Abstract
Plastid genomes of peridinin-containing dinoflagellates are unique in that its genes are found on multiple circular DNA molecules known as ‘minicircles’ of ∼2–3 kb in size, carrying from one to three genes. The non-coding regions (NCRs) of these minicircles share a conserved core region (250–500 bp) that are AT-rich and have several inverted or direct repeats. Southern blot analysis using an NCR probe, after resolving a dinoflagellate whole DNA extract in pulsed-field gel electrophoresis (PFGE), revealed additional positive bands (APBs) of 6–8 kb in size. APBs preferentially diminished from cells treated with the DNA-replication inhibitor aphidicolin, when compared with 2–3 kb minicircles, implicating they are not large minicircles. The APBs are also exonuclease III-sensitive, implicating the presence of linear DNA. These properties and the migration pattern of the APBs in a 2D-gel electrophoresis were in agreement with a rolling circle type of replication, rather than the bubble-forming type. Atomic force microscopy of 6–8 kb DNA separated by PFGE revealed DNA intermediates with rolling circle shapes. Accumulating data thus supports the involvement of rolling circle intermediates in the replication of the minicircles.
INTRODUCTION
Plastid genomes of most plants and algae usually carry about 120 genes on 120–200 kb linear DNA molecules (1,2). Satellite DNA from the dinoflagellate Heterocapsa triquetra was found to contain some 2–3 kb DNA as detected by plastid gene probes (3). Structural analysis by inward and outward PCR revealed that they were circular DNA molecules carrying single genes of plastid origin, named as minicircles. These 2–3 kb DNA molecules have since been found in several groups of peridinin-containing dinoflagellates, including several species of Heterocapsa (3–5) Amphidinium operculatum (6,7), Amphidinium carterae (4,8), Protoceratium reticulatum (4), Ceratium horridum (9), Adenoides eludens (10) and several strains of Symbiodinium isolated from various coral species (11,12). Apart from the usual unigenic minicircles, there are digenic and trigenic circles of about the same size in the two above-mentioned Amphidinium species (6,8). This may suggest the existence of a selection pressure towards small size minicircles, although there is an exception of large 5-kb unigenic circles and even 10-kb dimeric circles in A. eludens (10). An unusual architecture of minicircles in Gonyaulax polyedra shows that the psbA gene could be associated with DNA of roughly 50–150 kb (13). The size limit therefore appears to be species specific, and these minicircles, in respect to both their genome size and transcriptome, are regarded as the smallest plastid genome in nature (14). Recently, the copy numbers of minicircles during different growth phase was estimated, indicating that the total minicircle number changes dramatically from less than 100 copies during exponential growth stage, up to 5 × 103 during late growth phase (15). Researches regarding the localization of minicircles in dinoflagellates had also been done (16), however, many aspects of minicircle biology remains unclear.
Besides this unconventional genome organization, the gene content of the dinoflagellate plastid genome is greatly reduced. Typically, the plastid genome in higher plants and algae encodes over a hundred genes of various functions. To date, however, only about 14 plastid-derived genes are found to be carried in minicircles (17). These genes include ATP synthase, cytochrome complex, components involved in photosystems, as well as rRNAs. Several rRNA and tRNA genes have also been discovered by comparative analysis and reverse transcription-PCR (18,19). Overwhelming evidence support the mass transfer of protein-encoded genes from plastid to the nucleus in both higher plants and algae (20). In the case of the dinoflagellates, recent EST analysis indicates that at least 18 plastid-derived genes have been transferred to the nucleus (21). The minicircle genes also show a number of abnormal features. Several of them carry some unusual predicted translational start codons, e.g. the GUA in Amphidinium and AUA in Heterocapsa. Some genes display an accelerated rate of evolution compared with the same plastid genes in other organisms (22). For example, the similarity of putative minicircular 16S rRNA sequence to other 16S sequences is significantly low (17). Minicircular genes display a strong codon bias too, although the codon preference does not appear to be consistent (23). Moreover, some genes show several small deletions, particularly in the loop regions (7).
Minicircles of a given species have similar sizes but can carry more than one gene. Thus the size of the non-coding region (NCR) of the different circles has to vary. There are several further remarkable features of the NCR. An extremely conserved core region exists with specific motifs within these regions that are nearly identical (4). In general, each minicircle contains about three alignable motifs that make up around 250–500 bp of the NCR. The cores are always found in the same orientation with respect to the genes in the minicircles. However, although the core region is highly conserved among the minicircles of the same species of dinoflagellates, little similarities are shared between minicircles of different species (4,6). Therefore, an interspecific variability of NCR organization is expected. Another characteristic of the NCR is that, all the core regions are AT-rich and contain some direct and inverted repeated sequences. Several double hairpin elements and tandem repeats have been identified in an unusually large 5–6 kb minicircle (10). It is suggested that the core regions could function as the origin of replication or be involved in gene recombination (4,11). Lastly, at least 30 kinds of minicircles from different species of dinoflagellates have been identified and sequenced, so far, but no known origin of replication or promoter sequence has been identified yet. A linkage between dinoflagellate plastid DNA replication and transcription was proposed recently (24) and that the extended transcript may generate short fragment at the end and serve as primer for DNA synthesis. Nevertheless, the replication process still remains elusive.
Plastid DNA replication of photosynthetic organisms is rather unique when compared with other known replication systems. A general replication mechanism for plastid genome has been elucidated (25). Plastid DNA replication in a variety of plants and algae has shown to begin at specific regions referred to as the origins of replication (ori). At least two ori regions, the oriA and oriB, are found in plastid DNA and the locations of these ori have been identified in several plants including pea, Oenothera, tobacco, maize and soybean, and algae including Euglena and Chlamydomonas. Early studies using electron microscopy led to the observation of displacement loops (D-loops), which were 6–7 kb apart in the plastid DNA (26). Rolling circle-type intermediates were also observed in plastid DNA (25). Recent discovery of the linear nature of the plastid genomes, however, probably require a re-evaluation of its replication mechanism
Knowledge of the mechanism of replication would increase our understanding in the evolution of the plastid genomes. It is now appreciated that majority of the plastid genomes have been transferred to the dinoflagellate nuclear genomes, an understanding of minicircle's replication mechanisms will also give light on the phenomenon of the nuclear transfer of organelle genomes. The replication mechanism of a plastid-like genome has been studied in Plasmodium falciparum, an apicomplexan parasite which is the sister group to the dinoflagellates. This study suggests that twin D-loop initiation and rolling circle replication are involved during the replication (27). Whether minicircles are replicated by conventional replication bubble or by rolling circle remain to be seen. Another proposed replication mechanism for minicircles can be compared with the integron (17). Due to the fact that all the genes in minicircles possess same orientation with respect to the core region, and the similar structure between minicircles and bacterial integrons, it is suggested that minicircles may be integrated into the plastid/nuclear genome by recombination across a conserved core region. In this way the genes of minicircles could be replicated together with the plastid/nuclear genome, as well as driven by appropriate promoter during transcription initiation.
