Abstract
We present here a method to correlate tandem mass spectra of sample RNA nucleolytic fragments with an RNA nucleotide sequence in a DNA/RNA sequence database, thereby allowing tandem mass spectrometry (MS/MS)-based identification of RNA in biological samples. Ariadne, a unique web-based database search engine, identifies RNA by two probability-based evaluation steps of MS/MS data. In the first step, the software evaluates the matches between the masses of product ions generated by MS/MS of an RNase digest of sample RNA and those calculated from a candidate nucleotide sequence in a DNA/RNA sequence database, which then predicts the nucleotide sequences of these RNase fragments. In the second step, the candidate sequences are mapped for all RNA entries in the database, and each entry is scored for a function of occurrences of the candidate sequences to identify a particular RNA. Ariadne can also predict post-transcriptional modifications of RNA, such as methylation of nucleotide bases and/or ribose, by estimating mass shifts from the theoretical mass values. The method was validated with MS/MS data of RNase T1 digests of in vitro transcripts. It was applied successfully to identify an unknown RNA component in a tRNA mixture and to analyze post-transcriptional modification in yeast tRNAPhe-1.
INTRODUCTION
Accumulating evidence shows that diverse types of small RNAs generated from non-coding (nc) regions of the genome play pivotal roles in a variety of cellular processes, such as chromatin remodeling (1), transcriptional regulation (2), precursor mRNA processing (3), gene silencing (4), centromere function and translational regulation (5). In most cases, those nc RNAs function as a part of ribonucleoprotein (RNP) complexes and exert their activities on endonucleolytic RNA cleavage and ligation, site-specific RNA modulation, DNA methylation and telomere synthesis. ncRNAs frequently secure and position a nucleic acid target molecule for enzymatic activity that is catalyzed by an associated protein, and thus ncRNA activity is typically driven by base pairing (6,7).
Our knowledge of the biogenesis pathway of functional RNP complexes is still limited; however, studies of a number of RNP complexes that are fundamental for cell viability, such as the spliceosome, ribosome and RNA-induced silencing complex, suggest that the assembly of RNP complexes involves a complex series of events performed not only by the components of the final functional complex, but also by various additional ncRNAs and ‘trans-acting’ protein factors that regulate the intermediate processes of biogenesis and ensure the quality of the final products (8–12). The deregulation of RNP complexes often leads to severe pathology including tumorigenesis (13), tumor metastasis (14) and abnormal morphogenesis (15), indicating that the biogenesis pathway is under strict control (16). Thus, the analysis of the structure, function and biogenesis of functional RNP complexes is crucial for understanding normal and aberrant cellular processes.
Current mass spectrometry (MS)-based proteomics technology, coupled with various tagging technologies to isolate protein complexes, allows the comprehensive identification and quantitative analysis of protein components in many RNP complexes (11). The analysis of RNA components in RNP complexes, on the other hand, is currently carried out using mainly techniques based on genomics and RNA biochemistry, which include the process of reverse transcription from RNA to cDNA. Although this technique is useful for various aspects of RNA research, the method suffers from the relatively high error rate of the reverse transcriptase, which arises from the presence of both RNA secondary structure and base modifications, and from a lack of truly quantitative results because of the substrate specificity of the reverse transcriptase. In addition, the conventional approach does not provide the structural information on post-transcriptional modifications of nucleosides (17), which are common in tRNA and rRNA and are essential for their biogenesis and function (8,18–20). MS offers a sensitive method for the direct chemical analysis of RNA and therefore is ideally suited as a complementary method to conventional techniques.
There are numerous published papers that describe the development and application of MS for nucleic acids analysis (21). In particular, MS has been used in combination with biochemical techniques to identify various types of post-transcriptional modifications in tRNA, rRNA and ncRNA (18,22). For instance, Emmerechts et al. (23) applied liquid chromatography (LC)–MS analysis of oligonucleotides in combination with a reverse transcriptase assay and determined 11 modified nucleosides in Clostridium acetobutylicum 16S rRNA. Noma et al. (24) applied MS for modified nucleoside analysis and revealed a pathway to synthesize wybutosine (yW) from guanosine in eukaryotic tRNAPhe, an important post-transcriptional modification for the tRNA function. Ohara et al. (17) found 2′-O-methylation at the 3′-termini of Piwi-interacting RNAs, a germline-specific small ncRNA that is essential for spermatogenesis. MS has also been used for the chemical analysis of nucleic acids and oligonucleotides including RNA transcripts and synthetic RNA, as well as for the identification of oligonucleotides by MS-based fingerprinting (25). Mcluckey et al. (26) first described the collision-induced dissociation (CID) profiles of multiply charged anions of nucleic acids generated by electrospray ionization (ESI), and Schuerch et al. (27) studied the fragmentation profiles of mixed-sequence RNA/DNA oligonucleotides. Ni et al. (28) and Oberacher et al. (29) studied the fragmentation profiles of multiply charged oligodeoxynucleotide ions and developed algorithms for the interpretation of fragmentation ion spectra: Rozenski and McCloskey (30) and Oberacher et al. (31) implemented the algorithm into software to aid de novo sequencing of short oligonucleotides with approximately 15 nt. Thus, the previous studies demonstrated the potential of MS in various aspects of nucleic acids research, including automated tandem MS-based sequencing of short oligonucleotides and analysis of post-transcriptional modifications of RNA. There is, however, no method currently available that correlates a tandem mass spectrum with nucleotide sequences from a DNA/RNA database and allows tandem MS-based identification of RNAs in biological samples.
Here, we present a novel method for automated identification of RNA by using tandem MS data from RNA nucleolytic fragments to search against a DNA/RNA sequence database. The method uses (i) nucleolytic fragmentation of sample RNA with an RNase, such as RNase T1, that has defined cleavage specificity, (ii) LC–MS/MS analysis of the fragments and (iii) analysis of the MS/MS dataset by ‘Ariadne’, a database search engine that we developed. We examined the performance of the method with a number of applications to identify mRNA synthesized in vitro or small RNA isolated from biological samples. We show here that Ariadne is the first genome-oriented search engine for RNA analysis that could be equivalent to the many sequence search engines that are widely used for proteomics, such as SEQUEST (32) or Mascot (33). Furthermore, we show that the method described here is useful for MS-based identification and chemical analysis of small RNAs in biological samples, including RNA components in RNP complexes. Ariadne can be accessed across the World Wide Web at http://ariadne.riken.jp/.
