Abstract
The idea of identifying or characterizing an RNA molecule based on a mass spectrum of specifically generated RNA fragments has been used in various forms for well over a decade. We have developed software—named RRM for ‘RNA mass mapping’—which can search whole prokaryotic genomes or RNA FASTA sequence databases to identify the origin of a given RNA based on a mass spectrum of RNA fragments. As input, the program uses the masses of specific RNase cleavage of the RNA under investigation. RNase T1 digestion is used here as a demonstration of the usability of the method for RNA identification. The concept for identification is that the masses of the digestion products constitute a specific fingerprint, which characterize the given RNA. The search algorithm is based on the same principles as those used in peptide mass fingerprinting, but has here been extended to work for both RNA sequence databases and for genome searches. A simple and powerful probability model for ranking RNA matches is proposed. We demonstrate viability of the entire setup by identifying the DNA template of a series of RNAs of biological and of in vitro transcriptional origin in complete microbial genomes and by identifying authentic 16S ribosomal RNAs in a ‘small ribosomal subunit RNA’ database. Thus, we present a new tool for a rapid identification of unknown RNAs using only a few picomoles of starting material.
INTRODUCTION
It has become increasingly clear that RNA is involved in many cellular processes where RNA and protein form essential complexes, exemplified by the ribosome, the spliceosome and the signal recognition particle. Identification of the RNA species isolated from an RNA–protein complex during a biochemical research project may be done in different ways. One method relies on radiolabelling of the RNA followed by partial degradation by either nucleotide-specific RNases (1,2) or ditto chemicals (3,4). Direct sequencing is evidently the ultimate identification, but RNA sequencing requires a high level of expertise and typically tens of micrograms of material, which often necessitates extraordinary scale-up of the purification protocol. Alternatively, the RNA may be identified through hybridization to high-density oligodeoxynucleotide arrays. This method is primarily used for obtaining quantitative information on messenger RNA levels via production of labelled complementary DNA (5,6), but oligodeoxynucleotide arrays, in a few cases, have also been used for species identification via direct hybridization with labelled ribosomal RNA (rRNA) (7,8). Array technology will enable identification of an unknown RNA and of its genomic origin, provided the entire genome is appropriately represented on the array. However, delineation of the termini of an RNA molecule will only be possible in fortunate cases, and no information on posttranscriptional modifications can be obtained or included during array analysis. Furthermore, oligodeoxynucleotide arrays are costly, especially if an array representing the entire genome of a ‘non-model’ organism is required.
Proteins of unknown nature can be identified by the so-called peptide mass fingerprinting (PMF) approach (9–11). PMF relies on digestion of the isolated protein with an amino-acid-specific agent followed by accurate mass determination of the resulting peptides by mass spectrometry. The peptide masses constitute a fingerprint of the protein under investigation. Comparison of the experimentally obtained peptide mass fingerprint with an in silico digest of all entries in a protein database allows identification of best matching candidate and—in most cases—protein identification. The PMF approach has proven eminently successful over its 15 years of existence, partly because of the method's speed and sensitivity and partly because of the ever expanding number of entries in sequence databases. Searches can be performed in translated nucleic acid databases as well, and identification is possible for proteins from species with unsequenced proteins/genomes due to sequence similarity between homologous proteins. The mass spectrometric equipment used is essentially always matrix assisted laser desorption/ionization (MALDI) in combination with a time-of-flight (ToF) mass analyser, because this combination gives easily interpretable spectra dominated by singly charged ions, which are detected with high sensitivity.
An unknown RNA should be amenable to the same identification approach as PMF of proteins. The issues regarding use of nucleotide-specific digestion and mass spectrometry for identification purposes have been discussed thoroughly (12,13), including development of bioinformatics tools for identification (14), therefore a briefer account is given here. Because RNA consists of only four nucleotides, the risk that different sequences will give rise to similar digestion patterns is potentially greater than for peptides. A scenario can be envisioned where a database entry will give rise to a fragment mass-pattern that is very similar to one from the RNA in question, though there is no sequence homology between the two species. On the other hand, the presence of just four nucleotides in RNA warrants mass spectrometric analysis with relatively low mass accuracy, especially when the cleavage specificity of the RNase is taken into account (15): for example, the smallest RNase T1 digestion products that can occur within 0.01% in mass are A13G1 and C7U7G1 at around 4641 Da. The monoisotopic masses of the four RNA residues C, U, A and G are 305.04, 306.03, 329.05 and 345.05 Da, respectively. This 0.99 Da mass difference between C and U means that two RNA species differing by one C-to-U substitution will have overlapping isotope patterns in most mass spectrometers, a phenomenon that has to be taken into account in the interpretation of the mass spectrum. Wilson and co-workers have made in silico simulations of the possibility of identifying a specific 16S rRNA in a limited 16S rRNA database (13) and have made in silico studies on the degree of coincidence between mass spectra of RNase T1 digested sub-regions of 16S rRNA (14). The outcome of these calculations was clearly in favour of an RNase/mass spectrometry approach for RNA identification. Practical RNA characterization by nucleotide-specific cleavage followed by mass spectrometric analysis has been performed in several cases. The group of McCloskey and Crain first reported the use of nucleotide-specific RNase-digestions of rRNA in combination with electrospray ionization mass spectrometry (16) to search for posttranscriptional modifications in known sequences, and posttranscriptional modifications can likewise be studied by MALDI reflector ToF mass spectrometry (17). A combination of different nucleotide-specific enzymes can yield partial sequence information on RNA as demonstrated in a proof-of-principle work (18). This has been followed by applications of endonuclease digestion of RNA and subsequent MALDI ToF analysis for various typings based on a common strategy: a copy of the genomic region of interest is obtained by PCR followed by in vitro transcription into RNA. The transcribed RNA directly reflects the sequence of the original genomic region, and enzymatic digestion of the RNA will generate an origin-specific digestion pattern that can be visualised by mass spectrometry. This approach has been used for bacterial species determination using 16S rRNA as identifier (19–21), to study single nucleotide polymorphisms (22,23) and short tandem repeat polymorphisms (24). Later, Hossain and Limbach (25) reported proof-of-principle detection of individual tRNAs in complex mixtures through the masses of signature endonuclease digestion products, including known information on posttranscriptional modifications.
