crystal structure of LGP2 RD along with in vitro and in vivo functional analyses and a homology model of MDA5 RD. Although LGP2 RD is structurally related to RIG-I RD, we find it rather binds double-stranded RNA (dsRNA) and this binding is independent of 5′-triphosphates. We identify conserved and receptor-specific parts of the RNA binding site. Latter are required for specific dsRNA binding by LGP2 RD and could confer pattern selectivity between RIG-I-like receptors. Our data furthermore suggest that LGP2 RD modulates RIG-I-dependent signaling via competition for dsRNA, another pattern sensed by RIG-I, while a fully functional LGP2 is required to augment MDA5-dependent signaling.
INTRODUCTION
Upon pathogen infection the mammalian innate immune system represents the first line of defense. The response to pathogens by the innate immune system of mammals is initiated by the detection of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (1,2). Viral RNA is recognized in the endosome by membrane-bound Toll-like receptors (3), or in the cytoplasm by DExH box ATPases RIG-I (retinoic acid-inducible gene 1), MDA5 (melanoma differentiation-associated antigen 5) and LGP2 (laboratory of genetics and physiology 2) (4–6). RIG-I is a sensor for e.g. hepatitis C virus (7), Sendai virus (8), vesicular stomatitis virus (9), rabies virus (10), influenza virus and Japanese encephalitis virus (11), while MDA5 appears to detect picornaviruses and noroviruses (11,12). Overlapping roles of RIG-I and MDA5 were demonstrated for reovirus and dengue virus (13). LGP2 has not been shown yet to directly signal upon interaction with viral RNA but appears to have a regulatory role in virus sensing (6,14,15).
Upon recognition of cytosolic viral RNA, RIG-I and MDA5 bind to the adaptor protein MAVS (also known as CARDIF, IPS-1 or VISA), which is located in the outer mitochondrial membrane (9,16–18). The interaction of RIG-I or MDA5 with MAVS is mediated by two N-terminal caspase activation and recruitment domains (CARDs) of RIG-I and MDA5 (19,20) and initiates downstream signaling, resulting in an antiviral response by interferon α/β production and activation of interferon stimulated genes as well as NFκB target genes (21).
While the PAMPs for MDA5 are unclear, RIG-I senses 5′-triphosphates on viral RNAs (10,22,23), modifications that arise from RNA synthesis by many viruses, but are typically not found on normally capped, dephosphorylated or processed cellular RNA molecules. RIG-I also recognizes poly-U/UC rich regions in the HCV genome (24), 3′- and 5′-monophosphates of double-stranded RNA (dsRNA) (25) as well as RNA molecules generated by the activity of the antiviral 2′-5′ oligoadenylate-activated RNase L on host RNA molecules (26). The precise mechanism of PAMP sensing by RIG-I is still unclear. A C-terminal regulatory domain (RD) that is a 5′-triphosphate sensor recognizes 5′-triphosphate RNA (25,27). The DExH box ATPase domain of RIG-I is activated by dsRNA in vitro and may participate in dsRNA sensing (28,29). Current models suggest that PAMP binding results in RIG-I dimerization and together with ATP perhaps induces a conformation that leads to initiation of downstream events (27,30). The recognition of PAMPs and the initiation and time course of the antiviral response is highly regulated and involves RIG-I ubiquitinylation (31,32) as well as several positive and negative regulatory factors (33).
A poorly understood putative regulator of antiviral response is the third identified member of the RIG-I-like ATPase family, LGP2. LGP2 differs from RIG-I and MDA5 by the lack of the two N-terminal CARDs but shares the DExH box ATPase domain and a C-terminal RD. The exact function of LGP2 is controversial. It has been shown to inhibit RIG-I signaling and activity both in vivo and in vitro (6,14,15,34). However, it has been indicated that MDA5 signaling is stimulated in the presence of LGP2 (35). Additionally there is evidence that despite lacking the N-terminal CARDs LGP2 can interact with IPS-1, thereby competing with the downstream kinase IKKi/ɛ and inhibiting activation of transcription factor IRF-3 (interferon regulatory factor 3) (15).
Upon dsRNA binding LGP2 also forms dimers, suggesting an activation mechanism related to RIG-I (34). In support of this, the C-terminal RDs of all three RIG-I-like ATPases share sequence elements, in particular four invariant cysteine residues. The RD of RIG-I was found to be structurally related to GDP/GTP exchange factors of Rab-like small GTPases (36) and contains a zinc-binding site, formed by the four invariant cysteines, as a prominent feature. However many residues, shown to be crucial for 5′-triphosphate-dependent binding of RNA in RIG-I RD, vary amongst the other two representatives and may confer selectivity towards different RNA molecules (27).
To learn more about structure and function of the regulatory domains of RIG-I-like receptors, we determined the crystal structure of the RD of LGP2 as well as a solution structure of MDA5 RD, determined by small angle X-ray scattering. The crystal structure of human LGP2 RD revealed a zinc-binding site and the overall structure shares fold with RIG-I RD. However, we tested RNA binding selectivity of LGP2 RD and found a strong preference for dsRNA over single-stranded RNA (ssRNA), while 5′-triphosphates had no significant effect on RNA binding. This is a marked difference to RIG-I RD and argues that RDs are indeed pattern sensor sites. Using site-directed mutants, we derive commonalities and differences between RIG-I and LGP2 RDs. Our data suggest that RDs of RIG-I, LGP2 and possibly MDA5 share a common RNA binding site at the center of a concave surface, while a peripheral pocket and loop might confer specificity towards distinct patterns. Finally, in vivo analysis suggests that RD as well as DExH box domain can individually inhibit RIG-I-dependent signaling, but an intact LGP2 is necessary to augment MDA5-dependent signaling.