MATERIALS AND METHODS
Culture of dinoflagellates
Heterocapsa triquetra (CCMP 449) was obtained from the Provasoli-Guillard Culture Center for Marine Phytoplankton (CCMP) and maintained in a modified seawater medium (f/2) (28) at constant temperature (18 ± 1°C) in a 12-h light/12-h dark cycle using white fluorescent light. The culture was grown to a cell density of ∼15 000 cells ml−1 as measured by hemocytometer.
Nucleic acid extraction
Whole DNA from H. triquetra was prepared by a modified CTAB extraction method (29). Cells were harvested by low-speed centrifugation (1500 g, 10 min). The cell pellet was resuspended in CTAB extraction buffer (2% [w/v] CTAB, 2% [w/v] polyvinylpyrrolidone (PVP-Mr10 000), 2 M NaCl, 20 mM EDTA (pH 8.0), 100 mM Tris (pH 8.0), 10 μg ml−1 RNase A, 0.1 mg ml−1 Proteinase K, 5% [v/v] β-mercaptoethanol) and incubated in 60–65°C for overnight. The suspension was extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) (GE Health). After centrifugation (4000 g, 20 min), the aqueous phase was aliquoted into eppendoffs and added to 2× volume of 100% ethanol. DNA was precipitated by micro-centrifuge at 15 000g, 4°C for 20 min. The DNA pellet was dissolved in ddH2O and DNA concentration was measured by spectrophotometer (NanoDrop ND-1000, Coleman).
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) provides greater resolving power than conventional agarose gel electrophoresis. This method allows separation of a broad range of DNA molecules from ∼1 kb to ∼10 Mb in length (30,31). An unusual architecture of minicircles in G. polyedra shows that the minicircular gene could be associated with DNA of 50–150 kb (13); therefore, PFGE was performed to separate any possible minicircle-containing elements in dinoflagellate DNA. Linear DNAs from a commercial source (Invitrogen Corporation) were used.
Whole DNA samples from H. triquetra were resolved by PFGE (CHEF MAPPER, BIO-RAD). An in-built program ‘Auto-Algorithm’ was performed with inputs of the lower and higher molecular weights (1 and 10 kb, respectively), and the rest of the factors followed the program (calibration factor: 1.00, forward voltage gradient = 9.0 V cm−1, reverse voltage gradient = 6.0 V cm−1, Int. Sw. Tm = 0.05 s, Fin. Sw. Tm = 0.13 s, F. Ramp:a = linear, R. Ramp:a = linear), to run a 1% PFC (pulsed-field certified) agarose gel at 10°C for 16 h in 0.5× TBE buffer.
Cloning of psbA NCR, psbA gene fragments
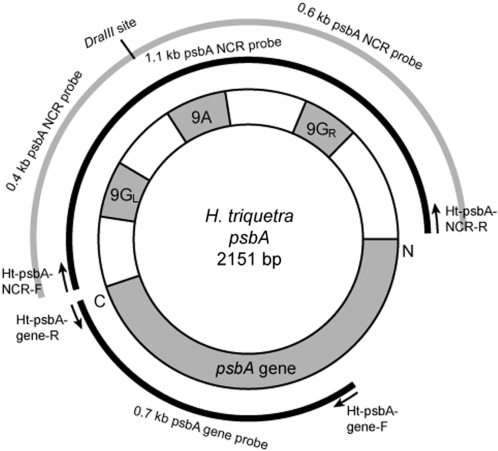
PCR amplifications were performed to generate minicircle NCR and gene fragments for Southern blot analysis of minicircles. Primer pairs were designed based on the H. triquetra psbA minicircle sequence obtained from GenBank (accession number: AF130‱033). The primer pairs for amplifications of the DNA fragments covered nearly the entire 1.1 kb psbA NCR and 0.7 kb conserved region of the psbA gene fragment are Ht-psbA-NCR-F1 (5′-TATATGCATTCATAAACCGTCGAAGC-3′) and Ht-psbA-NCR-R (5′-TAGAATGCAATAAAAATGAACCTAGCTTG-3′) and Ht-psbA-gene-F (5′-CAGTTTGCCAAGCTCTTGG-3′) and Ht-psbA-gene-R (5′-GCAAGATCAAGTGGGAAGTTG-3′), respectively (Figure 1). Using the H. triquetra whole DNA as template, the PCRs for generating psbA fragments were run as follows: 95°C for 3 min, 35 cycles of 95°C for 20 s, 54°C for 20 s, 72°C for 1 min and 72°C for 10 min. The PCR products of expected sizes were gel-purified. They were readily used as DNA probe in later experiments and cloned into the pGEM-T Easy vector (pGEM-T Easy Vector Systems, Promega) following manufacturer instructions. The DNA sequences were confirmed by using the ABI Prism BigDye Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems). Sequence analyses were performed by software MegAlign (DNAStar).
Figure 1.
Map of psbA minicircle in H. triquetra. The full psbA gene sequence is around 1 kb, while the rest is occupied by the NCR. The 9G-9A-9G region of around 450 bp within the NCR had similar amounts of different minicircles of one species. The full-length 1.1-kb NCR and partial 0.7-kb psbA gene, amplified with corresponding primer pairs, were used as probes for Southern blotting.
Southern blot analysis of minicircles
Minicircle DNA resolved in PFGE (as well as the 2D-gel electrophoresis, described below) was detected by ECL Nucleic Acid Labeling and Detection Kit (Amersham) using a PCR-amplified probe (either 1.1 kb psbA NCR or 0.7 kb psbA gene, or 0.4 kb and 0.6 kb fragments from the DraIII-digested 1.1 kb psbA NCR), following protocols provided by the manufacturer. The NCR contains species-specific core regions (4,6). Detection of more than one species of minicircles is expected. Hybridization signals were developed using Fuji Film and were measured and compared using a free-ware ImageJ 1.39v Software (National Institutes of Health, USA; http://rsb.info.nih.gov/ij/) (32).
In vitro topoisomerase II Assay
To test whether the presence of any catenated minicircles may have led to presence of large DNA species in dinoflagellates, an exogenous topoisomerase II (Topo II) was applied to the minicircle fraction. Topo II activity allows the separation of catenated circular form DNA into circular monomers by two steps: the introduction of a double-stranded nick on the molecule to generate a linearized DNA, and the resealing of the linear DNA to form an open circular or relaxed closed circular kinetoplastid DNA (33).
Whole DNA of H. triquitra was treated with Topo II enzyme (TopoGEN Corporation) in 1X Topo II assay buffer at 37°C for 1 h. The treated DNA was run in PFGE and detected with minicircle probes as described.
Aphidicolin treatment
Aphidicolin can inhibit DNA polymerase activity and lead to inhibition in replication intermediate synthesis (34). Heterocapsa triquetra culture was treated with aphidicolin to test the effect on minicircle DNA. Culture was incubated in 2.5 μg ml−1 aphidicolin (Sigma) at 18°C. Cells were collected at the time the drug added, and after 12 h and 18 h. Whole DNA samples were extracted and resolved in PFGE as described. Minicircular DNA was detected with Southern blot analysis.