The above examples all utilize specific cleavage and mass spectrometry to characterize the RNA in question, but the genetic origin of the RNA was known in all cases, which will evidently not be the case in an RNA identification strategy similar to the concept of PMF. We have developed bioinformatics tools to perform RNA identification via digestion/mass spectrometry and implemented an experimental approach for practical RNA mass mapping (RMM). We demonstrate the capabilities of this concept by correctly locating the DNA origin of a number of RNA molecules in complete microbial genomes. In addition, we are able to perform species identification based on digestion/mass spectrometry of authentic 16S rRNA.
MATERIALS AND METHODS
RNA preparations
Haloarcula marismortui 23S rRNA subfragments were from a previous study (26). Briefly, complementary DNA oligonucleotides were annealed to 23S rRNA regions flanking the subfragment of interest, and the RNA part of the DNA/RNA hybrid was digested with RNase H. The 23S rRNA subfragments were separated on a denaturing polyacrylamide gel and recovered by soaking of the relevant gel bands after visualization with ethidium bromide/UV-illumination.
Bacillus stearothermophilus 5S rRNA purification has been published previously (27).
Ribosomal subunits from Thermus thermophilus strain HB8, and Escherichia coli strain MRE 600, respectively, were purified via sucrose gradients as earlier described (28,29). The fractions containing the small ribosomal subunit were precipitated with 2.5 volumes of ethanol. The 16S RNA was purified by three rounds of extraction with a 1:1 mixture of phenol/chloroform and recovered by ethanol precipitation.
The in vitro transcript covering positions 2446–2632 of E. coli 23S rRNA was from a previous study (30) and the in vitro transcript of spot 42 RNA from E. coli was synthesized as described (31).
RNase T1 digestion and MALDI mass spectrometry
The RNase T1 digestion and the subsequent mass spectrometric analysis have been described in details elsewhere (32). In short, complete RNase digestion is obtained by a ratio of 0.1 µg of RNA to 10 U of RNase T1 (USB) followed by incubation at 37°C for 4 h. A typical digestion contained 1–2 pmols of RNA. MALDI mass spectrometry was done on a Perseptive Voyager STR instrument with 3-hydroxypicolinic acid matrix, detecting positive ions in a reflector ToF mass analyser.
Data processing
Mass spectrometric data were processed in the m/z free software (ProteoMetrics Inc.). RNA digestion spectra were smoothened and externally calibrated from spectra of synthetic oligodeoxynucleotides. Peaks were initially assigned by the software's ‘Auto label peaks’ function, and the assignments were subsequently evaluated manually. Mass lists as text files, containing on each line a mass peak were generated for import into the RMM program. RMM is a purposely adapted program based on VEMS (Virtual Expert Mass Spectrometrist) (33) developed in Delphi and run on Windows-based computers. It can be downloaded from http://yass.sdu.dk/RMM/ together with a tutorial. The following databases/sequences in FASTA format were used for the RMM searches: ‘The Ribosomal Database Project’ (http://rdp.cme.msu.edu/); E. coli genome, gi:48994873; Geobacillus thermodenitrificans genome, gi:138893679; H. marismortui genome, gi:55376942, gi:55380074, gi:55376579, gi:55376107, gi:55376187, gi:55376228, gi:55376280, gi:55376412 and gi:55376144. A 0.3 Da mass accuracy was used with no variable modifications specified for the searches presented in this manuscript. For genome searching, the approximate size of the RNA was an extra search parameter.
RESULTS
There are presently two reliable RNases available, which are useful for an RMM approach, namely RNase T1 (guanosine residue specific) and RNase A (pyrimidine nucleotide specific). Evidently, RNase T1 will in average generate longer digestion fragments that will have a higher value in an identification approach (13). We have consequently limited the current study to digestions with this enzyme.