MATERIALS AND METHODS
Chemicals
Standard laboratory chemicals and 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) were obtained from Wako Pure Chemical Industries (Osaka, Japan). High-performance liquid chromatography-grade methanol and acetonitrile were purchased from Kokusan Chemical Co. (Tokyo, Japan), triethylamine from Pierce (Thermo Fisher Scientific Inc., Rockford, IL, USA) and 2 M triethylamine acetate buffer (pH 7.0) from Glen Research (Sterling, VA, USA). A mixture of yeast tRNAs was obtained from Sigma (St Louis, MO, USA). Ribonuclease T1 (RNase T1; highly purified) was purchased from Worthington (Lakewood, NJ, USA) and purified by reversed-phase LC.
In vitro transcription of mRNAs
The coding region of Xenopus cyclophilin A (xCyPA) cDNA (NM_001089190.1) was inserted into pBluescript RN3 vector (34) (provided by Dr Makoto Asashima of The University of Tokyo) at BglII and EcoRI sites downstream of the T3 promoter. The globin-fused xCyPA mRNA with cap modification and poly(A) tail was synthesized from the template xCyPA-inserted RN3 vector (1 µg; linearized by cleavage at PstI) using an in vitro transcription kit (mMESSAGE mMACHINE, Ambion, Austin, TX, USA). After the digestion of template RN3 vector with DNase I, the synthesized mRNA was purified using the RNeasy kit (QIAGEN GmbH, Hilden, Germany). The purity of the mRNA as well as removal of the template RN3 vector was confirmed by 1% agarose gel electrophoresis.
Preparation of RNase T1 digests
RNase T1 digestion of RNA was performed in 10 mM sodium acetate buffer (pH 5.3) at 37°C for 30 min at an enzyme/substrate ratio of 1:500 (w/w). The digests were analyzed immediately by nanoflow LC–MS or were stored frozen at −20°C.
Nanoflow LC–MS/MS analysis
The LC system used was essentially as described (35) and consisted of a direct nanoflow pump with a pressure limit of ∼300 bars (LC assist, Tokyo, Japan), a ReNCon gradient device (35) and an injection valve (Cheminart C2-0006, Valco Instruments, Houston, TX, USA) for sample loading. The spray tip ESI column was prepared with a fused-silica capillary [150 µm (i.d.) × 375 µm (o.d.)] using a laser puller (Sutter Instruments Co., Novato, CA, USA). The column was slurry-packed with a reversed-phase material (Develosil C30-UG-3; particle size, 3 µm; Nomura Chemical, Seto, Japan) to a length of 50 mm and was connected to the LC line with micro-fingertight fittings via a metal anion (Valco Instruments) (12). High voltage for ionization in negative mode (−1.4 kV) was applied to the metal union, and the eluate from reversed-phase LC was sprayed on-line to an LTQ-Orbitrap hybrid mass spectrometer (model XL Thermo Fisher Scientific, San Jose, CA, USA) or to a quadrupole-time-of-flight hybrid mass spectrometer (Q-Tof Ultima, Waters, Bedford, MA, USA). Reversed-phase separation of oligoribonucleotides was performed at flow rates of 100 nl/min using a 60-min linear gradient from 10% to 40% methanol in 10 mM triethylamine acetate (pH 7.0) or in 0.4 M HFIP/10 mM triethylamine acetate (pH 7.0) (36). MS and MS/MS spectra of nucleotide anions were obtained in a data-dependent mode. The LTQ-Orbitrap mass spectrometer was set to acquire a full MS scan between m/z 500 and 1500 at the mass resolution of 30 000, followed by full MS/MS scans of linear ion-trap of the top two ions from the preceding MS scan at the mass resolution of about 2000. An MS scan was accumulated for 3 micro scans (about 3 s per scan), and an MS/MS scan for 5 micro scans (about 2.5 s per scan). Relative collision energy for CID was set to 35% with a 30-ms activation time. Dynamic exclusion was performed with a repeat count of 1, a repeat duration of 0.5 min, exclusion duration list size of 25 and exclusion duration window of 60 s. The instrument was calibrated externally with poly-tyrosine before LC–MS measurement. The Q-Tof Ultima mass spectrometer was set to acquire a full MS scan between m/z 500 and 1500 followed by full MS/MS scans of the top two ions from the preceding MS scan. The mass resolution of the instrument was 8000–10 000. An MS scan was accumulated for 2 s and an MS/MS scan for 3 s. Collision energy for CID was set as follows: 20 V for m/z 400−450; 21 V for m/z 450−500; 24 V for m/z 500−600; 28 V for m/z 700−800; 30 V for m/z 800−1000; 40 V for m/z 1200−1400; and 50 V for m/z 1400−1500. Dynamic exclusion was set to m/z 2.0 for 120 s.
Outline of the Ariadne database search
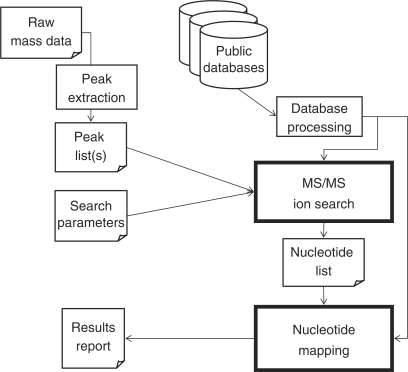
We designed the software Ariadne to assign a particular RNA identity by searching a set of MS/MS data resulting from the fragments produced by limited nucleolytic cleavage against a DNA/RNA database. Figure 1 illustrates the outline of the Ariadne search algorithm. To specify a particular RNA with limited MS/MS data from huge numbers of potential RNA species in the database, Ariadne evaluates the MS data in two steps. In the first ‘MS/MS ion searching’ step, the software compares the peak list extracted from raw MS data of a sample RNA and its RNase fragments with those obtained by in silico nuclease digestion of all RNA entries in the sequence database, evaluates the data matching by a quantitative score and selects candidate oligoribonucleotide(s). In general, however, a single oligonucleotide fragment produced by RNase digestion is assigned to multiple RNA entries in the database and cannot be used to specify a single RNA species. Thus in the second ‘nucleotide mapping’ step, Ariadne maps a set of nucleotide fragments identified in the sample RNA on all RNA entries in the database, evaluates the density of localization by a probability-based score and finds the RNA species with the highest score. It should be noted that these two evaluation steps were necessary for reliable RNA identification, essentially because RNA has fewer variable constituents than protein (4 versus 20) and thereby has a decreased likelihood of producing a unique fragment that can be attributed to a single RNA species upon RNase digestion. In addition, RNA is transcribed from relatively wide, undefined regions of the genome, whereas proteins are encoded by genes, which occur within relatively narrow, defined regions of the genome (see ‘Discussion’ section).
Figure 1.
Schematic diagram of the Ariadne database search program. Ariadne evaluates tandem MS data of nucleolytic fragments of RNA using a unique two-step algorithm, ‘MS/MS ion search’ and ‘nucleotide mapping’. See text for details.