2D agarose gel electrophoresis
To investigate the potential replication intermediates of minicircles, 2D agarose gel electrophoresis (2D-gel) was performed. The 2D-gel technique followed protocols from (42), with some modifications. For the first-dimensional electrophoresis, H. triquetra whole DNA [as well as exonuclease III (Exo III) treated DNA, described below] was first electrophoresed with a 0.35% agarose gel in 1× TAE buffer for 48 h at 0.8 V cm−1 at room temperature. The gel slab containing DNA was excised out and was turned by 90°, casting with 1% agarose in 1× TAE supplemented with 0.3 μg ml−1 ethidium bromide. This gel was then soaked for 1 h in TAE buffer that supplemented with 1 μg ml−1 ethidium bromide at 4°C. Second-dimensional electrophoresis was carried out at 8 V cm−1 for 3 hours at 4°C. Minicircles DNA was visualized by Southern blot hybridization using psbA NCR and gene as probes as described.
Exo III treatment
Replication intermediates produced by rolling circle contain a linearized form of DNA, resembling some circle-with-tail shaped molecules. To test whether minicircles contain linear DNA elements, DNA extract was treated with Exo III (NEB Inc.). Exo III catalyzes a stepwise removal of mononucleotides from the 3′-hydroxyl termini of linear duplex DNA. Heterocapsa triquetra whole DNA extract was treated with Exo III in 1× NEBuffer 1 for 1 h at 37°C. Exo III-treated DNA was then run in PFGE, 2D-gel and detected for minicircles by Southern blot hybridization.
Rolling circle intermediates may also contain nicks and single-stranded gaps (35). The trypanosomes kinetoplastic minicircles have two short gaps situated 180° apart and may represent the origins of replication (36). To investigate whether the minicircles contain nicks and gaps, minicircle DNA extract was treated with Klenow fragment and DNA ligase. Klenow is capable for polymerization of DNA and filling in single-stranded DNA gap. Potential single-stranded nick can be sealed with DNA ligase, therefore resistant to Exo III digestion. Whole DNA extract from H. triquetra was treated with large fragment of DNA polymerase I (Klenow; NEB Inc.) in 1× NEBuffer 1 supplemented with dNTP for 30 min at 25°C (followed with inactivation of Klenow at 75°C for 20 min). The DNA was then treated with T4 DNA ligase (NEB Inc.) in supplement of ATP for overnight at 16°C. Treated DNA was subjected to Exo III digestion. The effect of Exo III to the treated DNA was tested by PFGE and Southern blot hybridization as described.
Atomic force microscopy
Atomic force microscopy (AFM) was performed for direct imaging of the minicircle DNA from dinoflagellates. Protocols of AFM mainly relied on two previously published protocols (37,38). Minicircular DNA as detected after Southern blot analysis (including the APBs, discussed in later sections) was gel-purified. The DNA fragments were physically cut from the gel and purified by electroelution method (39) with dialysis tubing (Spectra 6-8000 m.w., Spectrum Lab). The DNA solution was dropped on freshly cleaved mica (AP-mica) and incubated for 10 min followed with rinsing in deionized water and a nitrogen blow-dry. Imaging was carried out on a scanning probe microscope in the Tapping-Mode in air using a coated silicon probe. To increase the yield of DNA immobilized on the mica, minicircle DNA in the gel slabs was directly transferred onto a 3-aminopropyl-triethoxysilane (APTES; Sigma) vapor-treated mica. This activated mica is positively charged, thus enabling attraction of negative charged DNA molecules. Images were carried out on a NanoScope III STM/AFM (Digital Instruments) in the Tapping-Mode using the commercially available Tapping-mode Etched Silicon Probe (TESP; Veeco Instruments Inc.). Images were analyzed with the program WSxM 4.0 Develop 10.0 Scanning Probe Microscopy Software (Nanotec Electronica, Madrid, Spain; http://www.nanotec.es).
RESULTS
Minicircle DNA of 6–8 kb (APBs) are identified in a pulsed-field gel
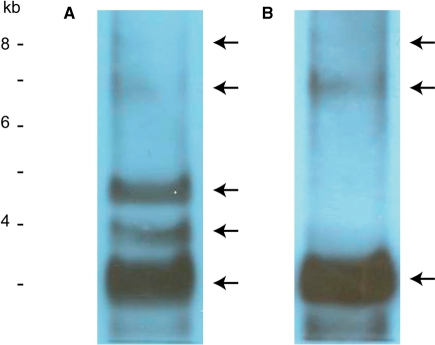
PFGE can resolve DNA ranged from ∼1 kb to ∼10 Mb (30,31). Southern blot analyses of H. triquetra whole DNA hybridized with the psbA NCR and the psbA gene fragments were performed to detect the location of minicircle DNA after PFGE. Five bands were detected with the psbA NCR fragment, including three strong bands at ∼3 kb, ∼4 kb and ∼4.6 kb, and two weak bands at ∼6.8 kb and ∼8 kb (Figure 2A). Hybridization signals resembling the ∼3 kb, ∼6.8 kb and ∼8 kb bands with similar intensities were obtained by using the psbA gene fragment for detection (Figure 2B). These sizes estimated were based on linear, unbranched, DNA markers.
Figure 2.
Southern blot analyses of minicircles from H. triquetra. Whole DNA extract from H. triquetra was subjected to PFGE. (A) Hybridization signals detected by 1.1-kb psbA NCR fragment. (B) Hybridization signals detected by 0.7-kb psbA gene fragment. DNA markers were linear DNAs from a commercial source (Invitrogen Corporation).
The psbA gene fragment specifically recognized DNA at around 3 kb, which is reasonably comparable with the psbA minicircle of 2151 bp. The slightly extended minicircle DNA distribution is perhaps due to its relaxed circular shape that leads to slower migration during electrophoresis. The psbA NCR fragment also hybridized to DNA at the same ∼3 kb location, with two extra DNA species at ∼4 kb and ∼4.6 kb. These two bands are likely due to cross-minicircle hybridization by the conserved core region of the psbA NCR which recognized other minicircle types. Hybridization signals for the three smaller bands are comparatively strong due to the high copy numbers of minicircles. In contrast, additional positive bands (APBs) of ∼6–8 kb were detected with both psbA NCR and gene fragments. This indicated that some rare DNA species of minicircular DNA exists in unexpected large sizes, which are larger than the well-known 2–3 kb minicircles. Detections of these APBs are unlikely due to other satellite DNA, such as mitochondrial DNA with sizes probably more than 20 kb (40). The 6–8 kb size is estimated based on linear DNA markers; the actual size could be smaller if APB were to contain branches.
The presence of these APBs has several possible explanations. They could be double-sized minicircles due to dimerization of 2–3 kb minicircles (10). Linking between two minicircles during a catenation process could also increase the mass of the molecule. DNA in open circular or relaxed circular forms could lead to retardation during electrophoresis too. Finally, this scantly detected minicircular DNA could be the replication intermediates of minicircles.