RNA identification in genomes
The chief aim of the present work was to show that we could identify the genetic origin of various RNAs. The algorithm used for the searching is based on the same combinatorial approach that has previously been described for VEMS using proteomics data (33), but we have developed a stand alone platform termed RMM.
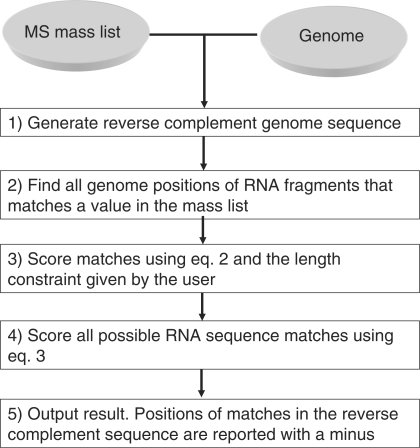
A crucial parameter in the genome search is to determine an approximate size of the RNA under investigation in order to give a window size that can be used as a search parameter. Size determination was done by running denaturing polyacrylamide gels of the purified fragment. In the first step, the mass list and genome sequence database are imported into RMM, and the reverse complement sequence of the genome is added to the sequence database (Step 1, Figure 1). The position of all RNA fragments in the genome that matches a mass peak in the given mass list is found (Step 2, Figure 1). Step 2 is computationally very fast and based on the same principles as the algorithm used for peptide mass mapping. Step 3 in Figure 1 consists of finding start and end boundaries of the RNA sequences in the genome. The current algorithm considers all possible starts and ends with the user-specified length constraint, which contains at least one RNA fragment match. Step 3 is done by a so-called brute-force search, since it considers all possibilities. However, the algorithm for defining the optimal boundary has a linear time complexity, which means that the time required for finding the optimal boundary is linearly correlated with the size of the genomic sequence and independent on the number of MS peak given in the input. This is achieved by using the fact that Si = Si−1 + log10(Pfrag, i−1) − log10(Pfrag, j), where Si [calculated by Equation (3) below] is the score obtained from the genomic start position i. Si−1 is the score obtained from the genomic start position i − 1. Pfrag, i−1 is the score of the digestion fragment in position i − 1 and Pfrag, j is the score of the last possible digestion fragment when starting from position i, both calculated by Equation (2) below. If j − i is larger than the user specified max length, then Pfrag, j = 0. Step 4 scores all the RNA sequences that contain at least one RNA fragment match using Equation (2) (see subsequent section for details on the ranking model). In other words, the algorithm reuses previous calculated scores for a genomic region to lower the computational time. In the final step, the program reports the top 20 RNA matches (matches on the reverse complement are indicated by a ‘minus’) together with their corresponding RNA fragment matches, scores, mass, deviation between observed and experimental masses (delta mass), sequence coverage and colour coding of the RNA fragment matches in the RNA sequence (Step 5, Figure 1).
Figure 1.
Bioinformatics strategy for identifying the genetic origin of an RNA molecule.
The scoring in Step 4 of Figure 1 is an intuitively simple but powerful probability model for ranking the RNA matches. All the RNA sequences that contain at least one RNA fragment mass match (only fragments larger than three nucleotides are considered here and in the subsequent steps) constitutes a database, which is converted on-the-fly into fragments based on the specificity of the enzyme used in the experiment, i.e., RNase T1. The a priori probability for an RNA digestion fragment to belong to a specific RNA is given by:
| 1 |
where n is the number of RNA fragments in a specific RNA and N is the total number of fragments in the database. On a given mass peak in the MS spectrum, we have Mi number of RNA fragments in the whole RNA database that matches the MS peak mass; Mi being dependent on the mass accuracy. In other words, Mi is the total number of RNA fragments present in the searched sequence database that has the mass mi ± Δm Da. In our case, Δm was equal to 0.3 Da. Each of the Mi can be considered as independent attempts to withdraw an RNA fragment that matches the specific RNA with the experimental mass mi. We can therefore estimate the probability that at least one of these RNA fragments extracted from a mass interval around a MS peak belongs to the specific RNA in question:
| 2 |
where Δm is the mass accuracy in daltons. This model takes the database size, the length of the target RNA and the mass accuracy into account. Since we assume that the correct RNA is present in the database, we do not apply any correction or penalty of unexplained peaks. In other words, our main goal is to find the target RNA in a given database that best explains the spectrum. The above probability values Pfrag is now used to calculate a ranking score Srank for the RNA matches:
| 3 |
where n is the number of matching peaks in the MS spectrum.
We use randomly calculated RNA masses as an attempt to address the question whether the correct RNA is present in the database. The result using the random RNA fragment masses is used to calculate a Z-score. The Z-score is calculated as the difference between the obtained score for a given match from the correct database minus the average of the top match from each of X iterations (user-defined number of iterations; 10 is used in the current work) of random RNA mass lists followed by division by the standard deviation of the scores of X random matches.
| 4 |
where Z is the Z-score and S is the raw score obtained by Equation (3) for the results originating from the experimental mass list. µ is the average of the raw scores from X top matches obtained from X database searches with randomly drawn RNA fragment masses and σ is the standard deviation of the scores of X random matches. Note that we use random in silico calculated RNA mass lists with the same number of masses as the experimental list for the searches against the database. Generating random masses from a uniform mass distribution would give too optimistic Z-scores. We find that two to five random iterative searches give fairly stable results for the Z-scores.
