Abstract
Direct cloning and parallel sequencing, an extremely powerful method for microRNA (miRNA) discovery, has not yet been applied to bacterial transcriptomes. Here we present sRNA-Seq, an unbiased method that allows for interrogation of the entire small, non-coding RNA (sRNA) repertoire in any prokaryotic or eukaryotic organism. This method includes a novel treatment that depletes total RNA fractions of highly abundant tRNAs and small subunit rRNA, thereby enriching the starting pool for sRNA transcripts with novel functionality. As a proof-of-principle, we applied sRNA-Seq to the human pathogen Vibrio cholerae. Our results provide information, at unprecedented depth, on the complexity of the sRNA component of a bacterial transcriptome. From 407 039 sequence reads, all 20 known V. cholerae sRNAs, 500 new, putative intergenic sRNAs and 127 putative antisense sRNAs were identified in a limited number of growth conditions examined. In addition, characterization of a subset of the newly identified transcripts led to the identification of a novel sRNA regulator of carbon metabolism. Collectively, these results strongly suggest that the number of sRNAs in bacteria has been greatly underestimated and that future efforts to analyze bacterial transcriptomes will benefit from direct cloning and parallel sequencing experiments aided by 5S/tRNA depletion.
INTRODUCTION
In bacteria, sRNAs that regulate diverse processes including quorum sensing, biofilm formation, stress responses, and virulence have been described (1,2). Most sRNAs characterized to date interact with specific mRNA targets, modulating mRNA stability or the efficiency of translation (3). While several experimental and bioinformatic approaches have proven useful in identifying sRNAs in diverse species, it is widely accepted that these approaches have yielded only a partial catalogue of these transcripts (4,5). Bioinformatic approaches for discovery of sRNA-encoding genes have often been limited to the subset of loci associated with predictable transcriptional signals and/or that are conserved in closely related species (6,7). Moreover, these computational screens have almost uniformly been limited to intergenic regions (IGRs) of the genome. Although most known bacterial sRNAs are encoded in IGRs, this does not preclude the possibility that there are sRNAs that are expressed within, or antisense (AS) to, protein coding sequences (4). Thus, even in Escherichia coli, the species in which the most extensive and diverse screens for sRNAs have been conducted, it has been suggested that the ∼80 known sRNAs represent only half of the entire sRNA repertoire (8).
Direct cloning, also known as shot-gun molecular cloning, is an approach by which small RNAs are identified through cDNA cloning of RNA transcripts. This method allows the identification of RNAs that are expressed in an organism under a given set of conditions regardless of whether they are encoded as distinct genetic elements or are generated via post-transcriptional processing. In recent years, direct cloning, in combination with parallel sequencing technology, has enabled researchers to ascertain the microRNA (miRNA) component of several eukaryote transcriptomes, such as those of Caenorhabditis elegans and Arabidopsis thaliana (9,10). In addition, using RNA-Seq methodology the transcriptome of Saccharomyces pombe, S. cerevisiae, A. thaliana, the laboratory mouse and human tissue have all been recently mined (11–15). These studies collectively demonstrate that previous transcriptome analyses have been incomplete and suggest that parallel sequencing is necessary for comprehensive identification of transcribed regions of the genome.
Massively parallel sequencing technology, or deep sequencing, has only recently been used to explore the transcriptomes of bacteria (16). In the lone example, only those transcripts that interacted with the sRNA-chaperone Hfq were sequenced. A major reason for this lag in prokaryotic transcriptome analysis through deep sequencing is the lack of a robust and readily adaptable method of removing highly abundant housekeeping RNAs, such as the tRNAs and rRNAs. Because they are significantly smaller than housekeeping RNAs, the direct cloning and sequencing of eukaryotic miRNAs is not hampered by abundant tRNA and rRNA sequences. In order to deplete tRNA and rRNA from eukaryote transcriptome analyses, the RNA-Seq technology uses poly-adenylated (PolyA) mRNA as starting material for direct cloning of mRNA populations. Bacterial sRNAs, which are not poly-adenylated, range from ∼30 to 300 nt in length, and are therefore not easily separated from the tRNAs (∼70 nt) and 5S rRNA (120 nt) species during RNA purification. Several commercial kits exist for the depletion of 16S and 23S rRNA from bacterial RNA preparations through either hybridization-based physical removal or ribonuclease activity. However, no methods are available for the removal of small subunit rRNA or bacterial tRNAs. A method was recently reported in which PCR products from direct cloning of Drosophila melanogaster small RNAs were screened prior to sequencing using a filter hybridization technique (17). This method, however, requires that each sequence be individually evaluated which may not be practical in massive-scale sequencing experiments. We therefore reasoned that a robust and unbiased method for the removal of bacterial tRNAs and 5S rRNA was needed to allow for more in depth analyses of prokaryotic transcriptomes, particularly the sRNA component.
To investigate the sRNA component of bacterial transcriptomes in an unbiased manner, we developed a method to directly clone and analyze whole populations of short bacterial transcripts, 14–200 nt in length, by parallel pyrosequencing (18). This protocol includes a treatment that depletes total RNA fractions of tRNAs and 5S rRNA, thereby enriching the starting pool for non-tRNA/rRNA transcripts. Because both the RNA species targeted for depletion and the size range of RNA to be sequenced are user-defined, sRNA-Seq represents a comprehensive cloning protocol that is versatile and readily applicable to the cloning of small RNAs of any size range from any organism. Here we present a proof-of-principle experiment in which we used sRNA-Seq to analyze Vibrio cholerae sRNAs. V. cholerae is a gamma-proteobacterium with a similar genome size and gene content as E. coli, and is a facultative human pathogen that causes the severe diarrheal disease cholera. Several sRNAs have been identified in V. cholerae through genetic screens and computational methods (19–21); it is likely, however, that more sRNAs, which are involved in diverse gene regulatory pathways, remain to be identified. Through the use of sRNA-Seq, we report the sequencing of 407 039 V. cholerae small transcripts, providing unprecedented coverage of the sRNA component of a bacterial transcriptome.