The APBs are not concatenated minicircles
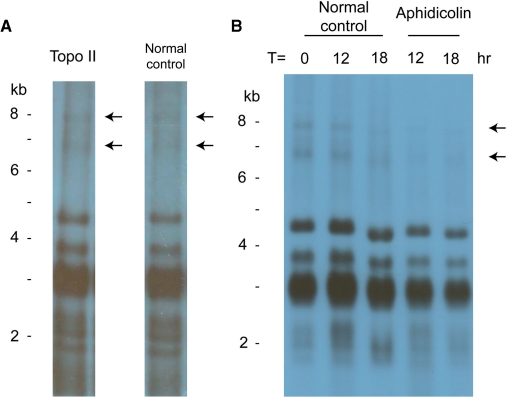
To verify whether the APBs are catenated minicircles, an in vitro Topo II assay was performed. Topo II is able to decatenate topologically linked circular DNA molecules by producing a transient double-stranded break (33). The decatenated products after resealing would be DNA in either the open circular or relaxed circular forms. Heterocapsa triquetra whole DNA was treated with Topo II and detected for minicircle DNA after PFGE. The APBs suspected to be linked minicircles should be removed due to decatenation by Topo II. In Figure 3A, however, the APBs were still detected in Topo II-treated DNA by the psbA NCR probe. This result indicates that the APBs are not catenated minicircles. Three extra bands with sizes around 2 kb were observed too, suggesting the presence of smaller minicircle DNA detected by the psbA NCR fragment.
Figure 3.
Effects of Topo II and aphidicolin on the minicircle DNA. (A) Topo II-treated whole-DNA extract from H. triquetra, and positive control of untreated whole DNA, were subjected to PFGE, and detected with 1.1-kb psbA NCR fragment. (B) Aphidicolin-treated whole-DNA extracts collected after 12- and 18-h incubation (with parallel untreated controls) were hybridized with the same probe after PFGE. DNA markers were linear DNAs from a commercial source (Invitrogen Corporation).
Nevertheless, this result cannot totally eliminate the possibility that the APBs are retarded minicircles due to their relaxed circular conformation. Indeed the Topo II enzyme is effective in relaxing supercoiled DNA into relaxed circle by simple cutting-and-resealing process. Dimerized minicircles leading to increase in mass cannot also be completely ruled out.
The APBs are sensitive to aphidicolin
DNA polymerase inhibitor aphidicolin can reduce replication intermediate synthesis (34). To test whether the APBs are potential replication intermediates of minicircles, H. triquetra culture was treated with aphidicolin and the change of minicircle DNA as detected in Southern blot analysis was investigated. After aphidicolin treatment, hybridization signals of minicircular DNA, especially the APBs, from both samples collected at 12 and 18 h were diminished (Figure 3B). These results showed that the APBs are sensitive to aphidicolin and they are likely to be replication intermediates produced from minicircles.
The APBs are not replication intermediates produced by bubble forming
As mentioned earlier, circular dimers and relaxed circles can lead to slower migration during electrophoresis. But both of them have defined molecular weights, unlike the replication intermediates that increase in sizes during replication. The 2D-gel is a well-developed tool to study the mechanisms of DNA replication (41). 2D-gel takes advantage of resolving DNA according to its size in the first dimension and to its shape in the second dimension. Rare replicating DNA species in the gel are detected by the Southern Blot method, and can produce several characteristic arc patterns (42). The mechanism of DNA replication can be deduced from these arcs. In general, replicative arc generated by bubble-forming type replication intermediates extend from one-unit mass (1n) to double-unit mass (2n) (43,44). Theoretical rolling circle replication intermediates contain branched tail with various lengths which sometimes can be larger than 1-U mass. Therefore, they produce a replicative arc resembling a curved diagonal and extending beyond the 2n of the non-replicating DNA (45,46).
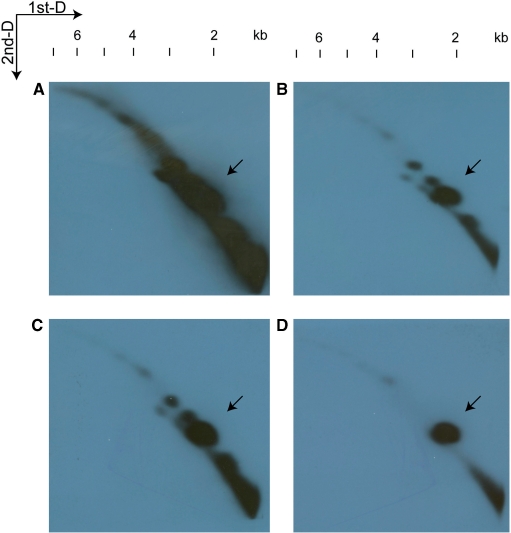
Heterocapsa triquetra total DNA was resolved in 2D-gel and detected with the psbA minicircle fragments (Figure 4A). The patterns of the replicative arcs resembled a diagonal with a slightly curved shape extending beyond 6 kb, as well as several strong signal spots. These spots were distributed across the diagonal. The full length 1.1 kb NCR probed more species of minicircular DNA, while the 0.4 kb and 0.6 kb NCR fragments detected less. The psbA gene fragment that specifically hybridize to the psbA minicircle showed one strong spot and two weak spots, resembling the observation in PFGE, as well as a very weak thread of signals joining these spots. Short curved diagonals up to 3 kb were also observed underneath the long diagonals, which contained spots of sizes similar to those in the long diagonals, but with slightly lower signal intensities. By comparing the four patterns, strong spots of minicircular DNA were observed at ∼1.6 kb, ∼2 kb, ∼2.5 kb and ∼3 kb, weaker spots were observed at ∼4 kb, ∼4.5 kb and ∼5.2 kb (Figure 4B).
Figure 4.
Patterns of minicircle DNA resolved in 2D-gel. Whole-DNA extract from H. triquetra was subjected to 2D-gel as described in the Methods section. The first dimension is represented horizontally (left to right) and the second dimension vertically (top to bottom). Minicircle DNA were detected with 1.1-kb (A), 0.4-kb (B) and 0.6-kb (C) psbA NCR, and 0.7-kb psbA gene fragment (D). Four images were aligned with respect to their DNA migration in both dimensions of electrophoreses. The putative psbA minicircle spots indicated by the arrows were perfectly aligned. The sizes of hybridization signals can be revealed by the size ladder on the top.
According to the pattern revealed by the psbA gene probe, it is conceivable that the ∼2 kb spot represents the psbA minicircle of 2151 bp. Minicircular DNA of dimer sizes at ∼4 kb and ∼4.5 kb were also detected, resembling the observation in minicircle hybridization after PFGE at ∼3 kb and 6–8 kb bands, respectively. Furthermore, the weak thread-like signals joining the ∼2 kb, ∼4 kb and ∼4.5 kb spots indicated the presence of minicircular DNA in a continuum of sizes, which were likely replication intermediates of the minicircles. However, a replicative arc resembling the bubble-forming mechanism was not observed in the 2D-gel (41,43). Although it is possible that these intermediate forms were too rare to be detected, the 2D-gel patterns were similar even after overnight exposure. Therefore, replication of minicircles is unlikely to involve any bubble-forming type intermediates.