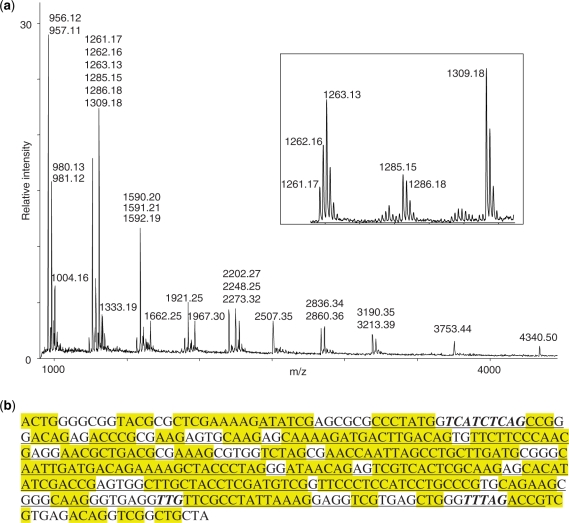
The overview of the entire bioinformatics workflow is depicted in Figure 1 and was tested with real data as described in the following. We have previously purified and MALDI mass spectrometry-analysed a series of defined subfragments of 23S rRNA from H. marismortui (26), and one of these spectra—covering approximately positions 2323–2630 of the 23S rRNA is displayed in Figure 2a. The assigned peaks constitute the mass list that was used to search the H. marismortui genome with a window size of 310 nucleotides. (Note that the RNase T1 digestion in this case produced almost exclusively the 2′–3′-cyclic phosphate versions of the digestion products (17), which was taken into account when creating the mass list). Mono- and di-nucleotide digestion products are not recorded, because these will be ubiquitous in essentially all RNAs and therefore have no value for identification. First round of peak assignment was performed by the data processing software, but we subsequently did a manual adjustment to assure labelling of the correct isotopic peaks, and to take into account partial signal overlap occurring due to the ∼1.0 Da mass difference between U- and C-nucleotides. The latter issue is illustrated in the insert of Figure 1. The peak cluster represented by assignment of the monoisotopic species at m/z 1309.18 has an intensity distribution that is close to the theoretically expected if only one analyte contributes to the peak cluster. The neighbouring peak clusters, on the other hand, has an intensity distribution that cannot be explained by a single analyte species; consequently, we chose to interpret these data as overlapping isotopic distributions of two (m/z 1285.15 and 1286.18) and three (m/z 1261.17, 1262.16 and 1263.13) species, respectively. We cannot always discern all species contributing to a given cluster, but the manual inspection of the peak intensity pattern in many cases allows indisputable assignment of additional signals from genuine digestion fragments.
Figure 2.
Mass spectrometry data and search result for a H. marismortui 23S rRNA subfragment. (a) Mass spectrum of H. marismortui 23S rRNA subfragment (around positions 2323–2630) digested with RNase T1. Assigned masses are from singly protonated digestion products, these masses were used in the subsequent genome search. Insert: zoom on peak clusters to illustrate the effect of digestion products with partially overlapping isotope distributions—see text for details. (b) Top scoring genomic region with flanks for RNA mass mapping of H. marismortui 23S rRNA subfragments 2323–2630. Underlined: identified sequence. Yellow highlight: RNase T1 digestion fragments with masses in peak list. Bold italic: RNase T1 digestion fragments with masses not present in peak list.
The search identified three genomic regions that code for the RNA in question (Table 1): the regions map to each of the three rRNA operons known in H. marismortui (34). Each of the rRNA operons resulted in numerous hits around the expected region, depending of the precise assignment of the termini; the search result with the highest score—positions 2305–2627 in the 23S rRNA gene together with flanking regions—is shown in Figure 2b. The digestion fragments, whose mass values are present in the mass list, are highlighted. This gives a good impression of the problem of precise assignment of the termini: the 310 nucleotide window will still comprise of observed digestion fragments if moved roughly 20 nucleotides upstream, without excluding large observed digestion fragments. Indeed, several such hits had identical scores and Z-scores. Two mass values in the mass list are not represented in the identified part of the 23S rRNA gene. One is at m/z 2248.25 corresponding to a posttranscriptionally modified fragment with the sequence U[Um][Gm]UUCG > p (26,35). The other is at m/z 1591.21 corresponding to an A1C2U1G1 > p composition, for which we cannot account. A full overview of peak assignment is given in Supplementary Table S1. Finally, three RNase T1 digestion fragments, expected from the identified 23S rRNA gene region, are not observed (bold italic in Figure 2b). The two larger ones are located where RNase H has digested during the purification of the RNA (see ‘Materials and methods’ section), whereas the smallest—UUG > p—is not produced by RNase T1, because of posttranscriptional modifications that results in the above mentioned composite fragment, U[Um][Gm]UUCG > p (26,35).