MATERIALS AND METHODS
Bacterial growth conditions
For sRNA-Seq and subsequent analysis of sRNA candidates, V. cholerae El Tor O1 clinical isolates N16961 and E7946, harboring derivatives of the pMMB67EH vector, were grown in Luria–Bertani (LB) broth at 37°C with aeration. For characterization of IGR7, V. cholerae N16961 was grown in LB or M9 minimal media supplemented with trace metals (1 ml/l of 5% MgSO4, 0.5% MnCl24H2O, 0.5% FeCl3, 0.4% trinitriloacetic acid) and either 0.4% glucose, 0.4% mannitol, or 0.4% glycerol. Arabinose was added at 0.02% final concentration to induce expression from the ParaBAD promoter. Antibiotics were added at the following concentrations: 100 µg/ml streptinomycin, 50 µg/ml ampicillin.
sRNA cloning
Each independent sample began with size-selecting RNA on preparative gels (Supplementary Figure 1A). For 14–60-nt transcripts (small fraction), 500 µg of total RNA, derived from phenol/chloroform extraction and isopropanol precipitation, was size-selected on a 15% denaturing polyacrylamide gel. For 60–200-nt transcripts (large fraction), 500 µg RNA was size selected on a 10% denaturing polyacrylamide gel. In each case, the extracted product was ligated to a 3′ linker (Linker 1, IDT, 5′-rAppCTGTAGGCACCATCATT/3ddC/-3′) as described (22,23). This was followed by a second gel purification; 1 nmol of the Oligo Mix was added to the extracted, linkered-RNA prior to ethanol precipitation. The Oligo Mix consisted of an equal molar mixture of 29 oligos complementary to either V. cholerae tRNAs or 5S rRNA (Supplementary Table 1). The precipitated mixture was resuspended in buffer (50 mM Tris–HCl, pH 7.8; 300 mM KCl; 10 mM MgCl2; 10 mM DTT) (24), heated to 65°C for 5 min, slow-cooled to 37°C, at which time 5 U RNaseH (NEB) was added. The reaction mixture was incubated at 37°C for 30 min. The depletion reaction was repeated for one additional cycle followed by another gel purification. The depleted, linkered-product was reverse-transcribed using Superscript II (Invitrogen) and the RT/REV primer (IDT, 5′-GATTGATGGTGCCTACAG) according to the manufacturer's instructions. ExoSAP-IT (USB) was added directly to the RT reaction, according to the manufacturer's instructions, to remove unused deoxynucleotides and primer. The reaction was terminated by phenol:chloroform treatment and ethanol precipitation, after which a second ligation was performed as above using another 3′ linker (Linker 2, IDT, 5′-rAppCACTCGGGCACCAAGGA/3ddC/-3) followed by another gel purification. The extracted material was amplified by PCR for 20 cycles with the following primers: Primer A: 5′-GCCTCCCTCGCGCCATCAGGATTGATGGTGCCTACAG-3′ and Primer B: 5′-GCCTTGCCAGCCCGCTCAGGTCCTTGGTGCCCGAGTG-3′ (sequences for 454 system underlined). The cDNA libraries were purified on a native gel. For each independent sample, the small and large fractions were combined in a 1:2 molar ratio, and the samples were submitted directly to the 454 Life Sciences Genome Sequencer 20 system for sequencing using 454 Primer B (18).
Analysis of sequencing data
The starting materials for the bioinformatics pipeline (Supplementary Figure 1B) were a set of four files in FASTA format, containing the results of the 454 sequencing runs for the four different sRNA-Seq samples. These files are provided herein as Supplementary Table 2. They contain a total of 681 205 reads.
Step 1: Removal of linker sequences and collapse of identical reads
Each read was examined in turn. If the sequence began with a complete 5′ linker sequence and ended with a complete 3′ linker sequence, the read was called a ‘perfect read’. Of the 681 205 total reads, 600 783 met this criterion. The remaining 80 422 reads were called ‘imperfect reads’, but most of these reads contained nearly complete linker sequences. The BLASTN algorithm (parameters: −W6 −S1 −e0.1) was used to identify the extent of the imperfect linker sequences in these reads. After linker sequences had been trimmed from all perfect and imperfect reads, a set of ‘unique sequences’ was created by collapsing together all reads with identical sequences. There were a total of 172 872 unique sequences.
Step 2: Identification of contaminant-derived sequences
Each unique sequence 14 nt or longer was compared using BLASTN (parameters: −e0.1) to a database containing the following sequences:
The genome of V. cholerae strain N16961 (Genbank accession numbers NC_002505 and NC_002506).
The genome of S. cerevisiae (NC_001133 − 001148 and NC_001224).
Sequence of cloning plasmid pMMB67EH.
Several miscellaneous sequences from other species, found to represent minor fractions of the contaminant pool.
The highest-scoring hit (first hit in the output list) was accepted as the source of the sequence. By setting an E-value cutoff of 0.1% in BLAST, any sequences 14 nt or shorter were not assigned to the V. cholerae genome; thus, sequences shorter than 15 nt were designated ‘short’. Sequences that failed to match any sequence in the database were designated ‘unidentified contaminant’.