MATERIALS AND METHODS
Protein production and purification
RDs of human LGP2 (residues 537–678, native C-terminus) were expressed in Escherichia coli Rosetta DE3 cells using a pET21a expression vector (Novagen). Cells were grown in shaking culture in Luria Bertani broth to an OD600 of 0.8 at 37°C. Protein production was started by the addition of 0.13 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) and the cells were shaken over night at 18°C. After harvest by centrifugation, cells were resuspended in lysis buffer (30 mM Tris–HCl pH 7.0, 100 mM NaCl, 2 mM dithiothreitol (DTT) and 10 µM ZnCl2) and disrupted by sonification. Proteins were purified using HiTrapSP cation exchange chromatography and Superdex S75 gel filtration, according to standard protocols (GE Healthcare). The purified protein [in 30 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT, 10 µM ZnCl2 and 5% (v/v) glycerol] was concentrated to 5–15 mg/ml, flash frozen in liquid nitrogen and stored at −80°C for further use. Site directed mutations were introduced using mutagenic primers and the Phusion® Flash High-Fidelity PCR Master Mix (Finnzymes). The resulting polypeptides were purified using the same procedure as for the wild-type protein.
RDs of human MDA5 (residues 897–1025, native C-terminus) and RIG-I (residues 801–925, native C-terminus) were produced with an N-terminal 6 × histidine tag in E. coli as described for LGP2 RD using a pET28 expression vector (Novagen). MDA5 and RIG-I RDs were purified by immobilized metal affinity, ion exchange chromatography and, subsequent to removal of the histidine tag with thrombin, by gel filtration chromatography. For SAXS measurements protein samples were concentrated to 2, 5 and 10 mg/ml, respectively (in 30 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT and 10 µM ZnCl2).
Crystallization, crystallographic data collection and structure determination
LGP2 RD was crystallized using hanging drop vapor diffusion by mixing 2 µl of protein solution at 13 mg/ml protein concentration with 2 µl of the reservoir solution [1.5 M (NH4)2SO4, 100 mM Tris–HCl pH 8.5]. Crystals grew after several weeks at 21°C. Prior to flash freezing in liquid nitrogen, the crystals were soaked with 2 mM mercury acetate for 10 min and then transferred for 5–10 s into the reservoir solution containing 2.5 M (NH4)2SO4. We collected the final diffraction data to 2.6 Å at ESRF beamline ID 14-4 (European synchrotron radiation facility, Grenoble, France). Diffraction data were processed with XDS (37). The crystals belong to space group P212121 and contain four molecules in the asymmetric unit. The structure was determined by molecular replacement with Phaser (38), using human RIG-I RD [PDB entry 2QFB, (27)] as a search model. The resulting 2Fo-Fc and Fo-Fc electron density allowed initial manual model building with Coot (39). The model was iteratively refined by cycles of bulk solvent correction, overall anisotropic B factor refinement, positional refinement and overall B factor refinement with CNS 1.2 (40) and Phenix (41). Initial NCS restraints were gradually removed in the final cycles of the refinement, to allow some structural variations. Manual model building was performed with Coot and 166 solvent atoms were added with CNS 1.2. Finally secondary structure was assigned with DSSP (42). Data collection and model refinement statistics are summarized in Supplementary Table 1.
RNA preparation
For binding assays, 5′-triphosphate containing RNA was either obtained by in vitro transcription using the Ambion MEGAshortscriptTM Kit or purchased from Eurogentec. RNAs from in vitro transcription mixes were purified using MicroSpinTM G-50 columns (GE Healthcare) and checked for purity by denaturing urea PAGE. RNAs were fluorescence-labeled by either incorporating Alexa Fluor 488-5-UTP during in vitro transcription or purchased from IBA with Alexa Fluor 488 as 5′-modification. Unmodified oligonucleotides were purchased from Metabion or biomers.net. RNAs used are listed in Supplementary Table 2.
Fluorescence anisotropy measurements
Fluorescence anisotropy experiments were performed with a FluoroMax-P fluorimeter (HORIBA Jobin Yvon), equipped with a Glan-Thompson prism polarizer and connected to a Haake F3 thermostat. Typically, 1 ml of buffer (30 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT and 10 µM ZnCl2) and 37 nM RNA (5′-Alexa Fluor 488-labeled/5′-OH dsRNA, 25 bp) were pre-equilibrated in a quartz cuvette at 12°C. Protein samples were added in a stepwise manner. After mixing and 4 min of re-equilibration, anisotropy data were collected using an excitation wavelength of 495 nm and monitoring the emission at 516 nm. The band pass was 5 nm for excitation and 5 nm for emission. A maximum number of 10 trials were performed until minimal deviation of the signal was reached. For the competition assays 800 µl of buffer, 40 nM of an in vitro transcribed hairpin RNA (Alexa Fluor 488-5-UTP incorporated, 18bp + 4n) and 470 nM protein were pre-equilibrated in a quartz cuvette at 12°C. Unlabeled RNA species (Supplementary Table 2) were added in a stepwise manner and the drop in anisotropy was monitored after mixing and 4 min of re-equilibration.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSA) were carried out in 10% (w/v) acrylamide native gels in Tris borate buffer (10.8 g/l Tris, 5.5 g/l borate). Samples contained 5% glycerol, 60 nM 5′ Alexa Fluor 488-labeled/5′-OH dsRNA (25 bp) and different concentrations of protein, diluted from 5 µM stock solutions (in 30 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT and 10 µM ZnCl2). Samples were incubated for 20 min at 4°C prior to electrophoretic analysis. Gels were analyzed with a TyphoonTM scanner (GE Healthcare).
Interferon-β reporter assay
HEK293 cells were cultivated in Dulbecco's; Modified Eagle Medium (DMEM) (PAA Laboratories) containing 10% fetal calf serum (FCS v/v, Biochrom KG), 1,5 mM l-glutamine (PAA Laboratories) and 8 µg/ml ciprofloxacin (Bayer Vital GmbH). To measure interferon-β induction, HEK293 cells were transiently transfected with 40 ng of a plasmid containing a firefly luciferase reporter gene under the control of an interferon-β promotor, 40 ng Renilla reniformis luciferase reporter gene for normalization and the indicated amount of expression plasmids using Genejuice transfection reagent (Novagen). After 24 h, cells were stimulated with the indicated RNA oligonucleotide for 18 h, as described above. In all experiments, cell lysates were prepared using passive lysis buffer (Promega) and the reporter gene activity was measured using firefly luciferase substrate (Biozym) and renilla luciferase substrate (Promega), respectively. Expression levels of the examined proteins were verified via western blot analysis (Supplementary Figure 2).