Table 1.
Top-scoring genomic regions for search with RNase T1 digested H. marismortui 23S rRNA subfragment (positions ∼2320–2630) against the H. marismortui ATCC 43049 genome
| Candidate region | Positions in gene | Score | Z-score | GenBank accession |
|---|---|---|---|---|
| 23S rRNA, rrnA operon | 2305–2627 | 380 | 9.9 | AY596297 |
| 23S rRNA, rrnC operon | 2305–2627 | 380 | 9.9 | AY596297 |
| 23S rRNA, rrnB operon | 2305–2627 | 348 | 9.5 | AY596298 |
Score is calculated according to Equation (3) and the Z-score according to Equation (4).
Table 2 summarizes the search results with other authentic RNA and in vitro transcribed species, which all showed the correct genomic location, but with varying precision in defining the termini. The identification of the 5S rRNA gene from B. stearothermophilus is particularly interesting (see Supplementary Table S2 for expected and observed peaks), because the genome of this species has not been sequenced. We chose to perform the search in the nearest relative that had a sequenced genome (36), namely Geobacillus thermodenitrificans. The genomic regions found were all perfectly within 5S rRNA genes of this species. G. thermodenitrificans has 10 copies of the 5S rRNA gene, but our search only identified nine of them with the same maximum score. A closer inspection of the unidentified 5S rRNA gene revealed it to be a sequence variant that results in an AAACACUCGp fragment (calculated m/z 2901.42) upon RNase T1 digestion instead of the observed digestion fragment (AAACACCCGp fragment; m/z 2900.41), resulting in a significantly lower score for this particular gene copy. The latter digestion fragment is predicted from all nine identified 5S rRNA gene copies. It was thus possible in the present case to identify the genomic origin of an RNA using genomic data from a related species.
Table 2.
Overview of top scoring genomic regions for various RNAs
| RNA | Position in gene, calculated | Position in gene, found | Sequence coveragea (%) |
|---|---|---|---|
| H. marismortui 23S rRNA subfragment | ∼530–697 | 531–702 | 92.4 |
| H. marismortui 23S rRNA subfragment | ∼681–969 | 685–935 | 98.8 |
| E. coli 23S rRNA subfragment; in vitro transcript | 2446–2632 | 2456–2621 | 100 |
| E. coli Spot 42; in vitro transcript | 1–109 | 4–97 | 100 |
| B. stearothermophilus 5S rRNA | 1–117 | 9–113 | 89.4 |
aThe sequence coverage is calculated as the percentage of the identified genomic region that is represented by masses from the peak list when considering that RNase T1 was used to obtain the peak list. Each mass may match several positions in the identified genomic region. Note that genome sequences that would result in mono- or di-nucleotides at the RNase T1 digestion level are not included in calculation of the sequence coverage.
Identification of 16S rRNA origin
A series of reports [e.g. (20,21)] have demonstrated bacterial identification by mass spectrometry of RNase-cleaved in vitro transcripts derived from PCR products of selected regions of the 16S rRNA gene. Our identification strategy relies on direct analysis of the authentic RNA, and it was therefore of interest to see if the correct 16S rRNA species could be identified in a database harbouring evolutionary related RNA species. For this purpose, we recorded mass spectra of 16S rRNA from T. thermophilus and E. coli and performed a search against an in silico digested database consisting of ∼165 000 prokaryotic 16S rRNA entries longer than 1200 nucleotides downloaded from ‘The Ribosomal Database Project’ (http://rdp.cme.msu.edu/).
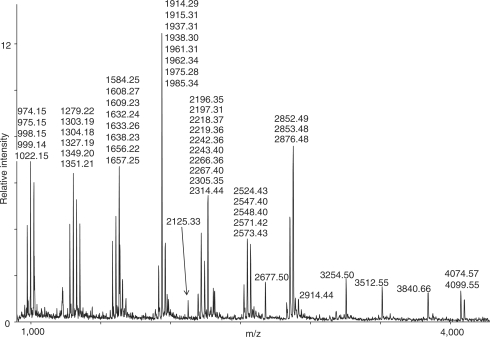
The MALDI spectrum of RNase T1 digested T. thermophilus 16S rRNA with ion signals used in the search is shown in Figure 3. The highest scores are presented in Table 3. It is obvious that we have an unambiguous identification of the correct species with all the 19 best matches being T. thermophilus 16S rRNA. The first match not specifically identified as a Thermus species comes as number 20 and is an uncultured species growing at 57°C with score and Z-score values of 313 and 43, respectively.
Figure 3.
Mass spectrum of T. thermophilus 16S rRNA digested with RNase T1. Assigned masses are of singly protonated digestion products, these masses were used in the subsequent database search.
Table 3.