A small fraction of the sequences were observed to have the unusual structure ‘BXA’, where the sequence ‘AB’ matched a contiguous stretch of the V. cholerae (or yeast) genome, and X was a short intervening sequence that did not match any sequence in either genome. These ‘shuffled’ sequences are believed to have formed from a circularized intermediate in the cloning process, although the details are unclear. For the purposes of the downstream analyses, the genomic match of the shuffled sequences was taken to be the full extent of ‘AB’, as if the sequences had had a ‘normal’ linear structure. The sequence of X, although usually very short (∼5 nt), was occasionally longer; however, a limit of 100 nt was imposed in the analysis.
Step 3: Genomic characterization of V. cholerae-derived sequences
Each of the 100 742 unique sequences that were found to match the V. cholerae genome was examined to determine what genomic features it overlapped with. The list of features in NC_002505 and NC_002506 was supplemented with the 20 known V. cholerae sRNAs that were previously confirmed by northern blot analysis (20,21). Most sequences overlapped only a single genomic feature or else lay entirely in intergenic regions (IGR). However, to deal systematically with cases of multiple overlaps, the following procedure was applied to all sequences. Each nucleotide along the length of the sequence was tabulated as being in one of the following nine classes: ORF, IGR, tRNA, rRNA, sRNA, or antisense to ORF, tRNA, rRNA or sRNA. Because some sets of overlapping sequences belonged to more than one of these nine classes, the class with the greatest tabulated count was accepted as the class of the sequence. For IGR sequences, note was taken of the closest upstream and downstream features and their distances.
Step 4: Merging of sequences with similar genomic endpoints into ‘transcripts’
Many of the 100 742 unique V. cholerae sequences were very similar to each other, only differing by one or two additional nucleotides at one or both ends. These differences may have resulted from minor transcriptional heterogeneity, and/or random events during the cloning. We wished to remove this source of variation from the data and unify similar sequences into putative ‘transcripts’. To accomplish this, the following steps were taken. First, each list of sequences was sorted by total number of reads, with the most frequent sequence at the top of the list. Then, starting with the sequence at the top of the list, all other sequences that mapped to the same genomic region (same strand of the same chromosome), with the starting and ending coordinates both matching within ±10 bp of that first sequence, were merged into a ‘transcript’ along with the first sequence, and their original entries were removed from the list. This process continued down the list until all sequences had been examined. At the end of this process, the original 100 742 sequences had been merged into 16 825 transcripts. For each transcript, we calculated the range (min and max), average, and standard deviation of the length, 5′ coordinate, and 3′ coordinate (Supplementary Table 3).
Step 5: Filtering of transcripts to create list of ‘candidate sRNAs’
We applied additional criteria to remove likely uninteresting entries from the transcript list. First, we removed any transcript that was observed in only a single sample out of the four independent samples. This reduced the size of the list to 2846 transcripts. Next, we removed transcripts of rRNA and tRNA, and transcripts originating from the intergenic regions between rRNA and tRNA genes. This further reduced the length of the list to 2140 candidates.
Candidate sRNA analysis
Total RNA was prepared by phenol/chloroform extraction and isopropanol precipitation and represented samples independent from the ones used in the sRNA-Seq experiment. Generally, northern blots were performed with RNA probes transcribed from PCR-derived templates (Supplementary Table 4) with T7 promoters using 32P-UTP and T7 polymerase (Promega) according to the manufacturer's instructions. In some cases, hybridizations were performed using DNA oligonucleotides (Supplementary Table 4) 5′-end-labeled with 33P-ATP. Low molecular weight DNA ladder (NEB) was run alongside RNA samples to provide estimations for the sizes of RNA bands. Total RNA (10–40 µg) was run on denaturing polyacrylamide gels (10% or 15%) and transferred to Hybond N+ membranes (Amersham) in 1× TBE using the Mini Trans-Blot Cell apparatus (Bio-Rad) according to the manufacturer's instructions. Blots were prehybridized in Ultrahyb or Ultrahyb-oligo buffer (Ambion) according to instructions.
Reverse transcription of total RNA was performed with reverse primers specific to the sRNA candidate of interest using the RETROscript kit (Ambion) according to the manufacturer's instructions. The cDNA generated, or N16961 genomic DNA, was used as a template for PCR with reverse and forward primers specific to each candidate. PCR reactions were performed with an initial denaturation step of 5 min at 95°C, followed by 26 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Control reactions were performed for each run and included RNA samples not treated with reverse transcriptase or samples lacking template DNA. In all cases, no band was observed in these controls. All primers used for this analysis are listed in Supplementary Table 4.
Analysis of IGR7
The IGR7 sequence and its antisense sequence were amplified by PCR (Supplementary Table 5) and cloned into the NheI and HindIII sites of pJML01. This plasmid, generated by site-directed mutagenesis (CTCCAT to GCTAGC), is a derivative of pBAD24 (25) with a new NheI site downstream of ParaBAD. sRNA genes cloned behind the ParaBAD promoter in pJML01 are expressed with the proper +1.
Growth of strains was determined by measured OD600 using a Bio-Tek microplate reader. For each strain, individual colonies were first grown in LB + arabinose for 2–3 h at 37°C, at which time the cultures were centrifuged, and the cell pellets washed and resuspended in M9 medium. The OD600 was adjusted to 0.01 in 200 µl of M9-glycerol or M9-mannitol, with arabinose. Bacteria were grown with aeration for 30 h at 37°C in the microplate reader. All growth experiments were performed in triplicate.