Small angle X-ray scattering measurements
SAXS data for MDA5 and RIG-I RDs were collected at DESY beamline X33 (Deutsches Elektronen-Synchrotron, Hamburg, Germany). Scattering patterns from solutions of the two constructs at concentrations of 2, 5 and 10 mg/ml were measured in buffer containing 30 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 µM ZnCl2 and 5 mM DTT. SAXS data were processed, evaluated and models built applying various programs from the ATSAS 2.1 software suite (43).
Coordinates and structure factors of LGP2 RD were deposited at the Protein Data Bank (PDB) with accession number 2W4R.
RESULTS AND DISCUSSION
Crystallization and structure determination
The expression construct for the RD of human LGP2 was designed using our previously determined crystal structure of the RD of RIG-I as guide for the domain boundary. LGP2 RD comprises residues 537–678 (native C-terminus). The protein was produced in E. coli. After purifying the protein by ion exchange and gel filtration chromatography, we were able to crystallize the protein in space group P212121 with four molecules per asymmetric unit. The crystals diffracted to 2.6 Å and we determined the structure by molecular replacement using the coordinates for RIG-I RD as search model. The four copies were initially refined with non-crystallographic symmetry restraints that were removed during final cycles of refinement. RMSD values for the Cα atoms of the four molecules in the asymmetric unit have been calculated by LSQMAN (44) with respect to molecule C and range from 0.6 to 0.7 Å with 117–122 of pairwise matched residues. A superposition of all four molecules is shown in Supplementary Figure 1A. Part of the final 2FOc-Fc electron density map is shown in Figure 1B. The final model spans residues 543 to 672, with the loop region between residues 593 and 601 not visible in the electron density map. The structure was refined to an R value of 21.5 (free R=26.6) with good stereochemistry (Supplementary Table 1).
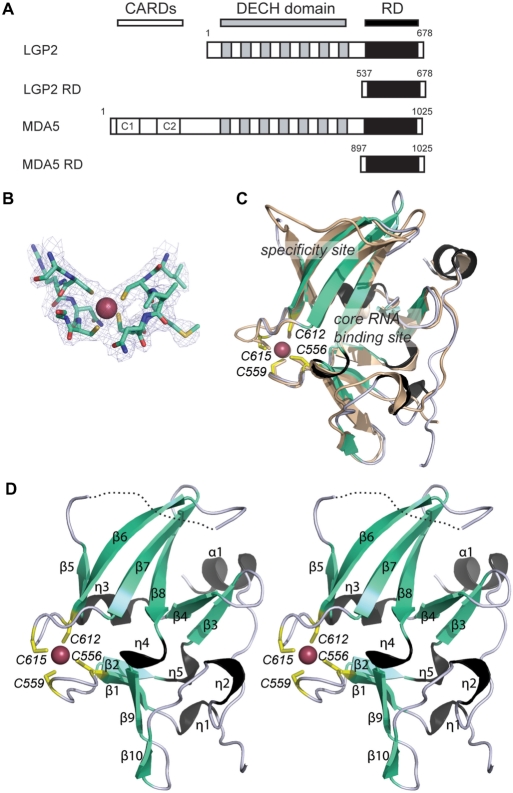
Figure 1.
Structure of the RD of human LGP2. (A) Scheme of the domain arrangement in LGP2 and MDA5. The two N-terminal CARD motives of MDA5 are depicted in white, the DExH box helicase domains are shown in gray and the RDs in black. Numbering of the RDs is according to the constructs used for structural and biochemical studies. (B) Portion of the final 2Fo−Fc electron density (1 s contour) at the mercury-binding site superimposed with the final model. (C) Superimposition of ribbon models of LGP2 RD (cyan, black and yellow) and RIG-I RD (wheat) with highlighted secondary structure and metal-binding site. Possible RNA binding and specificity sites are assigned. (D) Stereo image of a ribbon model of LGP2 RD with highlighted and annotated secondary structure, mercury ion (magenta sphere) and annotated mercury-coordinating cysteines (yellow sticks).
Overall structure
The ‘regulatory domain’ RD of LGP2 is a globular, slightly flattened domain with a concave and convex side and dimensions of ∼45 × 35 × 30 Å (Figure 1D). It is organized in three leaves, consisting of two four-stranded (β1, β2, β9, β10 and β5, β6, β7, β8) and one two-stranded (β3, β4) antiparallel β-sheet. Small 310 helical turns (η1–η5) connect the three β-sheets. The C-terminus contains a short α-helix (α1). The four-stranded β-sheets are laterally connected by two protruding loops, each containing two highly conserved cysteine residues (Cys556 and Cys559 as well as Cys612 and Cys615). Together, the four thiol groups of these cysteines coordinate the zinc ion (or mercury in our crystal structure) in each of the four molecules in the asymmetric unit (Figure 1D and B).
Comparison of LGP2 RD to RIG-I RD
With the exception of some differences in loop regions that connect secondary structure, the fold of LGP2 RD is highly related to the fold of RIG-I RD (Figure 1C). The RMSD, as calculated by LSQMAN (44), between the Cα atoms of LGP2 (molecule C) and RIG-I RD is 1.178 Å for 108 matched residues (Supplementary Figure 1B). In particular, the backbone geometry of the metal coordinating cluster is, with the exception of one loop insertion in RIG-I, conserved between RIG-I and LGP2 (Figures 1 and 2A). The metal in LGP2 is coordinated in a tetrahedral manner by the sulfur atoms of the cysteine cluster C556, C559, C612 and C615. Although we have a mercury ion in our crystal form we think the geometry is similar in the presence of zinc, since crystal structures of RIG-I RD in the presence of mercury and zinc turned out to be virtually identical (27). W619LGP2—conserved between LGP2, RIG-I and MDA5—bounds this cluster towards the core of the domain. Thus, correct formation of the metal coordination sphere is presumably necessary for fold of RDs, which might explain the severe effect of point mutation in the zinc-binding cluster on activity of RIG-I (27). In support of this, point mutation in metal coordinating cysteine residues resulted in an unstable LGP2 RD, which could not be over-expressed in E. coli in our hands. A notable difference in the metal binding site between RIG-I and LGP2 RD is the loop, which connects C612LGP2 and C615LGP2 (Figure 1C). In RIG-I this loop is two residues longer, forming a short β-turn. In MDA5, this loop is one amino acid shorter indicating that this region could account for functional differences between RIG-I-like helicases (Figure 2A).