Top scoring entries for search with RNase T1 digested T. thermophilus 16S rRNA in the RDP 16S rRNA database
| Rank | Score | Z-score | Organism | GenBank accession |
|---|---|---|---|---|
| 1 | 364 | 55 | T. thermophilus JN2 | AY554280 |
| 1 | 364 | 55 | T. thermophilus | AY497773 |
| 1 | 364 | 55 | T. thermophilus | No information |
| 1 | 364 | 55 | T. thermophilus E26 | DQ087525 |
| 5 | 362 | 54 | T. thermophilus HB27 | AE017221a |
| 5 | 362 | 54 | T. thermophilus HB8 | AP008226a |
| 5 | 362 | 54 | T. thermophilus HB27 | AE017221a |
| 5 | 362 | 54 | T. thermophilus HB8 | AP008226a |
| 9 | 360 | 54 | T. thermophilus L1 | AY788091 |
| 10 | 358 | 53 | T. thermophilus CS | AJ251938 |
| . . . | . . . | . . . | . . . | |
| 19 | 339 | 49 | T. thermophilus HB8 | X07998 |
| 20 | 313 | 43 | S000345626 uncultured bacterium; G24 | AF407704 |
The 10 highest scoring entries as well as number 19 and 20 are specified.
aEach identified rRNA represents one of the two copies present in the T. thermophilus genome.
If we make an in silico digestion of the top scoring 23S rRNA in Table 3 and compare the outcome with our experimental mass list, there are 29 theoretical digestion fragments, which do not occur in the mass list (data not shown). Only a single of the absent signals can be explained by posttranscriptional modifications (37), the rest either have m/z-values, which have overlap with the isotopic distribution of assigned signals or gave a mass spectrometric signal too weak for automatic detection by the software. There are 10 m/z-values in our mass list that do not appear in the in silico digestion of the top scoring 23S rRNA (Supplementary Table S3), three of which are explained posttranscriptional modifications (37). We can only speculate on the origin of the remaining observed signals, but conclude that these extra signals as well as missed assignment of true signals did not compromise a correct identification.
The outcome of a similar identification of E. coli 16S rRNA is presented in Table 4. At a first glance, the identification appears ambiguous, since 16S rRNA from two species—E. coli and Hafnia alvei were identified with identical scores followed by Shigella dysenteriae with a marginally lower score. However, sequence alignment of the three sequences (data not shown) reveal that the identified E. coli 16S rRNA is 98% identical to the H. alvei 16S rRNA and of identical length. The sequence identity is also 98% between the E. coli and S. dysenteriae 16S rRNAs, except for a 16 nucleotide extension in the 5′-end in the latter. Thus, the reason for the apparent ambiguous identification is an underlying de facto sequence identity between the 16S rRNA candidates, with differences only recognizable by complete sequencing. Nevertheless, this result suggests the possibility of identification of closely related species in case the sequence of the organism under study is not available.
Table 4.
Top three scoring entries for search with RNase T1 digested E. coli 16S rRNA in the RDP 16S rRNA database
| Score | Z-score | Description of organism | GenBank accession |
|---|---|---|---|
| 452 | 110 | Escherichia colia | Z83204 |
| 452 | 110 | Hafnia alvei | Z83203 |
| 450 | 110 | Shigella dysenteriaea Sd197 | CP000034 |
aThe subsequent seven identified 16S rRNA candidates originated from either E. coli or S. dysenteriae.
DISCUSSION
Since experimental data implicate RNA molecules as essential players in an increasing number of cellular processes, there is a need for reliable tools to identify novel RNAs. We demonstrate that it is experimentally possible to locate a purified RNA's genetic template in the appropriate genome by RMM.
To begin with, it was important to establish the efficiency and robustness of the entire set-up. Real mass spectral data are far from ideal, exhibiting missing peaks (due e.g. to modifications and low ionization ability) and artefact peaks (for example caused by contamination, adducts and noise), and therefore the practical usability of the method needed to be tested. Because we work with authentic RNA, the ∼1.0 Da mass difference between U- and C-nucleotides compelled the use of a reflector ToF mass analyser instead of a linear ToF ditto. This makes it possible to distinguish between masses from digestion products exhibiting a single pyrimidine nucleotide exchanges. Our mass accuracy is consistently better than 0.3 Da (often much better) after calibration, which in practice resulted in sufficiently accurate mass determination up to ∼6000 Da. As established in Figure 2, the resolution permitted the assigning of signals that differ by one m/z unit to different RNase digestion products in a manual interpretation of the mass spectra, which proved generally useful during the present work. However, digestion products differing in mass by a few daltons cannot always be identified, because they have overlapping isotope patterns. The isotope pattern of the peak cluster around m/z 1260 (Figure 2, insert) is so abnormal that it can only be explained by the presence of more than one digestion product; but often, the assignment of all but the lightest of the RNase digestion products in a peak cluster is difficult, depending on the number and stoichiometry of different digestion products contributing to a cluster. It might be attempted to get a better deconvolution of the digestion products contributing to a peak cluster by fitting the observed peak pattern to different combinations of theoretical isotope patterns for relevant potential digestion products.