For qRT–PCR analysis, bacteria were grown in LB or M9 media at 37°C with aeration. Total RNA was isolated from mid-exponential phase cultures using the RNeasy Mini Kit according to the manufacturer's protocols, with a modification to include sRNAs (<200 nt) in the final preparation (Qiagen). DNA was removed from all samples using the DNAfree kit, according to the manufacturer's instructions (Applied Biosystems). cDNA was made using ∼0.5 µg total RNA, specific primers, and the iScript Select cDNA Synthesis Kit, according to the manufacturer's instructions (BioRad). qPCR reactions were performed with iQ SYBR Green Supermix (BioRad) using Strategene Mv3005P equipment and MxPro qPCR software. At least three independent samples were tested for each condition and each template sample was tested in duplicate. In all cases, controls lacking reverse transcriptase were included and afforded results below the baseline of detection. All primers for qRT–PCR analysis are listed in Supplementary Table 5.
RESULTS
Direct cloning and parallel sequencing of V. cholerae sRNAs
Whereas miRNAs (20–22 nt) can be readily separated from tRNA and rRNA sequences, bacterial sRNAs often overlap in length with highly abundant tRNA and 5S rRNA transcripts. In order to address this problem, we developed a 5S/tRNA depletion step that results in an enrichment of sRNAs (23) (‘Materials and methods’ section and Supplementary Figure 1). Total RNA was isolated from exponential or early-stationary phase V. cholerae cultures. RNA (14–200 nt) was isolated and a specialized 3′ linker was ligated to the end of the transcripts. The linkered-RNAs were then annealed to a pool of oligodeoxynucleotides, each 29 nt, that were complimentary to V. cholerae 5S rRNA and the 3′-ends of V. cholerae tRNA (Supplementary Table 1); the resulting mixture was treated with RNaseH, which specifically targets DNA/RNA hybrids. This RNaseH treatment effectively removes tRNAs and 5S rRNA from downstream analysis by separating these transcripts from the 3′ linker that is necessary for amplification and addition of the 454 sequencing linkers. In addition, because the oligodeoxynucleotides and RNaseH are not consumed in the reaction, the depletion of 5S/tRNA proceeds enzymatically. The 5S/tRNA-depleted pool of RNA was then used to produce cDNA by reverse transcriptase and a second linker was ligated to the 3′-end of the cDNA products. The linkered-cDNA products were amplified by PCR and the amplicons submitted for sequencing with the 454 Life Sciences Genome Sequencer 20 system (18). Although the Solexa and SOLiD systems provide many more sequence reads, we reasoned that the longer (>100 nt) read lengths provided by the Genome Sequencer system would facilitate unambiguous identification of bacterial sRNAs that can range up to several hundred nucleotides.
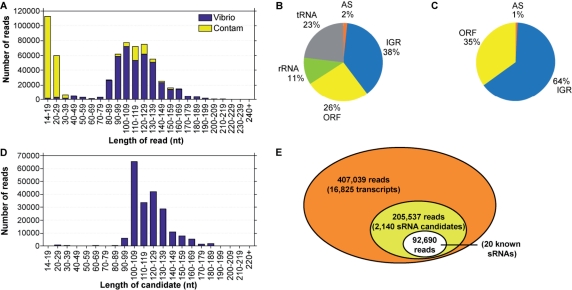
Four independent samples were prepared, affording a total of 681 205 sequence reads of which 88% were full length (with complete and perfect 5′ and 3′ linkers) (Supplementary Table 2). The resulting sequences trimmed of linkers had the length distribution shown in Figure 1A. Using the BLASTN algorithm (E-value cutoff of 0.1) on the 626 351 reads 15 nt and longer, we found that 407 039 of them matched the V. cholerae genome (Table 1). Based on analysis of all possible n-mers, of the 3066 unique 15 nt reads that we assigned as being V. cholerae transcripts, it is likely that at most 0.4% of them (∼12), are false positives. We also observed many small transcripts that mapped to the yeast genome (Table 1). We hypothesize that the media that the bacteria were grown in was the source of these contaminating sequences. Additional unidentified sequences may be the result of trace DNA contaminating materials used during the cloning process. Regardless of this contamination, the large majority of the reads >40 nt mapped to the V. cholerae genome. We observed that 73% of the V. cholerae reads matched the genome perfectly, 12% had one mismatch or gap, 6% had multiple mismatches or gaps and a small fraction (9%) represented transcript recombinations that were likely artifacts resulting from the cloning process (‘BXA’ reads; see ‘Materials and methods’ section). We then took all the 407 039 reads that mapped to the V. cholerae genome and grouped them into putative transcripts by combining sequences with genomic coordinates that agreed within ± 10 bp at each end. Based on this method of grouping reads together, the sequences that mapped to the V. cholerae genome represented 16 825 unique transcripts, 9% of which corresponded to rRNA or tRNA sequences. Overall, 34% of the total V. cholerae reads analyzed were derived from rRNA or tRNA (Figure 1B). Our previous attempt to directly clone 14–200 nt RNA resulted in 93% (180/193) 5S/tRNA sequences. The sRNA-Seq results therefore suggest that the 5S/tRNA depletion step was successful in eliminating the majority of these highly abundant transcripts.
Figure 1.