The APBs contain linear DNA
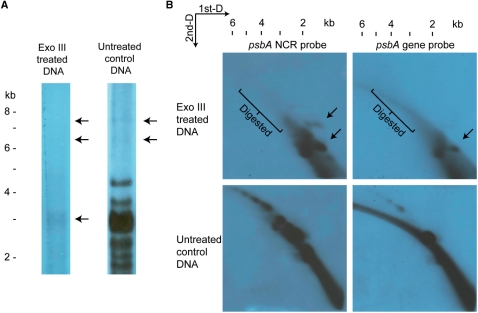
The mass of replication intermediates is always greater then the unreplicated parental DNA, regardless if it is produced by either bubble-forming or rolling circle mechanisms. One distinguishing feature of rolling circle intermediates is the presence of linear DNA, resembling a circle-with-tail structure. To test whether the APBs contains linear DNA, digestion of H. triquetra whole DNA by Exo III was performed. Exo III removes mononucleotides from 3′-hydroxyl termini of duplex DNA. Its preferred substrates are blunt or recessed 3′-termini, as well as nicks in double-stranded DNA. For a population of circular DNA undergoing rolling circle replication, Exo III digestion can lead to either the removal of linear tails on the intermediates, leaving single-stranded circular DNA molecules, or intact duplex circles that remain unattacked. Therefore, Exo III treatment on rolling circle DNA should lead to removal of the replicative arc, leaving 1n spot in the 2D-gel.
The Exo III-treated DNA was resolved after PFGE and 2D-gel electorphoresis, and detected with minicircle DNA by hybridization (Figure 5). In both gels, the large-sized minicircular DNA (6–8 kb in PFGE and 4–6 kb in 2D-gel) was removed after Exo III treatments. This indicated that these higher molecular mass DNAs were sensitive to Exo III digestion. Also, the putative psbA minicircles revealed as a ∼3 kb band in PFGE and ∼2 kb spot in 2D-gel showed a decrease in quantities. These results indicated that a fraction of the psbA minicircles exist as nicked open circular forms or contain single-stranded gaps that are sensitive to Exo III digestion. In the 2D-gels, extra signal spots of ∼1.5 kb masses were observed after Exo III treatments. They were potentially the single-stranded circular monomers produced after Exo III digestion.
Figure 5.
Exo III-treated minicircular DNA revealed after PFGE and 2D-gel electrophoresis. Exo III-treated whole-DNA extract from H. triquetra was subjected to PFGE (A) and 2D-gel (B), and detected with 1.1-kb psbA NCR fragment [and psbA gene fragment in (B) only]. (A) The 6–8 kb minicircular DNA bands (APBs) were removed in the Exo III lane, while the putative 3-kb psbA minicircle band was diminished in intensity. (B) Hybridization signals of larger than 2 kb were removed after Exo III treatment as revealed in both 2D-gels detected by psbA NCR and gene fragments. Extra spots indicated by the arrows were observed in these two images when comparing with the controls. DNA markers were linear DNAs from a commercial source (Invitrogen Corporation).
The APBs were removed after Exo III treatment, eliminating the possibilities of circular dimers and closed relaxed circles existing in the 6–8 kb region. Reviewing the results from 2D-gel electrophoreses, the diagonal-shaped pattern of minicircles DNA resembled the theoretical replicative arc produced by rolling circle mechanism (47,48). Exo III digestion could remove all larger molecular-sized psbA minicircular DNA down to ∼2 kb. These results are consistent with the rolling circle-type replication and those high molecular mass DNA are potential intermediates produced by this mechanism.
Minicircle DNA contains nicks
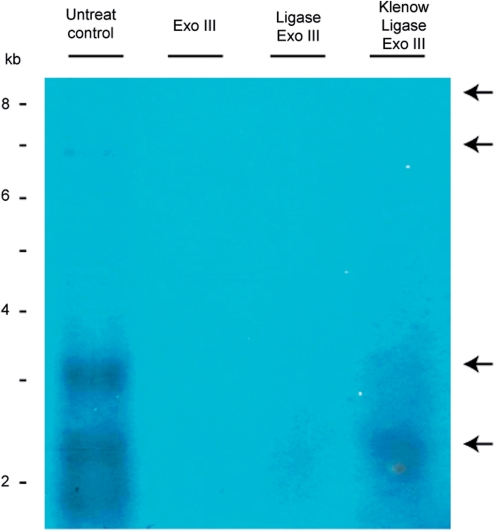
Replication of DNA is exclusively made possible with an exposed single-stranded DNA template. Bubble-forming replication involves the separation of double-stranded DNA during replication initiation, while rolling circle replication required a break on one of the single-stranded backbone (a nick) before this strand denatures from the opposite strand (35). Nicking is therefore necessary during rolling circle replication mechanism. To test whether minicircles contain nicks and gaps, total DNA extract was treated with DNA ligase and Klenow fragment. Klenow is able to fill in single-stranded DNA gap, while ligase can seal the single-stranded nick. Therefore, treated DNA was then tested on the sensitivity to Exo III.
Total H. triquetra DNA (which contained minicircle DNA as detected in the Southern hybridization) were sequentially treated with Klenow, T4 DNA ligase and Exo III. Presence of minicircle DNA was revealed by Southern blotting by the psbA NCR probe. The signals ∼3 kb corresponding to the monomer of psbA minicircle was retained under Klenow/ligase/Exo III treatment as compared with the hardly detected minicircle signals in the Ligas/Exo III-treated sample, as well as to the undetectable signal in the Exo III-treated sample (Figure 6). The results indicated that Klenow/ligase treatment can protect minicircle DNA from Exo III digestion, and inferred the presence of gap/nick on minicircles.
Figure 6.
Effects of DNA ligase/Klenow fragment on minicircular DNA. Total DNA from H. triquetra was treated with T4 DNA ligase, as well as Klenow fragment followed with T4 DNA ligase, prior to Exo III digestion. Treated DNAs were resolved by PFGE and detected the minicircular DNA signals by Southern Blot. The two upper arrows indicated APBs in the untreated control lane. The two lower arrows indicated the putative monomer of psbA minicircle signals of ∼2–3 kb. DNA markers were linear DNAs from a commercial source (Invitrogen Corporation).
The APBs were undetected in the Klenow/ligase/Exo III-treated samples. This also eliminates the presence of nick-containing dimerized minicircles. Linear DNA in the APBs were expected to join each other by Klenow/ligase, but they were still digested by Exo III. Opened termini or nicks in the ligated APBs could be one possibility that they were just ‘two circles connected with tails’ intermediates (45), therefore readily digested by Exo III. Nevertheless, minicircles contain nicks and this DNA structure is similar to the nick in rolling circle-type mechanism during replication initiation.