Mass spectrometric RMM exhibits very high sequence coverage, defined as percentage of sequence represented by signals in the mass spectrum, when comparing with PMF data. Based on our data, sequence coverage is typically 90% or better with RNAs up to 300 nucleotides (Table 2), whereas the sequence coverage in PMF is typically a factor of two lower. The main reason for the high sequence coverage is the relatively like physico-chemical properties of the nucleotides that make most RNA digestion products behave fairly similar during mass spectrometric analysis. We do observe low mass spectrometric response for some nucleotide compositions/sequences and in rare cases complete absence, particularly if the starting RNA is large and many uridines are present, but not to a degree where subsequent identification was impaired.
Tandem mass spectrometry may be used to generate sequence tags that would have very high values in an identification compared to the composition-based mass values of digestion fragments. The use of tandem mass spectrometry will be a further improvement of the identification approach, but not a trivial one to implement. RNA fragments not only in the backbone, but also through loss of nucleobases, which complicates tandem mass spectrum interpretation. In addition, tandem mass spectrometry on complex mixtures such as digestions will normally require online liquid chromatography—tandem mass spectrometry (LC-MS2), which is not routine for RNA analysis. The LC-MS2 approach is, however, appealing, because it would open for the identification of RNAs in a mixture. On the basis of previous experience with dynamic range of MALDI TOF analysis of RNA, we estimate that the sample has to be more than 80% pure for our current identification approach, but a robust LC-MS2 set-up would allow a higher throughput, because of less demand on sample purification.
The RRM program is able to include variable modifications (methylations being the most abundant by far) for RMM. However, taking modifications into consideration would only add little extra information in the identification, because any nucleotide may be methylated in RNA and the impact on the digestion cannot be predicted. Thus, the extra complexity in computation and output will not be justified by a marginal increase in identification score. tRNAs is a special case due to their extremely high degree of—and diversity in—modification. tRNA identification is likely only possible after tandem mass spectrometry of selected RNase digestion fragments (38).
Our results were obtained with RNA of prokaryotic origin. Prokaryotes in most cases have their genetic material on a single chromosome, and their genome size is typically orders of magnitude smaller than eukaryotes. The search time in the E. coli genome in forward and complement reverse direction is currently around 3 min on a standard desktop computer, and the search time will increase approximately linearly with the genome size. However, in future work, the computational time complexity of Step 3 in Figure 1 needs to be readdressed together with the problem of RNA splicing and exon/intron boundaries. We plan to do in silico simulations to address these issues and to implement the use of MS/MS spectra to obtain more significant genome position matches.
The B. stearothermophilus genome is not sequenced, but because the genome of the closely related Geobacillus thermodenitrificans was available, identification of the template for the B. stearothermophilus 5S rRNA was possible. Much more experience is needed to make a general statement about the possibilities for cross-species identification, but the important implication for our approach is that one can work with a much broader range of organisms than just the ones with a sequenced genome.
The identification of 16S rRNAs in a small ribosomal subunit RNA database was not surprising per se, because theoretical considerations have shown that the concept should be viable (13,14). The crucial point is that despite RNA's just four building blocks, the sequence differences are sufficient to give species-specific RNase digestion patterns that allow unambiguous identification. Several reports have also shown experimental proof of the concept (19–21), but these experiments were performed with in vitro synthesized RNA harbouring chemically modified nucleotides to optimize the experimental data for improved database identification. We demonstrate that identification is also possible using the rRNA with posttranscriptional modifications directly.
In conclusion, we have confirmed in practice that RMM is a powerful technique for RNA identification: given a few picomoles of purified prokaryotic RNA and an appropriate database, it is possible to identify the DNA template of an unknown RNA within a working day. Compared to previous reports, we have taken RNA mass-mapping algorithms a step further so it can also be applied for RNA genome searches. The tools—including software and robust experimental procedures—are available for a new and rapid identification of unknown RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Natural Science Research Council and Danish Biotechnology Instrument Centre. Funding for open access charge: University of Southern Denmark.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Anette Rasmussen for technical assistance and Jesper Johansen for supplying the spot 42 in vitro transcript. We also thank Peter Højrup for critical reading of the manuscript.