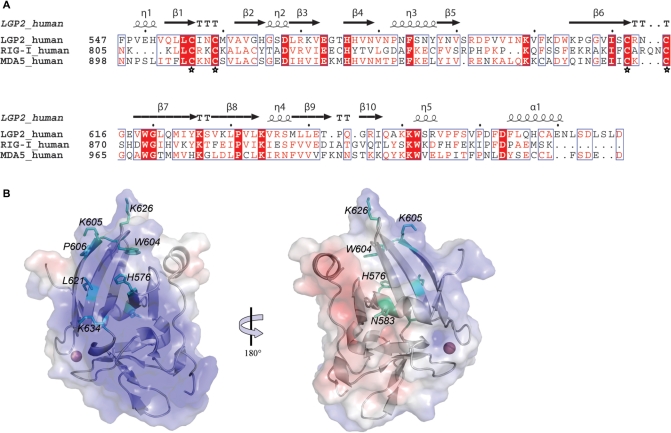
Figure 2.
Structure guided site directed mutagenesis of LGP2 RD. (A) Structure-based sequence alignment of the C-terminal regions of human LGP2, RIG-I and MDA5. Secondary structure elements corresponding to LGP2 RD are shown above the alignment. Invariant residues are highlighted with red background, conserved ones in red font. The zinc-coordinating cysteines are marked with asterisks. (B) Localization of mutations, shown in a cartoon representation with electrostatic surface potential (ranging from blue=5 kT/e to red=−5 kT/e). The model is shown in ‘standard view’, used in all other figures (left) and 180° rotated around the vertical axis. Mutated residues are highlighted with sticks (cyan).
The RD of RIG-I was shown to specifically bind to RNA with 5′-triphosphates and is suggested to be a main sensor site for 5′-triphosphate containing viral RNA. Key residues important for RIG-I RNA binding were mapped to a groove formed at the interface of the four- and two-stranded β-sheets. These include several lysines as well as a histidine, residues which could bind the RNA backbone and the 5′-triphosphate moiety. Intriguingly, two of these residues, K888RIG-I and H830RIG-I are conserved in LGP2 (K626LGP2 and H579LGP2, respectively), indicating that this region could also be an important functional site of LGP2 (Figure 2A and B). In LGP2 RD, the groove carries a strong positive electrostatic potential that might be suitable for RNA backbone interaction (Figure 2B). Furthermore, the groove is flanked by two conserved 310 turns (η2 and η4), which are suitable phosphate recognition sites (Figure 1D).
However, there are also important structural differences among RIG-I and LGP2 RD. K858RIG-I has been shown to be essential for 5′-triphosphate RNA interactions and this residue is not conserved in LGP2. In fact, LGP2 possesses a proline in this position (P606, Figure 2A and B). Based on these findings and subsequent mutational analysis (see subsequently), we propose that the region around P606 and the adjacent loop between β5 and β6 (Figure 1C) confer specificity in RNA binding. In sum, structural comparison reveals similarities but also differences between LGP2 and RIG-I in regions that were found to be important for RIG-I function.
LGP2 RD binds to dsRNA in a 5′-triphosphate-independent manner
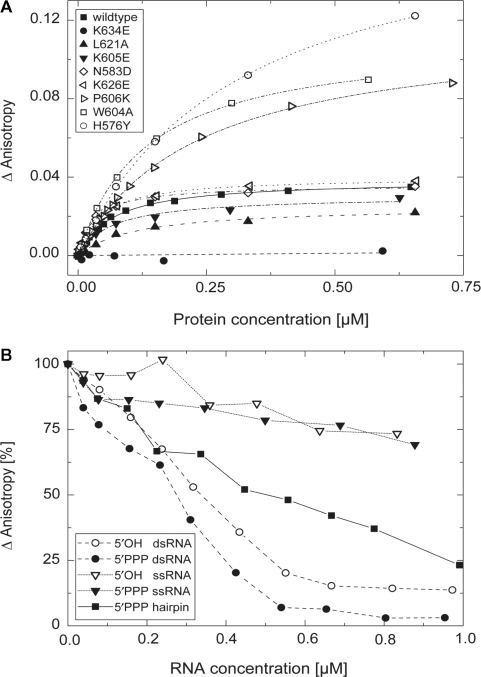
To learn more about the functional sites of LGP2 RD, we first analyzed whether it can directly bind RNA and what RNA structures or epitopes might contribute to binding. The physiological ligand for LGP2 is unknown. Its negative regulation of RIG-I-dependent signaling in vivo may result from competition for viral RNA and thus could be directed against two RIG-I PAMPs, 5′-triphosphate RNA and dsRNA. Since full-length LGP2 has been shown to bind to dsRNA with a higher preference than ssRNA, we hypothesized that RD of LGP2 might be one of its dsRNA recognizing elements. We tested several RNA oligonucleotides (Supplementary Table 2) in equilibrium binding experiments using fluorescence polarization anisotropy measurements. Indeed, we find that LGP2 RD binds to a dsRNA 25-mer with quite high affinity. We can fit the binding isotherm with a single site-binding model and using this model, obtain an apparent Kd of 68 ± 6 nM (Figure 3A).
Figure 3.