Results from V. cholerae sRNA-Seq experiment. (A) Length distribution of all 681 205 reads from 454 sequencing. Reads that match the V. cholerae genome are in blue; reads that represent contaminants are in yellow. (B) Breakdown of total V. cholerae reads from 454 sequencing based on their genomic origin (n = 407 039). ORF, annotated open reading frames; AS, transcripts antisense to ORFs; IGR, transcripts from intergenic regions. (C) Breakdown of sequencing reads that correspond to candidate sRNAs based on their genomic origin (n = 205 537). The IGR-derived candidates include the 92 690 reads of known or previously predicted V. cholerae sRNAs. (D) Length distribution of sRNA candidates. The 2140 sRNA candidates, corresponding to 205 537 total reads, are plotted based on the length of the most abundant sequence observed for each candidate. (E) Visual representation of the depth of the sRNA-Seq data. Approximately 50% of the reads mapping to the V. cholerae genome grouped into candidate sRNAs that were found in two or more samples (yellow). All the previously known and verified V. cholerae sRNAs (white) were amongst these candidate reads; this includes the 20 sRNAs of known function and the 5 sRNAs predicted and verified by northern blot analysis (20,21).
Table 1.
Breakdown of all sequences from 454 pyrosequencing
| N | Percentage | |
|---|---|---|
| V. cholerae | 407 039 | 60 |
| Yeast (rRNA) | 107 217 | 16 |
| Yeast (tRNA) | 31 586 | 5 |
| Yeast (RNA) | 27 227 | 4 |
| Other/unidentified | 57 448 | 8 |
| Short (<15 nt) | 50 688 | 7 |
| Total | 681 205 | 100 |
Identification of candidate V. cholerae sRNAs
The 16 825 unique transcripts were mapped to the V. cholerae genome and Figure 1B indicates the breakdown of all corresponding 407 039 reads, categorized by the region of the genome from which they are derived. Over one-fourth of the total reads correspond to annotated open reading frames (ORFs). These transcripts may represent the following: intact messenger RNAs (mRNAs) corresponding to small proteins; unstable degradation products from longer mRNAs; stable degradation or processed products from longer mRNAs; or functional non-coding transcripts whose promoters lie within existing ORFs. To differentiate between these possibilities, and to generally enrich our data set for candidates that may represent functional sRNAs, we filtered the data set and kept only those sequences that were observed in two or more of the independent samples submitted for 454 sequencing. We reasoned that this would remove the bulk of the transcripts that represented randomly derived RNA degradation products. We considered any transcript that passed these filtering criteria, and did not represent tRNA/rRNA or the intergenic sequences between tRNA/rRNA, to be an sRNA candidate.
Post-filtering, we identified 205 537 reads corresponding to 2140 sRNA candidates (Figure 1E and Supplementary Table 3). Importantly, of the sRNAs previously identified and verified in V. cholerae, all 20 were observed to meet our filtering criteria (20,21). Figure 1C contains the breakdown of the post-filter reads. IGR transcripts are abundant, representing 64% of the reads. IGRs represent 12% of the V. cholerae genome, indicating that IGRs are highly enriched for sRNAs over other regions of the genome. Our data set also indicates that small transcripts derived from the antisense strand of known ORFs or from within annotated ORFs represent a significant number of candidate non-coding RNAs. The candidates derived from within ORFs may represent functional sRNAs or they may encode small peptides (26). Together, these results suggest that many sRNAs are derived from the sense and antisense strands of ORFs, regions of the genome excluded from most previous bioinformatic sRNA screens.
Characterization of candidate sRNAs
Using our method of assigning putative transcripts by grouping sequences that agreed within ±10 bp at each end, we observed 500 previously unidentified sRNAs transcribed from IGRs (IGR-sRNAs) and 127 new sRNAs transcribed from the antisense strand of annotated ORFs (AS-sRNAs). The lengths of the final set of candidate sRNAs ranged from 16–225 nt (Figure 1D). To verify that these candidates indeed represent observable transcripts in vivo, we selected several candidates from each class of sRNA (IGR-, AS- and ORF-derived) to characterize further (Table 2). For six out of seven randomly chosen IGR-sRNA candidates, we were able to observe by northern blot analysis a transcript of expected size (Supplementary Figure 2). These results suggest that many of the sRNA candidates are components of the V. cholerae transcriptome. The northern blots also indicated that some of the observed sRNAs are likely transcribed as larger precursors (200–300 nt) that are processed into smaller species. We hypothesize that the 200 nt cutoff we used during the cloning process prevented us from observing such larger precursors in our 454 dataset.
Table 2.