Design of the Ariadne database search engine
Ariadne is a web application for interactive database searching. The user interface to Ariadne is a web browser, and the searches are defined using common gateway interface (CGI) forms. A form may be used to specify a peak list file and a search parameter file, which are then uploaded to the Ariadne server. Search parameters can also be specified using an interactive form. The Ariadne search engine is executed as a CGI program. Upon completion of a search, it calls a CGI script that reads the results from the internal database and returns an HTML report to the client browser. Links to additional pages generated on the fly by CGI scripts provide more detailed views of the results.
The peak list file, a set of queries for the search that contains pairs of a centroidal mass value of a precursor ion and lists of centroidal mass values of the corresponding product ions with associated signal intensities, is obtained from raw data by computational peak recognition. The monoisotopic mass value for multiply charged nucleotide ions can be picked up by measurement using a current high-resolution mass spectrometer. The precise determination of monoisotopic mass of the nucleotide ion can differentiate its base composition from that of other oligonucleotides and prevent the occurrence of false positives (FPs). For instance, the two major constituents of RNA, cytosine and uridine, have only a single mass-unit difference, and reduction of uridine to dihydrouridine brings alteration of only two mass-units in molecular mass. Thus, accurate and efficient peak picking and charge state determination are critical for the reliable identification and chemical analysis of RNA. In this study, a peak list was generated directly from raw MS data by the SpiceCmd program (Mitsui Knowledge Industry Co. Ltd., Tokyo, Japan), although Ariadne can also recognize the mascot generic format, a popular peak-list format used extensively in proteomics. A tool to allow for format interchanges will be supplied on the Ariadne web page.
A parameter file for Ariadne needs to contain a defined sequence database (see ‘Databases’ section) and specification of the cleavage method used to generate the RNA fragments (see ‘Enzyme’ section). Although the system contains the default values for other required parameters, we recommend specifying the following parameters for each specific experiment to ensure accurate identification:
species: specifies the species.
db_name: specifies the type of database, such as genome, refseq, snoRNA, etc.
enzyme_pattern_file: RNase T1, RNase U, etc. The file contains the substrate specificity of the enzyme or the reagent and the structure of newly generated termini, e.g. GN → G N (where N refers to any nucleotide); 5′: OH; 3′: 2′,3′-cyclic phosphate or 3′-phosphate for RNase T1.
5prime: specifies the 5′-terminal group of the database entry.
3prime: specifies the 3′-terminal group of the database entry.
Mass_tolerance: indicates the mass tolerance of precursor ions (fragments) in parts per million (p.p.m.). This parameter serves as a pre-filter for the product ion matching. We recommend setting the value, 125 for Q-Tof and 20 for Orbitrap analyzer, respectively, because too large mass tolerance, e.g. 1000 p.p.m., increases the candidates of fragments and thereby increases the chance of ‘incorrect identification’.
MS2_tolerance: indicates the mass tolerance of product ions in p.p.m. To obtain accurate search results, we recommend setting the value, 500 for Q-Tof and 750 for Orbitrap (Linear ion trap) analyzer, respectively.
max_missed_cleavages: specifies the upper limit of missed cleavages. The search engine considers less than or equal to the number of missed cleavages specified here.
max_mods: specifies the maximal number of modifications in an oligonucleotide. The search engine considers less than or equal to the number of modifications in a nucleotide specified here. Currently, methylation of four nucleotides and reduction of uridine to dihydrouridine are considered by the engine.
fragment_subset: indicates the number of fragments in a sub-entry of the database (see ‘Databases’ section).
base_number: nucleotide fragments equal to or longer than a length of ‘base_number’ + 1 nucleotides are considered in both ‘MS/MS ion search’ and ‘nucleotide mapping’ evaluation steps. A typical ‘base_number’ is 3.
Databases
Ariadne can use any FASTA-formatted DNA/RNA sequences as a database. Because most public sequence databases contain DNA sequences, the search engine is designed to transcribe DNA into RNA before the search; it can generate RNAs from both strands of genomic DNA. We considered that, although the genome contains extremely long DNA sequences, a relatively narrow sequence region should be specified for most biological applications. Thus, we designed the search engine to be able to divide the transcribed sequences into sub-entries as directed by search parameter ‘fragment_subset’ before the search process.
Enzyme
Because current LC–MS/MS techniques cannot measure large RNA molecules directly, the base-specific cleavage is essential for the detailed structural analysis. Among several endoribonucleases available to date, we used RNase T1 in this study. RNase T1 cleaves a phosphodiester bond at the 3′-end of guanine residue via a 2′,3′-cyclicphosphate intermediate, and the intermediate is subsequently hydrolyzed to generate a 3′-phosphate with the enzymatic and/or chemical reaction. Thus, the oligoribonucleotides generated by RNase T1 cleavage have 2′,3′-cyclicphosphate or 3′-phosphate or both types of phosphate. Ariadne takes the structural variation in the 3′-end into account upon database searching. The search engine reads the ‘enzyme_pattern_file’ to specify the cleavage profile and considers up to ‘max_missed_cleavages’ that result from incomplete hydrolysis (currently the upper limit of the parameter is 3).
MS/MS ion search
We first filtered the fragment mass of a query by matching with the masses of fragments generated by in silico nucleolytic digestion of all RNA entries in the specified database, and then evaluated the matched fragments by their product ions. For the evaluation, we defined the negative natural logarithm of the probability (P) calculated from the product of the relative frequencies of MS/MS ion peaks as the ‘nucleotide score’ for an RNA fragment. The concept for this scoring is that it is unlikely to find a high degree of resemblance between the experimental and theoretical spectrum, and thereby an ‘accidental’ match with low probability should have a high score. The score is calculated repeatedly for increasing numbers of the most intense product ions and the maximum score is finally selected as the ‘nucleotide score’ of the fragment. The assumptions that form the basis of this calculation are as follows: (i) a match between observed and theoretical product ion peaks is independent among product ion peaks, and (ii) the distribution of peaks is uniform and thus the probability for each matching is constant. Although these assumptions do not always fit the real spectra, we included them to maintain the simplicity of the search algorithm. Under these assumptions, the probability P is defined as Equation (1):
| 1 |
where n! denotes the factorial of n, x is the number of the matched peaks out of n trials and p is the probability of a match between the observed and theoretical product ion, as defined by the ratio of the mass tolerance to the mass range searched.