Binding of dsRNA to LGP2 RD. (A) Fluorescence anisotropy changes of a 5′-Alexa Fluor 488-labeled 25 bp RNA duplex (37 nM) in response to titration with wild-type LGP2 RD (filled square, Kd=68 ± 6 nM) and various mutants, respectively. Control mutation N583→D (open diamond, Kd=38 ± 4 nM) located on the convex site of RD and mutation K626→E (open left-facing triangle, Kd=51 ± 5 nM) do not show significantly altered dsRNA binding affinity. Affinities for mutants L621→A (filled up-facing triangle, Kd=165 ± 26 nM) and K605→E (filled down-facing triangle, Kd=140 ± 16 nM) are slightly decreased, while a mutation of K634→E (filled circle) completely suppresses binding. A decrease of binding affinity, but increase in maximum reached anisotropy signal is seen for P606→K (open right-facing triangle, Kd=230 ± 10 nM), W604→A (open square, Kd=136 ± 6 nM) and H576→Y (open circle, Kd=304 ± 10 nM). (B) Competition of binding of an Alexa Fluor 488-5-U-labeled hairpin RNA (in vitro transcriped, 40 nM) to LGP2 RD (470 nM) by stepwise addition of different non-fluorescent RNA species (synthetic 5′OH/5′OH dsRNA: 19 bp; 5′PPP/5′OH dsRNA: 19 bp; 5′OH ssRNA: 19n; 5′PPP ssRNA: 19n; and in vitro transcribed 5′PPP hairpin: 18 bp ± 4n) followed by fluorescence anisotropy. Data points were connected for better outline.
We then analyzed binding to other RNA structures using competition experiments (Figure 3B). In these experiments, a fluorescently labeled dsRNA hairpin is competed away from LGP2 RD with increasing amounts of unlabeled ssRNA or dsRNA ligands bearing or lacking 5′-triphosphates. Corresponding in length and sequence to one of the dsRNA strands of the labeled probe, 5′-OH ssRNA is a poor competitor, indicating that LGP2 RD does not efficiently bind to ssRNA. However, 5′-OH dsRNA with blunt ends is a very efficient competitor. Since dsRNA with two blunt ends binds more efficiently than a 5′-triphosphate hairpin of the same length, some RNA end structures could enhance specific dsRNA binding by LGP2 RD.
We next tested whether 5′-triphosphates are important for RNA binding by LGP2 RD (Figure 3B). We tested both ssRNA and dsRNA carrying a 5′-triphosphate. We did not see a substantial difference in binding of LGP2 RD to the corresponding RNAs with 5′-triphosphate compared to those without. This is in stark contrast to the 5′-triphosphate dependence of RNA binding by RIG-I RD. Taken together, it appears that RD is an important element in specific dsRNA recognition by LGP2 and that 5′-triphosphates are not central epitopes recognized by LGP2 RD.
The dsRNA binding site of LGP2
To identify the RNA binding site of LGP2 RD, we analyzed the electrostatic surface potential and introduced point mutations inside and outside of the positive groove that could form an RNA binding site (Figure 2B). RNA binding of the mutant RD variants was analyzed by fluorescence anisotropy (Figure 3A). Mutations in the convex, positively charged surface patch in general affect RNA binding, while mutations outside these regions did not result in decreased RNA binding. Most remarkably, K634→E completely abolished RNA binding. This residue is conserved between RIG-I-like helicases and situated at the N-terminus of a 310 turn (η4) at the center of the concave surface. The equivalent mutation K888RIG-I→E also had a severe effect on RIG-I activity (27). The conservation of this lysine residue in different RDs with evidently different RNA specificities (5′-triphosphate in RIG-I versus dsRNA in LGP2) argues that this site is not involved in distinguishing different RNA epitopes, but more likely is a central core RNA interaction site shared by all three RDs.
To our surprise, P606LGP2→K still bound RNA with fairly good efficiencies, while the equivalent K858RIG-I→A severely reduced RNA binding of RIG-I RD and abolished RIG-I activity in vivo. Other mutations in the same region (H576→Y, W604→A) also bind RNA with reasonable albeit reduced affinity (Figure 3A). Interestingly, although Kd values for dsRNA binding by these mutants are lower than for wild-type RD, we obtain much higher anisotropy changes compared to the wild-type RD, indicating that the RNA:protein complexes formed in the presence of these mutants are larger than those formed in the presence of wild-type RD. One likely explanation for this observation is that mutant proteins can bind to RNA in multimers or can bind simultaneously at multiple sites, while wild type binds more specifically to only one or a few sites.
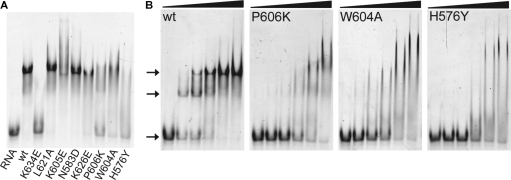
To test this idea further, we performed EMSAs with wild-type and mutant LGP2 RDs on dsRNA (Figure 4A and B). In general, the EMSAs confirm the results of the anisotropy measurements with respect to altered or retained dsRNA binding of site directed mutants (Figure 4A). Concentration-dependent analysis indicates that wild-type RD indeed shifts dsRNA to defined, specific bands. An initial shifted species with higher mobility is subsequently converted into a species with lower mobility (Figure 4B). These data suggest that LGP2 RD has two binding sites on the dsRNA ligand. One possibility is that RD specifically forms dimers on a single site on the dsRNA substrate. Since RD binds to dsRNA better than a hairpin of the same concentration, another explanation is that perhaps the RNA end structures contribute to binding and the two EMSA species are corresponding complexes with one and two RNA ends bound by protein. Dynamic light scattering, SAXS and analytical gel filtration suggest that RD is a monomer in the absence of RNA, and we do not see a convincing protein–protein dimer interface in the crystal lattice. In the presence of RNA our anisotropy studies, EMSAs as well as analytical gel filtration experiments can neither entirely eliminate nor verify the possibility of dimer formation. This is due to the resemblance in size between the RNA species and RD. Furthermore, judging from the dimension of the RD, more than one molecule could possibly bind to the 18–25 bp RNA duplexes used (Supplementary Table 2), independently of protein–protein interaction. Thus, analysis of an RNA-induced protein dimer formation must await further studies. In any case, we observe very defined species in the EMSAs, indicating a very specific interaction of LGP2 RD with the dsRNA substrate.
Figure 4.