AFM of replication intermediates
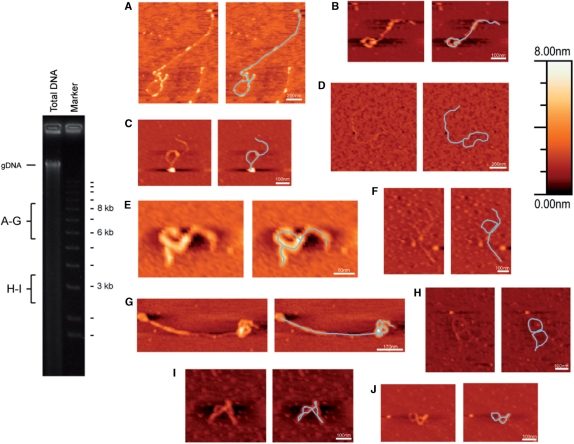
AFM is a useful tool to study topology of objects at the nanoscale. It has been used in biological research to image DNA and protein/DNA complexes (37,38). To directly investigate the shape of those double-sized minicircular DNA, AFM of the 6–8 kb fractions was performed. Minicircular DNA of the APBs as detected by Southern blot analysis was adsorbed on smooth surface mica and imaged with AFM. Figure 7 illustrates a number of images of thread-like structures, putative DNA species of minicircles.
Figure 7.
AFM images of minicircular DNA. Minicircular DNA of 6–8 kb fractions (A–G) and ∼3 kb fraction (H–J) were subjected to AFM. Images (A, E and G) were performed by scanning probe microscope from Veeco, while the rest (B–D, F and H–J) were by NanoScope III from Digital Instruments. The ethidium bromide stained gel on the left indicates the regions for DNA extractions (gDNA = genomic DNA of H. triquetra). The heights of objects in the AFM images can be denoted by the Z-scale on the right-handed top corner.
Dimensions of the thread-like structures were measured by the WSxM program. For the most common form of DNA, the B-form DNA, 3 kb is equivalent to 1 μm in length (distance between two neighboring base pairs at 0.34 nm) (49). From the APBs, circle-with-tail structures of 1.5–6 kb in total length were observed (Figure 7A–D). Circles with more than one tail were also found in this fraction (Figure 7E). Besides these, coiled linear structures (Figure 7F and G) were observed as well. In addition to the APBs, the ∼3 kb fraction suggested to be the psbA minicircle was subjected to AFM. Coiled circular structures of 1.2–2.1 kb were observed (Figure 7H–J). The areas of PFGE gel that were extracted for AFM analysis is indicated on the left-hand panel. Dinoflagellate mitochondrial DNA (40), as in the case of genomic DNAs, is much larger in size than the minicircles.
The AFM observations revealed that circle-with-tail(s) structures, as expected for rolling circle replication intermediates, exist in dinoflagellate DNA. Transient triple/double-sized rolling circle intermediates were deduced from the structures with tails of double (Figure 7A and B), or nearly the same length of the circle (Figure 7C and D). Circles with two tails (Figure 7E) are also possible during rolling circle replication (45). Although the sizes of some putative rolling circle intermediates were even less than the usual minicircles of 2–3 kb, much smaller ‘empty’ circles (8) with an open circular shape could result in extensively slower migration during electrophoresis, and thus, be detected in the APBs. This may also explain the sizes of the circular structures found from the ∼3 kb fraction (Figure 7H–J), which should correspond to monomers of minicircles. They were some ‘figure of eight’ shapes with DNA flipped over between the loops (50). A table summarizing the percentage of different forms is included in the Supplementary Data. Alternatively, the coiled linear structures from 6 to 8 kb fractions suggesting a complex organization of minicircles could be generated during recombination activity (Figure 7F and G). Nevertheless, these AFM results suggest that there are rolling circle intermediates in the APBs.
DISCUSSION
The dinoflagellates’ minicircles are the newest form of plastid genomes being reported and have been hailed as ‘the last frontier in understanding chloroplast evolution’ (51,52). These minicircles probably represent a penultimate terminus of the ever-decreasing organelle genomes; it is now known that many plastid genes have been transferred to the dinoflagellate nuclear genomes (21). Their mode of replication would thus be of interests to the study of organelle genomes’ evolution. The small size of the minicircles may facilitate fast genome duplication; the speed of genome replication could also be dictated by its mode of replication. The replication mechanism of the minicircles has not been previously reported. Results from this study showed that APBs consists of Exo III sensitive circle-with-tail DNA structures which resemble rolling circle replication intermediates. Despite the presence of rolling circle-type intermediates, as found in other plastid genomes, the replication of minicircles certainly differ from the two origin-mediated replication of conventional chloroplasts.
The rolling circle mode of replication is commonly used by Gram-positive bacteria carrying plasmid genomes of <10 kb and some viruses. However, there are known examples of plasmids from Gram-negative bacteria, including cyanobacteria, which involve a rolling circle mode of replication (53). Many strains of cyanobacteria contain endogenous plasmids of various sizes mostly <10 kb. A 2.4 kb plasmid from a cyanobacterium has been found to replicate by rolling circle mechanism and encode a Rep protein, which is likely to be involved in producing a nick at the origin of replication (52). Even though the relationship of a secondary endosymbiosis-acquired dinoflagellate plastid and a cyanobacterium remains ambiguous (54,55), the tiny minicircles in dinoflagellate plastids might use mechanisms consistent with that of cyanobacteria in regards to DNA replication.
Rolling circle replication allows rapid duplication of plasmid DNA. Although syntheses of plus strand and minus strand might be uncoupled, rolling circle replication could generate linear concatemers that produce multiple copies of the genome in a relatively short time. The replication protein complexes might stay on the DNA without the need of reassociation during this rolling circle replication, which leads to an efficient synthesis of DNA concatemers. It is also possible that more than one single-stranded opening could be generated when the origin of replication is reconstituted during rolling circle replications (45). With this R-shaped structure (a circle with two tails) rapid DNA duplication can be attained.
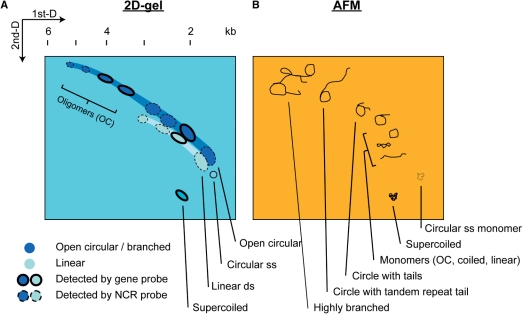
The 2D-gel and AFM experiments are most supportive in uncovering the replication mechanism of the minicircles. A continuous diagonal curve in a 2D-gel pattern indicates the presence of rolling circle replication intermediates (45,46). In our 2D-gel results, a continuous diagonal arc was observed (Figure 4). Signal spots along the diagonal arc which correspond to the replication intermediates of doubled or tripled common 2–3 kb minicircles are considered as open circular oligomers (illustrated in Figure 8A and B). In addition to the Exo III treatment which indicated the presence linear minicircle DNA (Figure 5), structures of these open potential circular oligomers were further confirmed as circle-with-tail shape by AFM (Figure 7). Various structures of minicircle intermediates are expected to be found in a 2D-gel (Figure 8B). Resealing of short gaps and nicks on minicircles by Klenow and ligase indicates the presence of nicked circular molecules (Figure 6), which correspond to rolling circle intermediates, too. Based on these findings, it is conceivable that minicircles contain rolling circle DNA. Furthermore, these rolling circle intermediates may form secondary structures possibly by DNA recombination (Figure 7F and G) (56,57).
Figure 8.