Analysis of candidate sRNAs
| Gene(s)a | Coordinatesb | ntb | NBc | Rfamd | blastne | Promf | Termf | |
|---|---|---|---|---|---|---|---|---|
| IGR | ||||||||
| IGR1 | VC0019<< VC0020<< | 16673–16801 (+) | 129 | + | – | ++g | + | – |
| IGR2 | VC1130<< VC1131>> | 1199011–1199144 (+) | 134 | +/− | – | + | – | – |
| IGR3 | VC2384<< VC2385>> | 2549444–2549562 (+) | 119 | + | msr | +g | – | + |
| IGR4 | VCA0518<< VCA0519>> | 449220–449323 (−) | 104 | + | – | Only Vc | + | – |
| IGR5 | VCA0279>> VCA0281>> | 300143–300253 (+) | 110 | + | – | +g | + | – |
| IGR6 | VC0175<< VC0176<< | 177132–177267 (−) | 136 | + | – | Only Vc | + | – |
| IGR7 | VCA1044>> VCA1045>> | 994875–994876 (−) | 120 | + | – | + | – | – |
| Antisense | ||||||||
| AS1 | VC0512 | 542160–542294 (−) | 135 | – | – | + | + | – |
| AS2 | VC0658 | 701570–701732 (+) | 163 | – | – | + | – | – |
| AS3 | VC2063 | 2217964–2218094 (+) | 131 | – | – | ++ | – | – |
| AS4 | VC2203 | 2349040–2349128 (−) | 89 | – | – | + | + | – |
| AS5 | VC1225 | 1301660–1301792 (−) | 133 | + | – | +g | – | – |
| AS6 | VC2332 | 2482288–2482403 (+) | 116 | – | – | + | + | – |
| AS7 | VCA0644 | 578617–578785 (−) | 169 | + | – | ++ | – | – |
| AS8 | VCA0676 | 611959–612076 (−) | 118 | +/− | – | + | + | – |
| AS9 | VC2269 | 2424422–2424525 (+) | 104 | + | – | ++ | + | – |
| ORF | ||||||||
| ORF1 | VCA1078 | 1031452–1031586 (+) | 135 | – | – | ++ | – | – |
| ORF2 | VC2540 | 2722407–2722525 (−) | 119 | + | – | ++ | – | – |
| ORF3 | VC0215 | 222692–222827 (−) | 136 | + | – | ++ | – | – |
| ORF4 | VC0913 | 974460–974582 (+) | 123 | + | – | + | – | – |
| ORF5 | VC1499 | 1611161–1611276 (+) | 116 | + | – | + | + | – |
| ORF6 | VC0689 | 736177–736300 (+) | 124 | – | – | + | + | – |
| ORF7 | VC0957 | 1021523–1021633 (+) | 111 | + | – | + | – | – |
aFor IGR-sRNA candidates, the gene numbers for the up- and downstream ORF are indicated. Genes encoded on the plus strand are denoted with >> and genes encoded on the minus strand are denoted with <<.
bCoordinates and length reported here represent the most abundant sequence corresponding to each candidate transcript. (+) plus strand; (−) minus strand.
cNorthern blot analysis; (+) band of expected size observed; (+/−) bands of significantly higher or lower molecular weight than expected observed; (–) no bands observed.
dCandidate was queried against the Rfam database (http://www.sanger.ac.uk/Software/Rfam/index.shtml); matches to known sRNAs are indicated.
eSequence conservation of candidate in other microbial organisms was queried using the BLASTN algorithm. (Only Vc) sequence was not observed in other bacterial species; (+) sequence was conserved primarily in Vibrionaceae; (++) sequence was conserved in many bacterial species.
fCandidates (including the 100 nt up- and downstream) were analyzed using BPROM and FindTerm software (Softberry, Mount Kisco, NY). (+) promoter/terminator was predicted for candidate; (–) promoter/terminator not predicted for candidate.
gMultiple hits within V. cholerae N16961 genome observed by BLASTN analysis.
We next performed northern blot analyses for the AS- and ORF- sRNA candidates. AS-sRNAs may pair with their partner mRNA and be rapidly degraded (3), thereby proving more difficult to observe as a full-length species. Nevertheless, northern blot analysis supported the identification of four out of nine randomly chosen AS-sRNA candidates; in some cases, multiple transcripts were observed by northern blot (AS5 and AS8) (Supplementary Figure 2). Similarly, five out of seven candidate sRNAs transcribed from within an annotated ORF (ORF-sRNA) were observed by northern blot analysis (Supplementary Figure 2). These results provide confirmation of these transcripts by means independent of the sRNA-Seq method. The ORF-sRNA candidates queried were chosen because they represented transcripts that were >15-fold more abundant than any other sRNA transcripts derived from the same ORF, in at least one of the four sRNA-Seq samples (Supplementary Figure 3). For comparison, the transcript abundance profile for 14 V. cholerae sRNAs of known function are shown in Supplementary Figure 4. Two of these ORF-sRNAs (ORF2 and ORF7) are transcribed within ORFs that are less that 500 bp long (VC2540, 393 bp; VC0957, 471 bp, respectively). These ORFs are annotated as encoding hypothetical proteins; however, in our northern blots we do not observe the full-length mRNA transcripts (Supplementary Figure 2). It is possible that under the conditions examined here, the mRNA is not expressed; alternatively, our deep sequencing results may have revealed regions of the genome that have been mis-annotated as coding sequences.
The sRNA-Seq methodology also provides information regarding the 5′ and 3′-ends of the sequenced transcripts (Supplementary Table 3). To investigate whether sRNA-Seq accurately maps the ends of sRNA transcripts, we performed reverse transcription on total V. cholerae RNA with specific primers for several candidate sRNAs. For each of five candidates analyzed, a PCR product of expected size was observed when the cDNA from the reverse transcription reaction was amplified with forward and reverse primers that annealed to regions within the putative sRNA gene (Supplementary Figure 5). When the forward primer was replaced with ones that annealed 20 or 50-nt upstream of the sequenced 5′-end of the sRNA, no PCR product was observed (Supplementary Figure 5). These results further suggest that the information provided by sRNA-Seq identifies candidate sRNA transcripts and accurately identifies the 5′-end of these transcripts.