Electrophoretic mobility shift assays of LGP2 RD and mutants with dsRNA. (A) Retardation of 5′-Alexa Fluor 488-labeled dsRNA (25 bp, 60 nM) in a 10% native polyacrylamide gel after incubation with 400 nM wild-type (wt) LGP2 RD or indicated RD mutants, respectively. (B) Retardation of 5′-Alexa Fluor 488-labeled dsRNA (25 bp, 60 nM) in a 10% native polyacrylamide gel after incubation with increasing concentrations (0, 0.4, 0.8, 1.6, 3.2 and 6.4 µM) of wt LGP2 RD and mutants P606→K, W604→A and H576→Y. For the wild-type RD two distinct bands appear shifted compared to free dsRNA at low protein concentrations already. The mutants show unspecific shifting and lower affinity to the RNA, indicated by remaining free dsRNA bands for all protein concentrations.
These defined bands are more or less lost when we analyze H576, W604 and P606 mutants of RD (Figure 4B). Instead, we observe a more unspecific distribution (‘smear’) of even lower mobility complexes. Taking into consideration that all tested mutants behave like the wild-type RD during purification and no tendency for enhanced aggregation was observed, our interpretation of these data is that these mutations do not disrupt RNA binding per se, but lower the affinity and lead to a more distributed binding. These mutant RDs could bind undiscriminatorily to many possible binding sites on the dsRNA ligand, possibly also as multimers at higher protein concentrations, which could account for the larger complexes observed in both EMSA and fluorescence anisotropy measurements. Taken together, these data argue that the residues around P606 form a specificity site on RD.
Solution scattering analysis and modeling of MDA5 RD
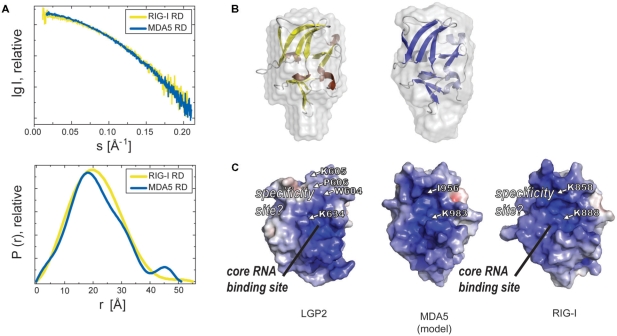
We have not been able to crystallize the RD of MDA5, so we used small angle X-ray scattering as well as homology modeling to generate a first structural draft of MDA5 RD (Figure 5). Our construct of MDA5 RD spans residues 897–1025, similar to the used LGP2 and RIG-I RDs. Small angle X-ray scattering data were collected at DESY beamline X33 and data processed with programs from the ATSAS 2.1 suite (43) (Figure 5A). We noticed some limited aggregation in RIG-I and MDA5 RD samples at higher concentrations, so very low-resolution data needed to be truncated. Using GASBORp, we generated ab initio reconstructions using the dummy residue approach (Figure 5B). In the case of RIG-I RD, the solution scattering data correlate well with size and shape of the respective crystal structures. Likewise, an ab initio dummy residue model matches the shape and size of the crystals structure to a high degree.
Figure 5.
Comparison of RDs of RIG-I-like helicases. (A) Small angle X-ray scattering curves of RIG-I RD and MDA5 RD (upper panel, merged from curves obtained with protein concentrations of 2, 5 and 10 mg/ml). The lower panel shows the pair distribution functions for RIG-I RD and MDA5 RD derived from raw data with Gnome (ATSAS 2.1). (B) Superimposition of SAXS surface models of RIG-I (left) and MDA5 RD (right) with crystal structures of RIG-I RD (yellow, brown) and a model of MDA5 RD (model based on RIG-I and LGP2 RD crystal structure, blue, light blue), respectively. (C) Electrostatic surface potential (ranging from blue=9 kT/e to red=−9 kT/e) of LGP2 RD, RIG-I RD (from crystal structure) and a model of MDA5 RD. Residues crucial for general RNA interaction and either 5′-triphosphate RNA specificity in RIG-I (K888, buried K858) or dsRNA specificity in LGP2 (corresponding K634, P606 and additionally buried W604 and K605) are highlighted as well as corresponding residues for MDA5 RD (K983, buried I956).
MDA5 RD produces highly similar scattering data to RIG-I RD. The resulting ab initio models exhibit a similar shape and size compared to RIG-I RD and overall match the geometry of the available RD crystal structures (Figure 5B). Together with the sequence conservation of key residues such as core tryptophanes and metal coordinating cysteines (Figure 2), MDA5 RD likely possesses a similar overall structure as RIG-I and LGP2 RDs. We generated a homology model of human MDA5 RD with MODELLER (45) using coordinates of RIG-I and LGP2 RD as base and compared the molecular model to the solution structure of MDA5 RD. Although the model will presumably differ in the precise side chain and loop geometries from an experimental structure of MDA5 RD, several important conclusions can be drawn at this stage, including analysis of the surface potential. Indeed, the concave surface, identified in RIG-I and LGP2 RDs as important RNA recognition site, also carries a highly positive electrostatic potential in MDA5 RD (Figure 5C). In addition, a lysine residue, K984MDA5, is situated at the same position as K888RIG-I and K634LGP2, both of which are key elements of RNA binding by RIG-I and LGP2 RDs, respectively. In contrast, MDA5 possesses an isoleucine in the place of P606LGP2 or K858RIG-I, and the proposed ‘specificity site’ differs in all three RDs. Taken together, MDA5 RD could share with RIG-I and LGP2 RD a common RNA binding site at η4, although the nature of the PAMP sensed by MDA5 and perhaps MDA5 RD remains to be identified.