Schematic diagrams showing the observed spots from 2D-gel and some DNA topology observed in AFM studies. By compiling the 2D-gel results that looked for psbA minicircles (Figure 4 and 5) and DNA structures revealed by AFM (Figure 7), the 2D-gel pattern (A) in corresponding to the minicircle structures (B) are illustrated. Major signal spots representing psbA minicircle DNA are drawn in the schematic 2D-gel (A), while possible corresponding DNA structures are outlined in the schematic AFM images (B). Extensive recombination activity may produce highly branch structures.
Considering the arrangement of the conserved non-coding cores, as well as direct and inverted repeats, secondary structures which may reflect probable functions regarding DNA replication are expected. Indeed, the replication mechanism for mitochondrial genome (mtDNA) of P. falciparum (a group phylogenetically related to dinoflagellates) involves the recombination of DNA molecules (56). A recombination-dependent replication in P. falciparum mtDNA, in which the linear concatemerized 6-kb genome bends on itself and undergoes recombination, generates circular monomer for subsequent rolling circle replication. This mode of replication is apparently consistent with the findings of 6–8 DNA conformation observed under AFM. Although recombining minicircle intermediates at the repeated region was not directly determined, AFM images depicting coiled linear ∼2.6 kb and ∼3.4 kb DNA molecules (Figure 7F and G, respectively) provide plausible evidence for minicircle recombination events. Recombination-dependent process was also suggested to be the origin of inversion isomers of inverted repeats in higher plants’ chloroplast DNA (1,58). In addition, the resemblance of the dinoflagellate plastid genome to the cassettes of bacterial integrons was suggested (23). Circular molecules of bacterial integrons carry an open reading frame and conserved 59-bp core regions are inserted into the bacterial chromosome by recombination across a similar sequence in the chromosome. A similar proposal that active transposition of minicircles occurs at the putative replicon origin on the NCR is also postulated (4). Recombination among minicircles may also explain the discovery of rare digenic/trigenic minicircles in which the two genes are generally not adjacent in other plastid genomes (8).
The results in this study support the hypothesis that plastid minicircle replication involve rolling circle mechanisms. The minicircle genomes may exist in circular, concatemerized linear and branched forms. Structurally, it is dissimilar to the plastid DNA molecules from various higher plant species which appear in forms of linear and branched molecules undergoing replication (2). The origin of replication on minicircles is estimated at an approximate location on the NCR. Minicircles are apparently producing rolling circle-type intermediates during replication. However, this rolling circle-type replication may involve other mechanisms, for instance, transposition of DNA molecules, which is required to complete the full cycle of replication. The possibility of a mixed type of plastid genome replication should not be excluded.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CERG grant from Research Grant Council of Hong Kong (HKUST6406/05M to J.T.Y.W.). Funding for open access charge: University Grant Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Jie Xhie (Materials Characterization and Preparation Facility, The Hong Kong University of Science and Technology) and Mr Man-nin Yeung (The Hong Kong Polytechnic University Materials Research Centre) for their help with AFM analyses. The authors wish to thank Dr. Alvin Kwok and Miss MingHua Ju for their help in the revision of the manuscript.
REFERENCES
- 1.Oldenburg DJ, Rowan BA, Zhao L, Walcher CL, Schleh M, Bendich AJ. Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta. 2006;225:41–55. doi: 10.1007/s00425-006-0329-6. [DOI] [PubMed] [Google Scholar]
- 2.Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004;16:1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Cavalier-Smith T, Green BR. Evolution of dinoflagellate unigenic minicircles and the partially concerted divergence of their putative replicon origins. Mol. Biol. Evol. 2002;19:489–500. doi: 10.1093/oxfordjournals.molbev.a004104. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Cavalier-Smith T, Green BR. A family of selfish minicircular chromosomes with jumbled chloroplast gene fragments from a dinoflagellate. Mol. Biol. Evol. 2001;18:1558–1565. doi: 10.1093/oxfordjournals.molbev.a003942. [DOI] [PubMed] [Google Scholar]
- 6.Barbrook AC, Symington H, Nisbet RE, Larkum A, Howe CJ. Organisation and expression of the plastid genome of the dinoflagellate Amphidinium operculatum. Mol. Genet. Genomics. 2001;266:632–638. doi: 10.1007/s004380100582. [DOI] [PubMed] [Google Scholar]
- 7.Barbrook AC, Howe CJ. Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol. Gen. Genet. 2000;263:152–158. doi: 10.1007/s004380050042. [DOI] [PubMed] [Google Scholar]
- 8.Hiller RG. ‘Empty’ minicircles and petB/atpA and psbD/psbE (cytb559 alpha) genes in tandem in Amphidinium carterae plastid DNA. FEBS Lett. 2001;505:449–452. doi: 10.1016/s0014-5793(01)02871-x. [DOI] [PubMed] [Google Scholar]
- 9.Zauner S, Greilinger D, Laatsch T, Kowallik KV, Maier UG. Substitutional editing of transcripts from genes of cyanobacterial origin in the dinoflagellate Ceratium horridum. FEBS Lett. 2004;577:535–538. doi: 10.1016/j.febslet.2004.10.060. [DOI] [PubMed] [Google Scholar]
- 10.Nelson MJ, Green BR. Double hairpin elements and tandem repeats in the non-coding region of Adenoides eludens chloroplast gene minicircles. Gene. 2005;358:102–110. doi: 10.1016/j.gene.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Moore RB, Ferguson KM, Loh WK, Hoegh-Guldberg O, Carter DA. Highly organized structure in the non-coding region of the psbA minicircle from clade C Symbiodinium. Int. J. Syst. Evol. Microbiol. 2003;53:1725–1734. doi: 10.1099/ijs.0.02594-0. [DOI] [PubMed] [Google Scholar]
- 12.Barbrook AC, Visram S, Douglas AE, Howe CJ. Molecular diversity of dinoflagellate symbionts of Cnidaria: the psbA minicircle of Symbiodinium. Protist. 2006;157:159–171. doi: 10.1016/j.protis.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Morse D. The plastid-encoded psbA gene in the dinoflagellate Gonyaulax is not encoded on a minicircle. Gene. 2006;371:206–210. doi: 10.1016/j.gene.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Morse D. Rampant polyuridylylation of plastid gene transcripts in the dinoflagellate Lingulodinium. Nucleic Acids Res. 2006;34:613–619. doi: 10.1093/nar/gkj438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koumandou VL, Howe CJ. The copy number of chloroplast gene minicircles changes dramatically with growth phase in the dinoflagellate Amphidinium operculatum. Protist. 2007;158:89–103. doi: 10.1016/j.protis.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Laatsch T, Zauner S, Stoebe-Maier B, Kowallik KV, Maier UG. Plastid-derived single gene minicircles of the dinoflagellate Ceratium horridum are localized in the nucleus. Mol. Biol. Evol. 2004;21:1318–1322. doi: 10.1093/molbev/msh127. [DOI] [PubMed] [Google Scholar]