For all 16 candidate sRNAs confirmed by northern blotting, as well as the 20 known sRNAs, we visually inspected their sequences and could find no ribosome-binding site with an appropriately spaced start codon. The conservation of the candidate sRNA sequences was also investigated. While some sRNAs, such as IGR7, were highly conserved in other bacteria, particularly other Vibrionaceae, several sRNAs (IGR4, IGR6) were not identified in other bacteria by BLASTN analysis (Table 2). In addition, we analyzed the candidate sRNAs for nearby promoters and Rho-independent terminators using BPROM and FindTerm software available from Softberry (Mount Kisco, NY). For most candidate sRNAs we were unable to identify putative promoters and terminators using such prediction algorithms (Table 2). Taken together, our findings suggest that computational approaches for sRNA identification that rely on primary sequence conservation and/or on promoter and Rho-independent terminator detection are likely effective in identifying only a small subset of non-coding transcripts.
sRNA candidate IGR7 is involved in carbon metabolism
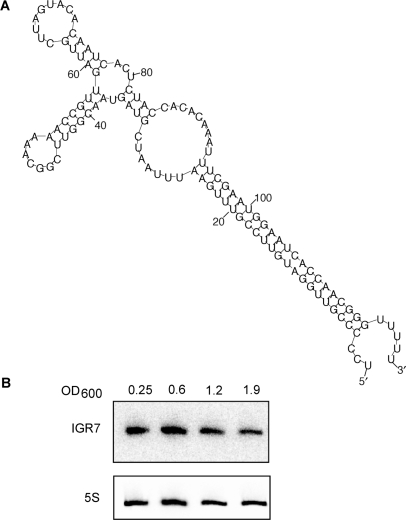
Preliminary functional characterization was also performed on one of the candidate sRNAs, IGR7 (Table 2 and Figure 2A). Northern blot analysis suggested that this transcript is constitutively expressed as a 120-nt transcript throughout V. cholerae growth in rich medium (Figure 2B). This candidate sRNA is transcribed antisense to the 5′-untranslated region (UTR) of mtlA (VCA1045), which encodes a transporter for mannitol, a naturally occurring six-carbon sugar alcohol. Using the sRNA-Seq data, we were able to map the 5′ and 3′-ends of IGR7; the transcript begins five nucleotides upstream of the mtlA start codon and ends 120-nt downstream after a series of five Us. The sequencing data strongly suggests that this 120-nt transcript is the predominant sRNA transcribed from this region of the V. cholerae genome.
Figure 2.
sRNA candidate IGR7. (A) Predicted secondary structure of candidate IGR7 (27,28). (B) Northern blot analysis of IGR7 expression (top panel) throughout growth of V. cholerae in LB medium. Total RNA samples were prepared at the indicated OD600. 5S rRNA is shown as a loading control (bottom panel).
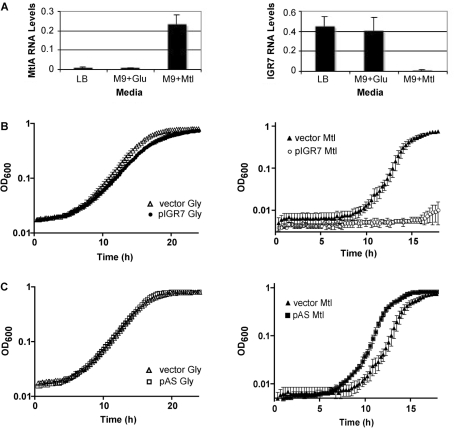
We reasoned that in V. cholerae IGR7 might be involved in regulating the expression of this sugar transporter and chose to investigate, quantitatively, the expression of IGR7 and mtlA in V. cholerae grown in different conditions. Similar to rich medium growth, in glucose minimal medium, IGR7 is expressed at high levels and mtlA mRNA levels are very low (Figure 3A). Conversely, in mannitol minimal medium, mtlA mRNA levels increase ∼20-fold and there is a reciprocal 20-fold decrease in IGR7 expression (Figure 3A). Under these conditions, the intracellular abundance of IGR7 had no effect on the expression of VCA1044, the gene upstream of IGR7 and mtlA and which codes for a hypothetical protein (data not shown). When IGR7 was overexpressed in trans from a plasmid, growth of V. cholerae was abrogated in mannitol medium (Figure 3B, right panel). This effect was specific to growth on mannitol since overexpression of IGR7 had no effect on growth in glycerol medium (Figure 3B, left panel). A knock-down of IGR7 through overexpression of an antisense RNA, AS-IGR7, resulted in a more rapid start of growth in mannitol medium (Figure 3C, right panel), suggesting that the cells were primed for growth in this carbon source. AS-IGR7 had no affect on bacterial growth in glycerol medium (Figure 3C, left panel). Collectively, these observations indicate that IGR7 is involved in the regulation of mannitol metabolism in V. cholerae likely through control of mtlA expression. In addition, our results demonstrate that sRNA-Seq was successful in identifying a functional and previously unknown sRNA.
Figure 3.
Analysis of IGR7 sRNA. (A) Intracellular abundance of mtlA mRNA and IGR7 sRNA, relative to 4.5S, as measure by qRT-PCR. Error bars represent standard deviations of three or more independent trials. (B) Growth curves of derivatives of V. cholerae N16961 harboring empty vector (pJML01) or plasmid expressing IGR7 from an arabinose-inducible promoter (pIGR7). All samples were grown in M9 media supplemented with 0.4% glycerol (Gly) or 0.4% mannitol (Mtl). Arabinose was added at a final concentration of 0.02% to all samples. All samples grown without arabinose grew similar to each other and to V. cholerae without plasmid (data not shown). (C) Growth curves of derivatives of V. cholerae N16961 harboring empty vector (pJML01) or plasmid expressing AS-IGR7 from an arabinose-inducible promoter (pAS). Growth conditions were the same as in (B). Error bars represent standard deviation of three independent trials.