Effect of LGP2 DExH domain and RD in modulating RIG-I- and MDA5-dependent signaling
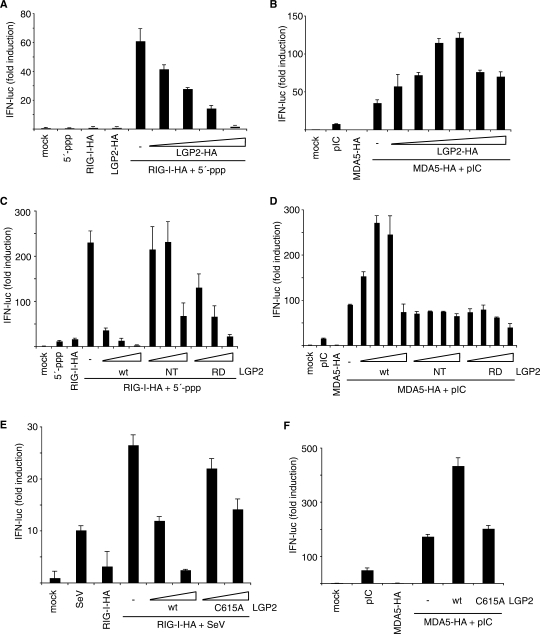
To elucidate the mechanisms by which LGP2 differentially regulates RIG-I and MDA-5 and the role of its RD in these processes, we expressed increasing amounts of LGP2 together with RIG-I or MDA5 in Hek293 cells and monitored activation of an interferon-β reporter system after stimulation with the appropriate ligand (Figure 6). As expected, when we co-expressed LGP2 together with RIG-I, loss of interferon-promoter activation was observed proportional to the level of LGP2 over-expression, consistent with previously reported data (14,35). This suggests a competitive mechanism at the level of the ligand or at the level of downstream signaling molecules as it has already been proposed (15). Interestingly, when we co-expressed MDA5 with different amounts of LGP2 protein in the presence of the dsRNA analog poly(I:C), we found that optimal activation was observed at LGP2 expression levels similar to those of MDA5. This balance-type behavior suggests that the mechanism of MDA5 activation by LGP2 is mechanistically different and might involve the formation of a defined hetero-complex that contains the ligand, which is supported by our finding that the LGP2 RD similar to MDA5 binds dsRNA.
Figure 6.
LGP2 represses RIG-I and activates MDA5 by different mechanisms. (A) Hek293 cells were transfected with plasmids coding for RIG-I-HA (10 ng plasmid), different amounts of LGP2-HA (2, 5, 10 and 50 ng plasmid) and plasmids encoding for a luciferase-based interferon-β reporter assay system. After 24 h, transfected cells were stimulated using 5′-triphosphate RNA or left unstimulated as a control. Relative luciferase activities were determined 18 h after stimulation. The values indicated are normalized to an unstimulated control that expresses only the assay system components (mock). (B) Hek293 cells were transfected with plasmids coding for MDA5-HA (1 ng plasmid) and different amounts of LGP2-HA (0.05, 0.1, 1, 5, 50 and 100 ng plasmid) and stimulated with poly(I:C) 24 h after transfection. Luciferase activities were measured und are represented as in (A). (C) Hek293 cells were transfected with RIG-I-HA and different amounts of wild-type (wt), amino-terminal (NT, amino acids 1–468) or carboxy-terminal (RD, amino acids 476–678) constructs of LGP2-HA and assayed for interferon-β reporter activity. (D) Hek293 cells were transfected with MDA5-HA and different amounts of wild-type (wt), amino-terminal (NT, amino acids 1–468) or carboxy-terminal (RD, amino acids 476–678) constructs of LGP2-HA and assayed for interferon-β reporter activity. (E) Hek293 cells were assayed for suppression of RIG-I signaling by wild-type LGP2 or a C615A mutant of LGP2 (10 ng and 100 ng) as in (A) using Sendai virus (SeV) infection as a stimulus. (F) Hek293 cells were assayed for activation of MDA5 signaling by wild-type LGP2 or a C615A mutant of LGP2 as in (B).
To clarify the relative contributions of the two LGP2 functional domains, we examined RIG-I repression and MDA5 activation in the presence of different amounts of either full-length LGP2 or one of the two functional domains, DExH box domain (NT) or RD. Expression levels for all constructs were shown to be regular when analyzed by western blots (Supplementary Figure 2A and B). Similar to what has been observed before, we find that the LGP2 RD domain is able to mediate RIG-I repression albeit weaker than the full-length (46). Consistently, we do observe a contribution of the DExH box to LGP2-mediated repression of RIG-I, which would be in line with a model in which the helicase domain is also involved in competitive binding to either the RIG-I ligand or downstream signaling molecules. Instead, analysis of LGP2-mediated MDA5 activation showed that full-length LGP2 activated MDA5 as expected and no activation could be observed expressing different amounts of either LGP2 DExH or regulatory domain. This indicates that neither the LGP2 DExH domain nor the LGP2 RD alone are sufficient to mediate the synergistic effect on MDA5 activation and that this might involve more complex protein–protein and protein–ligand interactions.
To examine the importance of the zinc-binding motif for LGP2's function in repressing RIG-I signaling or synergizing with MDA5, we co-expressed wild-type LGP2 or a mutant lacking one of the conserved cysteines (C615A) in the motif together with RIG-I or MDA5. The respective expression levels in human cells were analyzed in western blots (Supplementary Figure 2C), showing similar protein levels for the C615A mutant compared to the wild-type proteins. When we analyzed LGP2-mediated repression of RIG-I signaling in Sendai virus infected cells, a partial loss-of-function phenotype for LGP2 C615A was observed, which fits well with our finding that both the RD and the DExH domain contribute to RIG-I repression. Interestingly, when we analyzed MDA5 activation by LGP2 the loss-of-function phenotype was nearly complete, indicating that the integrity of the LGP2 RD is essential for full MDA5 function.
CONCLUSIONS
We established that although the RD of LGP2 has a structure related to the RD of RIG-I, it exhibits remarkably different RNA binding specificities. Thus, our results suggest that RDs of RIG-I-like helicases confer pattern specificity. LGP2 RD specifically binds dsRNA in a 5′-triphosphate-independent manner, perhaps recognizing also some dsRNA end structures. The binding affinity of LGP2 RD to RNA duplexes is similar or even higher than the affinity of RIG-I RD for 5′-triphosphate RNA, indicating a physiologically relevant interaction. We have not found an increased interaction in the presence of 5′-monophosphates (data not shown), and conclude that LGP2 specifically senses duplex RNA. We see high binding affinities for both 25 bp and 19 bp duplexes (Figure 3A and B), which are sequence unrelated. It is therefore unlikely that the RNA sequence is critical for binding by LGP2 RD. However, we do not want to rule out that perhaps some aspects of RNA sequence, structure or length significantly influences binding affinity and it will be necessary to address these points in future studies.