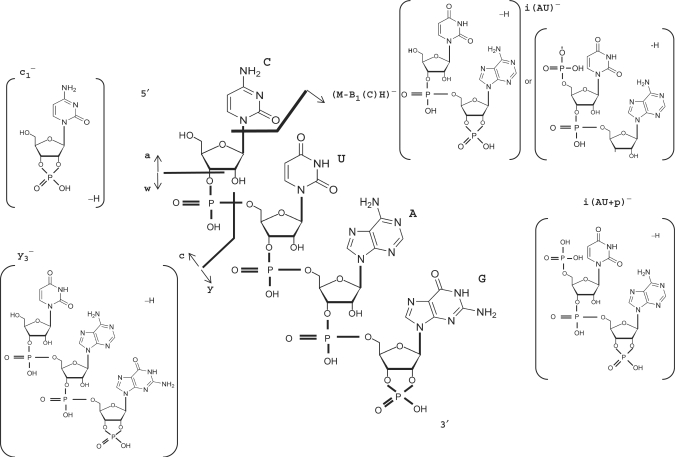
To implement the MS/MS spectral information of oligoribonucleotides into Ariadne, we studied low-energy CID profiles of multiple charged negative ions of synthetic RNAs generated by ESI (Taoka,M. et al., manuscript in preparation). As noted previously by Schuerch et al. (27) and Tromp and Schuerch (37), we detected mainly the c/y series of product ions and the w1 ion in many CID spectra, as well as weaker signals of a/w ions. We also observed internal fragment ions that arose from double-backbone fragmentation and fragment ions that had lost nucleotide bases. Although the nomenclature of internal fragment ions has not yet been defined, we tentatively assigned those ions in our search engine as shown in Figure 2. Thus, we implemented the assignments of a, c, w and y series of sequence ions, the internal fragmentation ions and base loss from the molecular ions into our search engine Ariadne (Figure 2).
Figure 2.
Low-energy CID pattern of deprotonated ion of RNA. The fragmentation pattern is illustrated by an oligoribonucleotide with a sequence 5′-OH-CUAG-cyclic-phosphate-3′. The nomenclature of sequence ions is in accordance with McLuckey et al. (26). The structures of c/y ions are according to Tromp and Schuerch (37). The tentative structures of internal fragment ions produced by the double-backbone cleavage are designated as i(AU) and i(AU+p) for the structural variants shown in the figure. In this example, i(AU) contains two isomers that cannot be discriminated according to their mass.
Because the ‘nucleotide score’ is independent on the size of the database and does not necessarily represent the statistical significance of a search result, we defined an additional parameter, score threshold (T), for quantitative measure of the significance. T is defined by Equation (2).
| 2 |
where p is the probability of the significance level, and n is the number of oligonucleotide fragments that have molecular masses within a mass tolerance window previously specified by the ‘Mass_tolerance’ parameter. Usually, a p-value of 0.05 was considered statistically significant.
RNA modification
The search engine considers post-transcriptional modifications in the nucleotide bases and/or the riboses of nucleotides. In addition to hundreds of post-transcriptional modifications, such as methylation, reduction of uridine to dihydrouridine and pseudouridilation, which occur naturally in RNA, there are a number of modifications introduced deliberately during sample handling, such as cyanoethylation of uridine. Except for mass-silent pseudouridilation, these modifications cause characteristic mass shifts with respect to the original unmodified sequences, so that tandem MS can detect and specify the positions of those modifications. In many cases, the modifications are not quantitative even if the modification sites are described in the literature, so that the search engine needs to generate all permutations of potential modification sequences. For example, if we assume that a pentaribonucleotide might be methylated maximally at two positions, the search engine must test for a match with the experimental data for that oligonucleotide containing 0-, 1- or 2-methylated nucleotides. This greatly increases the target sequence from 1 to 16 (1 + 5 + 10) oligonucleotides, resulting in a much longer search time and reduced specificity. For this reason, we have designed Ariadne, which considers methylation of all four nucleotides (A, C, G and U) and reduction of uridine to dihydrouridine to a maximal number of ‘2’ at this stage of software development. Ariadne can also consider the terminal groups of RNAs and their nucleolytic (and possibly chemically cleaved) fragments; namely, it postulates OH and H2PO3 for RNA and OH, HPO2 (cyclic phosphate intermediate) and H2PO3 for oligoribonucleotide fragments.
Nucleotide sequence mapping
Because a single RNase T1 fragment assigned by ‘MS/MS ion search’ is often insufficient to specify a single original RNA species, multiple fragments generated from a sample RNA are evaluated quantitatively by an additional probability-based scoring algorithm. This ‘mapping score’ for a database entry is thus defined as the negative natural logarithm of the entry's probability calculated from the product of relative frequencies of all fragments that appear in the entry. Even if a certain sequence appears multiple times in more than two fragments in an entry, the sequence is counted only once to calculate the mapping score of the entry. Without this unification, ‘repeat’ regions tend to provide higher scores that result in FPs. If a fragment sequence is identified by multiple MS/MS queries, the search engine counts the sequence only once. And if a query identifies multiple fragments, the search engine regards them as a meta-sequence, a set of sequences for the query. For example, if a query identifies both ACUG and CAUG, and their relative frequencies are 0.1 and 0.2, respectively, then the search engine counts the summed relative frequencies as 0.3 (0.1 + 0.2) for the meta-sequence {ACUG, CAUG}. Thus, the score for nucleotide mapping (Sm) is defined by Equation (3).
| 3 |
where t is a total number of the unique sequences identified by MS/MS ion search, pi (i = 1, 2,…, t) is a relative frequency of each sequence, N is a total number of the unique sequences in the (sub-)entry and pnothit is 1 − (p1 + p2 + … + pt). The threshold for Sm can be determined by Equation (2), reading n as a total number of (sub-)entries.
In addition, when searches against DNA databases are carried out, the information on the chemical modifications of a candidate fragment identified by MS/MS ion search is removed, and the resulting unmodified sequence is mapped on the entries.
RESULTS
Validation of the Ariadne search engine
Because Ariadne is the first search engine to correlate tandem-MS data of the RNA and genome/RNA databases, we first performed a simple proof-of-principle experiment to assign a known RNA. Thus, we designed a cDNA construct containing a full-length sequence of xCyPA sandwiched [with the sequences of the Xenopus globin untranslated region (UTR) and used it as a template to transcribe the corresponding mRNA in vitro (see Materials and Methods section). After purification, the resulting 788-nt RNA, which consisted of the xCyPA mRNA connected with globin UTRs by linker sequences, and a 3′-poly(A) tail, was digested with RNase T1 and analyzed by a direct nanoflow LC–MS system to generate the MS and MS/MS spectra (see ‘Materials and Methods’ section and Supplementary Figure S1). Subsequently, the resulting 427 MS/MS spectral data were submitted as queries to Ariadne for the identification of nucleotide fragments by searching against a ‘database’ consisting solely of the xCyPA mRNA sequence. In this experiment, Ariadne replied to 112 queries and identified 67 of the 71 fragments of 4 nt or longer that are expected from the RNase T1 cleavage of xCyPA mRNA (71% sequence coverage; Supplementary Table S1 and Figure S2). The remaining four fragments were not identified: one of the fragments contained heterogeneous poly(A) tails at the 3′-end with undefined large molecular masses and was thereby not detected by MS. The other three fragments, a large fragment with 32 nt (spanning nucleotides 694–725, see Supplementary Figure S2) and two small fragments, AACUG and ACUG, were detected by MS but were not selected for data-dependent MS/MS analysis because those ions were excluded from MS/MS analysis by a mechanism of dynamic exclusion (38). Thus, Ariadne correctly identified all of the oligonucleotides that generated MS/MS signals.