- 17.Koumandou VL, Nisbet RE, Barbrook AC, Howe CJ. Dinoflagellate chloroplasts–where have all the genes gone? Trends Genet. 2004;20:261–267. doi: 10.1016/j.tig.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Barbrook AC, Santucci N, Plenderleith LJ, Hiller RG, Howe CJ. Comparative analysis of dinoflagellate chloroplast genomes reveals rRNA and tRNA genes. BMC Genomics. 2006;7:297. doi: 10.1186/1471-2164-7-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson MJ, Dang Y, Filek E, Zhang Z, Yu VW, Ishida K, Green BR. Identification and transcription of transfer RNA genes in dinoflagellate plastid minicircles. Gene. 2007;392:291–298. doi: 10.1016/j.gene.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett JD, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Nosenko T, Bhattacharya D. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr. Biol. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Green BR, Cavalier-Smith T. Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: a possible common origin for sporozoan and dinoflagellate plastids. J. Mol. Evol. 2000;51:26–40. doi: 10.1007/s002390010064. [DOI] [PubMed] [Google Scholar]
- 23.Howe CJ, Barbrook AC, Koumandou VL, Nisbet RE, Symington HA, Wightman TF. Evolution of the chloroplast genome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:99–106. doi: 10.1098/rstb.2002.1176. discussion 106–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisbet RE, Hiller RG, Barry ER, Skene P, Barbrook AC, Howe CJ. Transcript analysis of dinoflagellate plastid gene minicircles. Protist. 2008;159:31–39. doi: 10.1016/j.protis.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Kolodner RD, Tewari KK. Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature. 1975;256:708–711. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- 26.Kolodner R, Tewari KK. Presence of displacement loops in the covalently closed circular chloroplast deoxyribonucleic acid from higher plants. J. Biol. Chem. 1975;250:8840–8847. [PubMed] [Google Scholar]
- 27.Williamson DH, Preiser PR, Moore PW, McCready S, Strath M, Wilson RJ. The plastid DNA of the malaria parasite Plasmodium falciparum is replicated by two mechanisms. Mol. Microbiol. 2002;45:533–542. doi: 10.1046/j.1365-2958.2002.03033.x. [DOI] [PubMed] [Google Scholar]
- 28.Guillard RR, Ryther JH. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 29.Puchooa D. A simple, rapid and efficient method for the extraction of genomic DNA from lychee (Litchi chinensis Sonn.) Afr. J. Biotechnol. 2004;3:253–255. [Google Scholar]
- 30.Carle GF, Olson MV. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984;12:5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu G, Vollrath D, Davis RW. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 32.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- 33.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decker RS, Yamaguchi M, Possenti R, DePamphilis ML. Initiation of simian virus 40 DNA replication in vitro: aphidicolin causes accumulation of early-replicating intermediates and allows determination of the initial direction of DNA synthesis. Mol. Cell Biol. 1986;6:3815–3825. doi: 10.1128/mcb.6.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan SA. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birkenmeyer L, Ray DS. Replication of kinetoplast DNA in isolated kinetoplasts from Crithidia fasciculata. Identification of minicircle DNA replication intermediates. J. Biol. Chem. 1986;261:2362–2368. [PubMed] [Google Scholar]
- 37.Chan YH, Wong JT. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acids Res. 2007;35:2573–2583. doi: 10.1093/nar/gkm165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE, Jacobs BL, Oden PI, Lindsay SM. Atomic force microscopy imaging of double stranded DNA and RNA. J. Biomol. Struct. Dyn. 1992;10:589–606. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 40.Nash EA, Barbrook AC, Edwards-Stuart RK, Bernhardt K, Howe CJ, Nisbet RE. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol. Biol. Evol. 2007;24:1528–1536. doi: 10.1093/molbev/msm074. [DOI] [PubMed] [Google Scholar]
- 41.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 42.Dandjinou AT, Larrivee M, Ralf E W, Wellinger RJ. Two-dimensional agarose gel analysis of DNA replication intermediates. In: Xiao W, editor. Yeast Protocols. 2nd edn. Vol. 313. Totowa: Humama Press; 2006. pp. 193–208. [DOI] [PubMed] [Google Scholar]
- 43.Kunnimalaiyaan M, Shi F, Nielsen BL. Analysis of the tobacco chloroplast DNA replication origin (oriB) downstream of the 23 S rRNA gene. J. Mol. Biol. 1997;268:273–283. doi: 10.1006/jmbi.1997.0972. [DOI] [PubMed] [Google Scholar]
- 44.Han Z, Stachow C. Analysis of Schizosaccharomyces pombe mitochondrial DNA replication by two dimensional gel electrophoresis. Chromosoma. 1994;103:162–170. doi: 10.1007/BF00368008. [DOI] [PubMed] [Google Scholar]
- 45.Backert S. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 2002;21:3128–3136. doi: 10.1093/emboj/cdf311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belanger KG, Mirzayan C, Kreuzer HE, Alberts BM, Kreuzer KN. Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen S, Agmon N, Yacobi K, Margarita M, Segal D. Evidence for rolling circle replication of tandem genes in Drosophila. Nucleic Acids Res. 2005;33:4519–4526. doi: 10.1093/nar/gki764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson DH, Janse CJ, Moore PW, Waters AP, Preiser PR. Topology and replication of a nuclear episomal plasmid in the rodent malaria Plasmodium berghei. Nucleic Acids Res. 2002;30:726–731. doi: 10.1093/nar/30.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Virnik K, Lyubchenko YL, Karymov MA, Dahlgren P, Tolstorukov MY, Semsey S, Zhurkin VB, Adhya S. ‘Antiparallel’ DNA loop in gal repressosome visualized by atomic force microscopy. J. Mol. Biol. 2003;334:53–63. doi: 10.1016/j.jmb.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 51.McFadden G. Chloroplasts Ever decreasing circles. Nature. 1999;400:119–120. doi: 10.1038/22011. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, McFadden BA. A small plasmid, pCA2.4, from the cyanobacterium Synechocystis sp. strain PCC 6803 encodes a rep protein and replicates by a rolling circle mechanism. J. Bacteriol. 1993;175:3981–3991. doi: 10.1128/jb.175.13.3981-3991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keeling P. A brief history of plastids and their hosts. Protist. 2004;155:3–7. doi: 10.1078/1434461000156. [DOI] [PubMed] [Google Scholar]
- 55.Lockhart PJ, Steel MA, Barbrook AC, Huson DH, Charleston MA, Howe CJ. A covariotide model explains apparent phylogenetic structure of oxygenic photosynthetic lineages. Mol. Biol. Evol. 1998;15:1183–1188. doi: 10.1093/oxfordjournals.molbev.a026025. [DOI] [PubMed] [Google Scholar]
- 56.Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, Strath M, Williamson DH. Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 1996;15:684–693. [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson DH, Preiser PR, Wilson RJ. Organelle DNAs: the bit players in malaria parasite DNA replication. Parasitol. Today. 1996;12:357–362. doi: 10.1016/0169-4758(96)10053-3. [DOI] [PubMed] [Google Scholar]
- 58.Kunnimalaiyaan M, Nielsen BL. Chloroplast DNA replication: mechanism, enzymes and replication origins. J. Plant Biochem. Biotechnol. 1997;6:1–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.