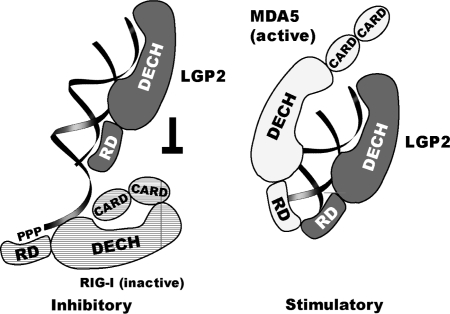
The function and mechanism of LGP2 in antiviral innate immune response is puzzling because recent analysis of LGP2 deficient mice has uncovered a surprising dichotomy of LGP2 function with respect to regulation of RIG-I and MDA5 activity. Whereas LGP2-deficient mice showed reduced susceptibility to viruses recognized by RIG-I, these mice show enhanced susceptibility to challenge with MDA5-specific viruses like EMCV (35). Complex formation between LGP2 and these receptors might account for some of these activities. For instance, a complex of a functional LGP2 with MDA5 could specifically detect longer dsRNA stretches, explaining why a fully functional and intact LGP2 is necessary to augment polyI:C-dependent MDA5 signaling, whereas isolated DExH domain and RD have no effect on MDA5-dependent signaling. In contrast, inhibition of RIG-I-dependent signaling does not require an intact LGP2, but is also observed for the individual DExH domain and RD. Part of this inhibition could involve protein–protein interactions (15). However, we have not been able to detect any protein–protein interactions of LGP2 RD with RIG-I so far, so a plausible explanation for the inhibitory effect of RD is that due to its strong dsRNA binding it can compete with RIG-I for dsRNA. RIG-I can sense different viral RNA patterns, including RNA duplex regions as well as 5′-triphosphate moieties on RNA. Our results of the 5′-triphosphate-independent interaction of LGP2 RD with RNA make it unlikely that LGP2 competes with RIG-I for 5′-triphosphate RNA. Rather, our results suggest that LGP2 RD could compete with RIG-I for RNA duplex regions, another efficient activator of RIG-I, which presumably binds at the dsRNA-activated DExH box domain. Since duplex regions are abundant in many cellular RNA species, LGP2 might help to prevent false positive signaling of non-viral dsRNA by RIG-I, but could help to detect dsRNA by MDA5. A model of the competitive and augmentative role of LGP2 is shown in Figure 7.
Figure 7.
Model for regulation of RIG-I and MDA5 by LGP2. Model for LGP2 interaction with dsRNA and proposed regulation of RIG-I- and MDA5-dependent signaling. Stoichiometry and interaction sites are simplified and aspects of the model are speculative. dsRNA binding by both RD and DExH box domains could compete with dsRNA-dependent activation of the RIG-I DExH box domain. A fully functional LGP2 might together with MDA5 sense dsRNA and augment MDA5-dependent signaling.
Our data suggest that RDs of all three RIG-I-like helicases are RNA binding elements with a common core RNA binding site but specific adaptation to their specific patterns. In general, the binding site corresponds well with NMR titration experiments on RIG-I RD (25). We propose that the specificity loop is involved in the differential sensing of RNA ligands. The pattern for MDA5 and its RD needs to be identified, but positive augmentation by LGP2 with its dsRNA binding activity suggests that RNA duplexes play a role in MDA5-dependent signaling, consistent with recent in vivo data on MDA5-dependent sensing of long RNA duplexes (47). In this regard, our results establish a framework for the detailed analysis of RNA selectivity by regulatory domains of RIG-I-like helicases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Program Project 455 to K.-P.H.); German Excellence Initiative (Center for Integrated Protein Science Munich and Munich Center for Advanced Photonics to K.-P.H.); grants from the Deutsche Forschungsgemeinschaft (RO 2525/3-1 to S.R, GK 1202 to J.C.H., S.R. and A.K.). Funding for open access charges: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sophia Hartung for advice on SAXS data interpretation, Gregor Witte for advice on anisotropy measurements, Wolfgang Hamm for support with RNA preparation and Johannes Soeding for help with molecular modeling. We also thank the staff of the European Synchrotron Radiation Facility (beamline 14-4) for help with diffraction data collection and staff of Deutsches Elektronen-Synchrotron (beamline X33) for help with scattering data collection and the beamline agencies for generous collection time. This work is part of the thesis of D.A.P., A.K. and J.C.H. at the University of Munich.
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 5.Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 7.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Chen Z, Kato N, Gale M, Jr., Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 9.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 10.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 11.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 12.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 15.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 17.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Vitour D, Meurs EF. Regulation of interferon production by RIG-I and LGP2: a lesson in self-control. Sci. STKE. 2007;2007:pe20. doi: 10.1126/stke.3842007pe20. [DOI] [PubMed] [Google Scholar]
- 20.Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct. Biol. 2008;8:11. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CL, Gale M., Jr. CARD games between virus and host get a new player. Trends Immunol. 2006;27:1–4. doi: 10.1016/j.it.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 23.Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gee P, Chua PK, Gevorkyan J, Klumpp K, Najera I, Swinney DC, Deval J. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 2008;283:9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 32.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl Acad. Sci. USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, Kao CC. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J. Biol. Chem. 2008;283:15825–15833. doi: 10.1074/jbc.M800542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, Schreiber SL. Structure of guanine-nucleotide-exchange factor human Mss4 and identification of its Rab-interacting surface. Nature. 1995;376:788–791. doi: 10.1038/376788a0. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve PD, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 43.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Cryst. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleywegt GJ, Jones TA. A super position. CCP4/ESF-EACBM Newsletter on Protein Crystallogr. 1994;31:9–14. [Google Scholar]
- 45.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl Acad. Sci. USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl Acad. Sci. USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.