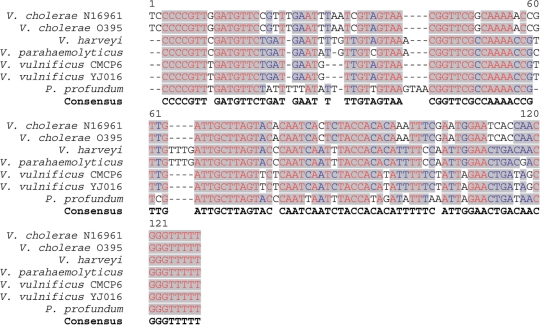
The sequence of IGR7 is highly conserved in all Vibrionaceae, as well as in Photobacterium profundum (Figure 4); in all these species, the sRNA gene is present upstream of the mtlA homolog. In addition, the predicted secondary structures of the IGR7-homologs in these other species are all similar to the one shown in Figure 2A: a long stem with one to two bulges and two stem-loops (data not shown). Although E. coli has an mtlA homolog, there was no homologous IGR7 sequence upstream of this gene. We analyzed the E. coli transcriptome by northern blot analysis and did not observe an sRNA transcribed antisense to the 5′-UTR of the mtlA gene (data not shown). This observation suggests that Vibrio and Photobacterium species may be unique in their use of an sRNA to regulate mannitol metabolism.
Figure 4.
Alignment of the IGR7 homolog sequences.
DISCUSSION
Our understanding of non-coding RNAs and gene regulation networks, in both prokaryotes and eukaryotes, has grown exponentially in the past 20 years. Remarkable advances in biotechnology in recent years have further fueled our attempts to understand the complexity of genomes and their programmed transcriptomes. Most studies on sRNAs in prokaryotes have focused on the E. coli transcriptome; collectively, ∼80 sRNAs have been identified in this model organism. Bioinformatic analysis and genetic screens have also led to the identification of sRNAs in many other bacteria, including V. cholerae. Nevertheless, the findings we report here suggest that the transcriptomes of bacteria remain ill-defined and that the number of sRNAs in bacteria has been greatly underestimated.
Prior to this work, the use of direct cloning and massively parallel sequencing had been generally limited to miRNAs that are much smaller than the abundant ‘housekeeping’ RNAs, including tRNAs and rRNAs, or Poly(A) mRNA. The 5S/tRNA depletion method we present here should allow for more in-depth investigations of a variety of small RNAs in a wide array of organisms. The sRNA-Seq approach we developed in this work is readily applicable to other prokaryotic and eukaryotic species and we anticipate that improvements in parallel sequencing technology will allow even more extensive and economical explorations of transcriptomes to be performed. The 5S/tRNA depletion step, moreover, can be extended to the removal of any RNA of known sequence based on the principle of separating the unwanted RNA from the necessary 3′ linker. For example, future efforts may include removal of additional housekeeping sRNAs such as the 4.5S and 6S RNAs. Additional studies to further improve upon this new depletion technique are currently underway. In addition, it should be possible to precede the sRNA-Seq direct cloning protocol with treatment of total RNA with commercial kits for the removal of the 16S and 23S rRNAs; the resulting cDNA libraries could be used for general transcriptome analyses.
Our sRNA-Seq results suggest that while most sRNAs are indeed expressed from intergenic sequences, a significant portion of bacterial sRNAs is derived from protein-encoding regions of bacterial genomes, regions excluded from nearly all previous computational screens for sRNA-encoding loci. Through northern blot analysis, we were able to confirm the expression of most sRNA candidates queried. In addition, our investigations into the function of these sRNAs led to the discovery of a novel regulator of carbon metabolism in V. cholerae. This sRNA, IGR7, is abundant during growth in rich medium and appears to negatively regulate a partially overlapping mRNA encoding a mannitol transporter. The sequencing data also allowed us to map the 5′- and 3′-ends of IGR7 and other candidate sRNAs. Collectively, these results suggest that sRNA-Seq studies can complement existing techniques used to identify sRNAs. We anticipate that sRNA-Seq can also be used to analyze changes in transcriptomes as bacteria are subjected to different growth conditions. Since sRNA-mediated regulation plays a central role in a myriad of biological processes, the comprehensive and unbiased profiles of sRNA repertoires afforded by sRNA-Seq will likely yield important insights that will advance our understanding of gene regulation networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [AI45746 to A.C.]. A.C. and M.K.W. are investigators of the Howard Hughes Medical Institute. National Institutes of Health/NIGMS fellowship [to J.M.L.]. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gottesman S. Micros for microbes: non-coding regualtory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Romby P, Vandenesch F, Wagner EGH. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 7.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Gene Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang Z, Ling L, Shi B, Chen R. Conservation analysis of small RNA genes in Escherichia coli. Bioinformatics. 2004;20:599–603. doi: 10.1093/bioinformatics/btg457. [DOI] [PubMed] [Google Scholar]
- 9.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 10.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C-elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 11.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Meth. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 15.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A Global View of Gene Activity and Alternative Splicing by Deep Sequencing of the Human Transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 16.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan GZ, Klambt C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 2003;31:2495–2507. doi: 10.1093/nar/gkg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Livny J, Fogel MA, Davis BM, Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. 2005;33:4096–4105. doi: 10.1093/nar/gki715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 23.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C-elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Lee HJ, Corn RM. Fabrication and characterization of RNA aptamer microarrays for the study of protein-aptamer interactions with SPR imaging. Nucleic Acids Res. 2006;34:6416–6424. doi: 10.1093/nar/gkl738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-Bad promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadler CS, Vanderpool CK. A dual function for a bacteria small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves predicition of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold web server for nucleic acid folding and hyrbidization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.