For the second attempt to validate the performance of Ariadne, we searched the same dataset against a merged database consisting of a large dataset of human reference sequences (hrefseq; 2008/3/10, 45 892 entries) spiked with the sequence of xCyPA mRNA. We evaluated the results of an Ariadne search semi-quantitatively by following two parameters, sensitivity and specificity, as defined by Kapp et al. (39). Sensitivity is the ability to correctly identify RNA fragments irrespective of the quality of the data and is defined by Equation (4).
| 4 |
where true positive (TP) is a query that gives rise to a correct reply with a score above a threshold, and false negative (FN) is a query that gives rise to a correct reply with a score below a threshold.
Specificity is the ability to calculate low-ranking scores for random (incorrect) matches and is defined by Equation (5).
| 5 |
where true negative (TN) is a query that gives rise to an incorrect reply with a score below a threshold, and FP is a query that gives rise to an incorrect reply with a score above a threshold. It is often difficult to validate whether the reply to a query is correct or not; however, in our validation scheme, we can easily distinguish correct-hit queries that identified oligonucleotides in xCyPA mRNA and incorrect-hit queries that identified oligonucleotides originating from RNAs in the hrefseq database. Thus, TP, FN, FP and TN can be defined as follows (also see Figure 3):
TP: a query that gives rise to a correct hit when searching against both xCyPA and the merged database;
FN: a query that gives rise to a correct hit when searching against xCyPA but produces a score below the threshold (non-hit) when searching against the merged database;
FP: a query that gives rise to an incorrect hit when searching against the merged database; and
TN: a query that gives rise to a non-hit when searching against both xCyPA and the merged database.
Furthermore, FP can be classified into type 1 (FP1; hit for xCyPA database) and type 2 (FP2; non-hit).
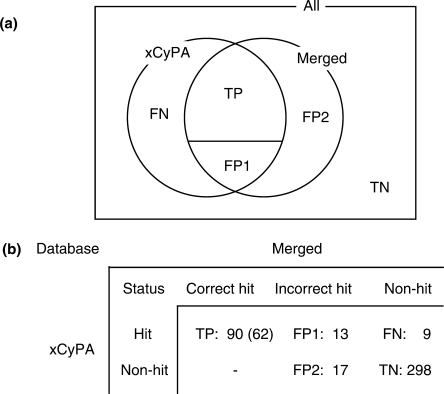
Figure 3.
Classification of MS/MS ion search results of RNase T1-digested xCyPA. The MS/MS data were searched by Ariadne against a ‘database’ consisting only of xCyPA mRNA sequence (xCyPA) or a ‘database’ consisting of human refseq and xCyPA mRNA (merged), and each result was classified as a TP, FP type 1 (FP1) or type 2 (FP2), FN or TN. (a) A Venn diagram of the classification, and (b) the classified search results. Details are provided in the text. Number in parenthesis in (b) indicates the number of unique sequences.
Of 112 queries obtained as ‘hits’ by searching a database consisting solely of the xCyPA sequence, 90 queries (62 sequences) were also correctly identified by searching against the merged database and were thereby classified as TPs (Figure 3). The analysis of 13 FP1s and 9 FNs shown in Figure 3 suggests that FP1 occurred frequently by mis-identification of incorrect sequences with the same base composition and FN occurred when the scores did not exceed the thresholds of the search. On the other hand, 10 of 17 FP2s arose from oligonucleotide fragments generated by non-specific RNase T1 cleavage of xCyPA mRNA (Supplementary Table S1). Thus, the sensitivity and specificity of the Ariadne search engine were estimated to be 0.91 and 0.91, respectively. This is an acceptable level of performance as compared with the equivalent search engines used for proteomics such as Mascot and SEQUEST (39).
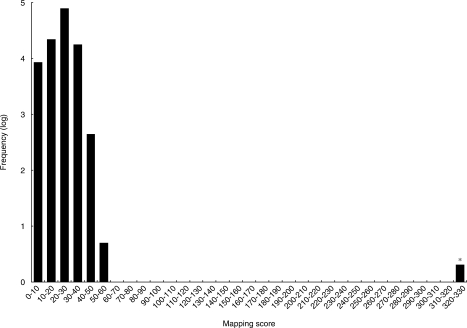
Ariadne has a unique second-step evaluation algorithm called ‘nucleotide mapping’, whereby a set of oligonucleotide fragments identified in the first ‘MS/MS ion-searching’ step is mapped on all RNA entries in the database. Ariadne then evaluates the density of the oligonucleotide localization by a probability-based score and presents the RNA species with the highest score. To assess the performance of this ‘nucleotide mapping’ procedure, all oligonucleotide fragments containing both 62 correct and 74 incorrect sequences derived from the analysis of xCyPA mRNA (Supplementary Table S1) were mapped on all entries in the merged database. Each entry was scored, and the entries were then sorted by this score in descending order. As shown by the histogram in Figure 4, the xCyPA mRNA sequence was easily singled out because of its significantly higher score from all other entries in the database, which consisted of sequences numbering in the several tens of thousands. Thus, the two-step scoring algorithm of Ariadne provided confident identification of a single RNA species from among a large number of potential candidate RNAs in a database.
Figure 4.
Mapping score histogram of the xCyPA mRNA search results. Scores for all entries in the database are summarized in the histogram. Frequencies of entries within a 10-point scoring range were counted, converted to common logarithm of frequency +1 and plotted. Note that the histogram shows a ‘hit’ for the query, as indicated by a distinctly high score.