REFERENCES
- 1.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acid Res. 1977;11:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupp G, Krupp HJ. Rapid RNA sequencing: nucleases from Staphylococcus aureus and Neurospora crassa discriminate between uridine and cytosine. Nucleic Acid Res. 1979;6:3481–3490. doi: 10.1093/nar/6.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peattie DA. Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldmann R, Gross HJ, Krupp G. Protocol for rapid chemical RNA sequencing. Nucleic Acid Res. 1987;15:7209. doi: 10.1093/nar/15.17.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 6.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridisation to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 7.Adamczyk J, Hesselsoe M, Iversen N, Horn M, Lehner A, Nielsen PH, Schloter M, Roslev P, Wagner M. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 2003;69:6875–6887. doi: 10.1128/AEM.69.11.6875-6887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsholz B, Worl R, Blohm L, Albers J, Feucht H, Grunwald T, Jurgen B, Schweder T, Hintsche R. Automated detection and quantitation of bacterial RNA by using electrical microarrays. Anal. Chem. 2006;78:4794–4802. doi: 10.1021/ac0600914. [DOI] [PubMed] [Google Scholar]
- 9.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl Acad. Sci. USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann M, Højrup P, Roepstorff P. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 11.Pappin DJ, Højrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 12.Kirpekar F. RNA mapping. In: Caprioli RM, editor. The Encyclopedia of Mass Spectrometry. Biological Applications, Part B: Carbohydrates, Nucleic Acids and other Biological Compounds. Vol. 3. Oxford, UK: Elsevier; 2006. pp. 10–14. [Google Scholar]
- 13.Zhang Z, Jackson GW, Fox GE, Willson RC. Microbial identification by mass cataloging. BMC Bioinform. 2006;7:117. doi: 10.1186/1471-2105-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson GW, McNichols RJ, Fox GE, Willson RC. Bacterial genotyping by 16S rRNA mass cataloging. BMC Bioinform. 2006;7:321. doi: 10.1186/1471-2105-7-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomerantz SC, Kowalak JA, McCloskey JA. Determination of oligonucleotide composition from mass spectrometrically measured molecular weight. J. Am. Soc. Mass Spectrom. 1993;4:204–209. doi: 10.1016/1044-0305(93)85082-9. [DOI] [PubMed] [Google Scholar]
- 16.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirpekar F, Douthwaite S, Roepstorff P. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA. 2000;6:296–306. doi: 10.1017/s1355838200992148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahner S, Lüdemann H-C, Kirpekar F, Nordhoff E, Roepstorff P, Galla H-J, Hillenkamp F. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI) of endonuclease digests of RNA. Nucleic Acids Res. 1997;25:1957–1964. doi: 10.1093/nar/25.10.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Wintzingerode F, Böcker S, Schlötelburg C, Chiu NHL, Storm N, Jurinke C, Cantor CR, Göbel UB, van den Boom D. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc. Natl Acad. Sci. USA. 2002;99:7039–7044. doi: 10.1073/pnas.102165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefmann M, Honisch C, Böcker S, Storm N, von Wintzingerode F, Schlötelburg C, Moter A, van den Boom D, Göbel UB. Novel mass spectrometry based tool for genotypic identification of mycobacteria. J. Clin. Microbiol. 2004;42:339–346. doi: 10.1128/JCM.42.1.339-346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson GW, McNichols RJ, Fox GE, Willson RC. Universal bacterial identification by mass spectrometry of 16S ribosomal RNA cleavage products. Int. J. Mass Spectrom. 2006;261:218–226. [Google Scholar]
- 22.Hartmer R, Storm N, Boecker S, Rodi CP, Hillenkamp F, Jurinke C, van den Boom D. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res. 2003;31:e47. doi: 10.1093/nar/gng047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krebs K, Medugorac I, Siechter D, Förster M. RNaseCUT: a MALDI mass spectrometry-based method for SNP discovery. Nucleic Acids Res. 2003;31:e37. doi: 10.1093/nar/gng037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siechter D, Krebs S, Förster M. Rapid and accurate characterization of short tandem repeats by MALDI-TOF analysis of endonuclease cleaved transcripts. Nucleic Acid Res. 2004;32:e16. doi: 10.1093/nar/gnh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirpekar F, Hansen LH, Rasmussen A, Poehlsgaard J, Vester B. The archaeon Haloarcula marismortui has few modifications in the central parts of its 23S ribosomal RNA. J. Mol. Biol. 2005;348:563–573. doi: 10.1016/j.jmb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Douthwaite S, Christensen A, Garrett RA. Binding site of ribosomal proteins on prokaryotic 5S ribonucleic acids: a study with ribonucleases. Biochemistry. 1982;21:2313–2320. doi: 10.1021/bi00539a007. [DOI] [PubMed] [Google Scholar]
- 28.Douthwaite S, Powers T, Lee JY, Noller HF. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- 29.Mengel-Jørgensen J, Jensen SS, Rasmussen A, Poehlsgaard J, Iversen JJL, Kirpekar F. Modifications in Thermus thermophilus 23 S ribosomal RNA are centered in regions of RNA-RNA contact. J. Biol. Chem. 2006;281:22108–22117. doi: 10.1074/jbc.M600377200. [DOI] [PubMed] [Google Scholar]
- 30.Treede I, Jakobsen L, Kirpekar F, Vester B, Weitnauer G, Bechthold A, Douthwaite S. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol. Microbiol. 2003;49:309–318. doi: 10.1046/j.1365-2958.2003.03558.x. [DOI] [PubMed] [Google Scholar]
- 31.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirpekar F, Douthwaite S. Identifying modifications in RNA by MALDI mass Spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 33.Matthiesen R, Trelle MB, Højrup P, Bunkenborg J, Jensen ON. VEMS 3.0: algorithms and computational tools for tandem mass spectrometry based identification of post-translational modifications in proteins. J. Proteome Res. 2005;4:2338–2347. doi: 10.1021/pr050264q. [DOI] [PubMed] [Google Scholar]
- 34.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA. 2002;8:202–213. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS, et al. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 37.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006;45:4888–4999. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 38.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homologue Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.