Identification of yeast tRNAPhe-1 and its post-transcriptional modifications by searching against the genome database
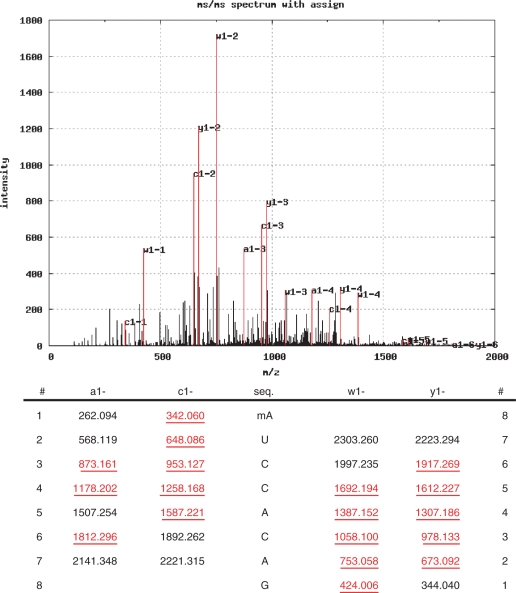
To evaluate whether Ariadne can be used to identify post-transcriptional modifications in RNA, we analyzed the RNase T1 digest of tRNAPhe-1 (Supplementary Figure S3), and the MS/MS data were used to search against a Saccharomyces cerevisiae tRNA database transcribed from tDNA sequences [The Genomic∼tRNA Database, http://lowelab.ucsc.edu/GtRNAdb/(40)] under a search condition that considers up to two modifications and permits up to two missed cleavages. After carrying out automated two-step evaluations of the MS/MS data against all entries in the database, Ariadne clearly identified tRNAPhe-1 with seven oligonucleotides and assigned five of the seven oligonucleotides with the methylated nucleotides and/or dihydrouridines (Figure 5). In this application, Ariadne discriminated tRNAPhe-1 from its structural homologue tRNAPhe-2 by the fragment AAUUCG, which is unique to tRNAPhe-1; however, more detailed analysis indicated that the tRNAPhe-1 preparation used in this study contained small amount of tRNAPhe-2 (Taoka et al., manuscript in preparation). Figure 6 illustrates the identification of monomethylated adenosine (mA) in the sequence 5′-OH-mAUCCACAG-cyclic phosphate-3′ (indicated by an arrow in Supplementary Figure S3). In fact, we could identify all monomethylated nucleotides in the yeast tRNAPhe-1 by the automated LC–MS/MS–Ariadne analysis, except for the hypermodified oligonucleotide containing yW and the intron sequence.
Figure 5.
Nucleotide sequences of the mature tRNAPhe-1 (Mature tRNA) and the equivalent transcript identified in the yeast tRNA database (Database). The identified RNase T1 fragments are underlined in the upper half of the figure and are listed in the lower half. Lower case letters indicate the methylated forms of the corresponding unmodified nucleotides (e.g. g for methylated G), and ‘D’ indicates dihydrouridine. Other symbols for modified nucleotides are according to Limbach et al. (47): ‘L’, 2-methylguanosine; ‘D’, dihydrouridine; ‘R’, N2,N2-dimethylguanosine; ‘B’, 2′-O-methylcytidine; ‘#’, 2′-O-methylguanosine; ‘Y’, wybutosine; ‘P’, pseudouridine; ‘?’, 5-methylcytidine; ‘7’, 7-methylguanosine; ‘T’, 5-methyluridine; and ‘"’, 1-methyladenosine. The boxed sequence indicates an intron.
Figure 6.
Identification of post-transcriptional modification in yeast tRNAPhe-1 by MS/MS ion search. A typical search result obtained by the analysis of the doubly charged ion with m/z 1282.7, identified as the fragment 5′-OH-mAUCCACAG-2′,3′-cyclic phosphate-3′ spanning nucleotides 76−83 in the tRNAPhe-1 sequence, is shown. The product ions are assigned as indicated in the MS/MS spectrum, and the masses of each ion are underlined in the table. Of 470 total signals detected, 15 hits of the most intense 38 signals gave the highest score. The result was visualized by Ariadne.
To further examine whether Ariadne can identify tRNAPhe-1 and its post-transcriptional modifications by directly searching the genome database, the same MS/MS dataset was searched against the S. cerevisiae genome database. This resulted in correct identification of seven oligonucleotides that included the five containing the same modified nucleotides as described above (Figure 5). Furthermore, these oligonucleotides could be mapped to a very discrete chromosomal locus in the yeast genome; namely, there were only 10 sub-entries from the yeast genomic sequences with clearly distinctive mapping scores, all of which encode for tRNAPhe-1 and tRNAPhe-2 (data not shown). Finally, Ariadne assigned most of the RNase T1 fragments to tRNAPhe-1 and identified more than half of the known mass-sensitive modifications by the genome-wide searching without previous knowledge of modification. tRNAs undergo a complex series of post-transcriptional modifications during biogenesis, such as splicing out the intron sequence (41), transformation from G to yW (24) and the CCA addition to the 3′-end by the template-independent CCA-adding enzyme (42), and the resulting mature tRNA carries a variety of modified nucleotides; however, these modifications were not considered in the current stage of software development (see ‘Discussion’ section).
Identification of small RNA in a tRNA mixture by searching against the yeast genome database
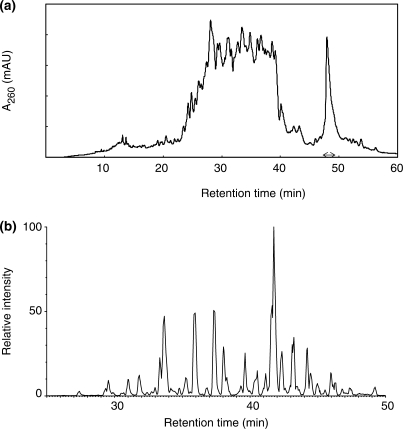
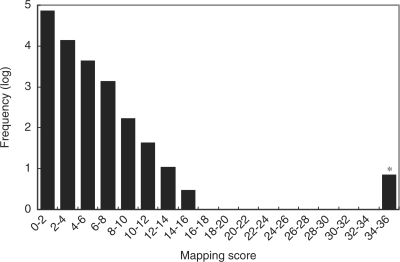
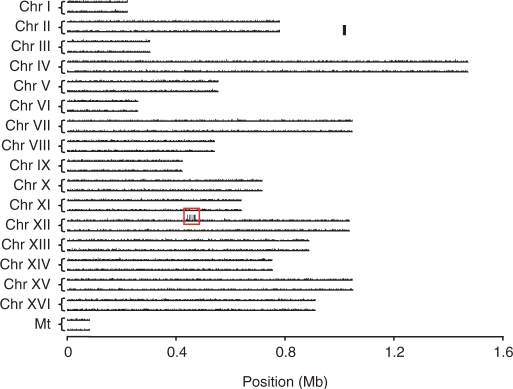
During the purification of a commercial yeast tRNA preparation by anion-exchange chromatography, we found that an RNA component(s) eluted significantly later than the bulk of tRNAs (Figure 7a). This component had a greatly decreased mobility as compared with yeast tRNAs on polyacrylamide gel electrophoresis (PAGE) under denaturing conditions (data not shown), suggesting that it might be an extra RNA component that was larger than typical yeast tRNAs. To examine whether Ariadne could identify this unknown RNA component, we purified the RNA by reversed-phase LC (Figure 7a) and digested it with RNase T1. The digest was then analyzed by nanoflow LC–MS/MS (Figure 7b), and the resulting MS/MS data were used to search against the genome database of S. cerevisiae. As shown in Figures 8 and 9, six genomic regions on yeast chromosome XII were identified based on their significantly high scores. These regions had nucleotide sequences in common with one another and contained eight RNase T1 fragments of the sample RNA identified by MS/MS ion search (Figure 10). The BLAST search of this nucleotide sequence against the yeast genome database clearly indicated that it encodes 5S rRNA, a 120-nt RNA that is slightly larger than typical tRNAs (which are about 75 nt). The MS/MS-based de novo sequencing of the major fragment of this RNA confirmed this conclusion (data not shown). The genome-sequencing study of Goffeau et al. (43) showed that the right arm of yeast chromosome XII contains approximately 140 tandem copies of a 9.1-kb sequence encoding rRNAs. However, the currently available S. cerevisiae genome database excludes most of the repeated sequence and contains only 1.1 Mb of the 2.4-Mb sequence of the total length of chromosome XII, including six copies of 5S rRNAs (43). It should be noted that the six chromosomal loci that were recognized by Ariadne include all of the genomic loci that encode 5S rRNA in the S. cerevisiae genome database. Thus, Ariadne could be useful for MS-based identification of unknown RNA components in biological mixtures.
Figure 7.
Characterization of an unknown small RNA in yeast tRNA preparation. (a) Isolation of an unknown small RNA by anion-exchange chromatography. A mixture of yeast tRNA (tRNA typeX, Sigma R9001; 100 μg) was applied to a TSKgel DNA-NPR column (4.6 × 75 mm; Toso) and eluted with an 80-min gradient of NH4Cl (0.1–1.0 M) in 25 mM Tris–HCl buffer (pH 9.0) at a flow rate of 0.5 ml/min at 60°C. The fraction indicated by a double-headed arrow was collected, purified by reversed-phase chromatography (data not shown) and subjected to the LC–MS analysis after digestion with RNase T1 (see Materials and Methods section). (b) Base peak chromatogram of the RNase T1 digest of the small RNA.
Figure 8.
Score histogram of the search results for an unknown small RNA. Scores for all sub-entries in the yeast genome database are summarized in the histogram. Frequencies of entries within a 10-point scoring range were counted, converted to common logarithm of frequency +1 and plotted. Note that the histogram shows a ‘hit’ for the query.
Figure 9.
Score distribution of the unknown RNA results using a search of the S. cerevisiae whole genome. Scores for all sub-entries in the yeast genome database were mapped on chromosomes I–XVI (Chr I–XVI) and on the mitochondrial genome (Mt). A bar scale in the figure indicates score as 50. Note that six genomic loci clustered on chromosome XII were identified as the unknown RNA with significantly high scores (boxed in the figure).
Figure 10.
Nucleotide sequences of six chromosomal loci identified for the unknown small RNA. The genomic sequences are shown with their positions on yeast chromosome XII. The sequences found in common among all genomic regions are indicated in bold, and the sequences of RNase T1 fragments identified in the unknown RNA are underlined. A BLAST search of the sequence revealed that all of those chromosomal loci encode 5S rRNA.
DISCUSSION
Here, we describe a method that correlates tandem MS data of RNA and the DNA/RNA sequence database. The method consists of LC–MS/MS analysis of multiple oligonucleotide fragments generated by limited nucleolytic cleavage of sample RNA and fully automated retrieval of the MS/MS dataset with the database-searching software Ariadne. A method with two distinct principles is currently available for proteomics: peptide mass fingerprinting (PMF) identifies a protein based on a set of peptide masses produced by limited proteolysis, and MS/MS ion searching identifies a protein based on MS/MS data of proteolytic fragment(s) (32,33). PMF is most commonly used for the identification of a relatively pure protein such as that which results from 2D PAGE; whereas the MS/MS ion searching method is frequently used for ‘shotgun proteomics’, a technology for MS-based large-scale protein identification of complex biological mixtures (44,45). In any case, the software algorithm used in proteomics to correlate tandem MS data of peptide fragments and the protein sequence database relies on the variety of masses of amino acid constituents of a protein, i.e. in a typical example; the MS/MS ion searching method can specify a particular protein in a genome-wide database even with MS/MS data of a few proteolytic fragments. Our goal was to develop software for the MS-based analysis of RNA that is equivalent to the one used in proteomics; however, we found that the software algorithms for the MS-based analysis of proteins were not necessarily sufficient for the analysis of RNA, essentially because oligoribonucleotide fragments are much less variable than peptide fragments in composition and molecular mass. Thus, we designed Ariadne to evaluate the MS data of oligoribonucleotide fragments derived from limited hydrolysis of an RNA sample in two steps: the identification of nucleotide fragments based on MS/MS ion search and the mapping of identified nucleotides on all RNA entries in a database. The results presented here suggest that the software is capable of identifying a single particular RNA molecule from a large number of candidates in the yeast genome or human refseq database.
Post-transcriptional modification is essential for the structure and function of RNAs. MS is ideal for the analysis of modifications because almost all modifications are accompanied by characteristic alterations in intrinsic molecular mass. In addition, tandem MS provides a powerful tool to analyze the types and sites of modifications in target molecules. As described in this article, Ariadne could detect some of the modifications that occurred in RNA (Figures 5 and 6), although the software needs further improvement to be able to detect an increased number of and much more modifications with higher reliability. This will require higher specification hardware to accelerate the search speed for huge possibility of structural variations and refinement of the algorithm used in the search engine. This study does, however, hint at the potential of MS-based comprehensive chemical analyses of RNA, because Ariadne, which uses relatively simple probability-based algorithms, was able to detect RNA modifications by searching the MS/MS data against a relatively large database such as the whole yeast genome.
One of the difficulties encountered in the software development reported here was the lack of established RNA databases. This is in contrast to the field of proteomics, which has many databases containing gene sequences or expression tags that are readily available. Less than 2% of the human genome is translated into protein, yet >40% of mammalian genomes might be transcribed into RNA (46). In addition, protein-coding genes are well defined as open reading frames in the genome; whereas we cannot predict RNA-coding regions at present, nor can we predict processing events of ncRNAs during biogenesis. Although the current development of RNA MS is hindered by this lack of information, the rapid growth of the RNA world combined with the capability of MS, leads us to hypothesize that future studies will solve most of these problems and accelerate the MS-based analyses of RNA.
CONCLUSION
This article provides a novel method for the MS-based identification and chemical analysis of RNAs in biological samples using DNA/RNA sequence databases as a reference. The method was validated by a number of proof-of-principle experiments and was subsequently used to identify RNA species in a yeast tRNA preparation, as well as to analyze the post-transcriptional modification of tRNAPhe-1. These applications suggested that the method could be used for the direct chemical analysis of RNA and would thereby serve as a complementary tool to the conventional techniques based on RNA biochemistry and molecular biology. One of the goals of our study was to develop an MS-based technology similar to that used in ‘shotgun proteomics’ that would allow simultaneous analysis of multiple RNA species in biological mixtures. Ariadne has the potential to serve as the software for this purpose, because it uses a unique algorithm including two quantitative evaluation steps of MS/MS signals of nucleolytic fragments of RNA. In fact, our preliminary examination suggests that Ariadne allows the ‘shotgun’ identification of multiple RNA components in several RNP complexes following affinity purification from cultivated cells. For example, we could identify U4, U5, U6 snRNAs in the yeast Lsm3 associated pre-spliceosomal complex by a single LC–MS/MS–Ariadne analysis (Taoka,M. et al., manuscript in preparation). Although the development of ‘shotgun ribonucleomics’, simultaneous RNA analysis in more complex biological mixture, will certainly require the surmounting of several technical challenges and software improvements, we expect the current study to be a first step toward this goal.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Core Research for Evolutional Science and Technology (CREST); Japan Science and Technology Agency. Funding for open access charge: CREST, Japan Science and Technology Agency.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms M. Koike for help in MS data analysis, and Mr G. Terukina for synthesizing xCyPA mRNA.
REFERENCES
- 1.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer SE, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455:491–496. doi: 10.1038/nature07274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 5.Bryan RC. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Huttenhoffer A, Schatner P. The principles of guiding by RNA; chimeric RNA-protein enzymes. Nat. Rev. Genet. 2006;7:475–482. doi: 10.1038/nrg1855. [DOI] [PubMed] [Google Scholar]
- 7.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessions from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 8.Venema J, Tollervey D. Ribosome synthesis in Saccharomyses cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 9.Valadkhan S. The spliceosome: caught in a web of shifting interactions. Curr. Opin. Struct. Biol. 2007;17:310–315. doi: 10.1016/j.sbi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Neuenkirchen N, Chari A, Fisher U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi N, Isobe T. Proteomic analyses of ribonucleoprotein complexes formed at various stages of ribosome biogenesis in mammalian cells. Mass Spec. Rev. 2003;22:287–317. doi: 10.1002/mas.10057. [DOI] [PubMed] [Google Scholar]
- 12.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J. Biomed. Biotechnol. 2006;1:1–20. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 15.Chien KR. MicroRNAs and the tell-tale heart. Nature. 2007;447:389–390. doi: 10.1038/447389a. [DOI] [PubMed] [Google Scholar]
- 16.Stone MD, Mihalusova M, O'connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 18.Païs de Barros JP, Keith G, El Adlouni C, Glasser AL, Mack G, Dirheimer G, Desgres J. 2′-O-methyl-5-formylcytidine (f5Cm), a new modified nucleotide at the ‘wobble’ position of two cytoplasmic tRNAsLeu(NAA) from bovine liver. Nucleic Acids Res. 1996;24:1489–1496. doi: 10.1093/nar/24.8.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas B, Akoulitchev AV. Mass spectrometry of RNA. Trends Biochem. Sci. 2006;33:173–181. doi: 10.1016/j.tibs.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Mengel-Jorgensen J, Jensen SS, Rasmussen A, Poehlsgaard J, Iversen JJL, Kirpekar F. Modifications in Thermus thermophilus 23S ribosomal RNA are centered in regions of RNA-RNA contact. J. Biol. Chem. 2006;281:22108–22117. doi: 10.1074/jbc.M600377200. [DOI] [PubMed] [Google Scholar]
- 23.Emmerechts G, Barbe S, Herdewijn P, Anne J, Rozenski J. Post-transcriptional modification mapping in the Clostridium acetobutylicum 16S rRNA by mass spectrometry and reverse transcriptase assays. Nucleic Acids Res. 2007;35:3494–3503. doi: 10.1093/nar/gkm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 27.Schuerch S, Bernal-Mendez E, Leumann C. Electrospray tandem mass spectrometry of mixed-sequence RNA/DNA oligonucleotides. J. Am. Soc. Mass Spect. 2002;13:936–945. doi: 10.1016/S1044-0305(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 28.Ni J, Pomerantz SC, Rozenski J, Zhang Y, McCloskey JA. Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry. Anal. Chem. 1996;68:1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 29.Oberacher H, Wellenzohn B, Huber CG. Comparative sequencing of nucleic acids by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2002;74:211–218. doi: 10.1021/ac015595a. [DOI] [PubMed] [Google Scholar]
- 30.Rozenski J, McCloskey JA. SOS: a simple interactive program for ab initio oligonucleotide sequencing by mass spectrometry. J. Am. Soc. Mass Spectrom. 2002;13:200–203. doi: 10.1016/S1044-0305(01)00354-3. [DOI] [PubMed] [Google Scholar]
- 31.Oberacher H, Mayr BM, Huber CG. Automated de novo sequencing of nucleic acids by liquid chromatography-tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:32–42. doi: 10.1016/j.jasms.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 33.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Lemaire P, Garrett N, Gurdon JB. Expression cloning of siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 35.Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- 36.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal. Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 37.Tromp JM, Schuerch S. Gas-phase dissociation of oligoribonucleotides and their analogs studied by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 38.Gatlin CL, Eng JK, Cross ST, Detter JC, Yates JR., III Automated identification of amino acid sequence variations in proteins by HPLC/microspray tandem mass spectrometry. Anal. Chem. 2000;72:757–763. doi: 10.1021/ac991025n. [DOI] [PubMed] [Google Scholar]
- 39.Kapp EA, Schutz F, Connolly LM, Chakel JA, Meza JE, Miller CA, Fenyo D, Eng JK, Adkins JN, Omenn GS, et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: Sensitivity and specificity analysis. Proteomics. 2005;5:3475–3490. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 40.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valenzuela P, Venegas A, Weinberg F, Bishop R, Rutter WJ. Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc. Natl Acad. Sci. USA. 1978;75:190–194. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- 43.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 44.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 45.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 46.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 47.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.