Abstract
Helicobacter pylori colonizes the human gastric mucosa and this can lead to chronic gastritis, peptic and duodenal ulcers, and even gastric cancers. The bacterium colonizes over one-half of the worlds population. Nickel plays a major role in the bacteriums colonization and persistence attributes as two nickel enzyme sinks obligately contain the metal. Urease accounts for up to 10% of the total cellular protein made and is required for initial colonization processes, and the hydrogen oxidizing hydrogenase provides the bacterium a high-energy substrate yielding low potential electrons for energy generation. A battery of accessory proteins are needed for maturation or activation of each of the apoen-zymes. These include Ni-chaperones and GTPas-es, some of which are unique to each Ni-enzyme and others that are individually required for maturation of both the Ni-enzymes. H. pylori’s need for some conventional hydrogenase maturation proteins playing roles in urease maturation may have to do with the poor nickel-sequestering ability of the UreE urease maturation protein compared to other systems. H. pylori also possesses a NixA nickel specific permease, a nickel dependent regulator (NikR), a recently identified nickel efflux system (CznABC), and a histidine-rich heat shock protein, HspA. Based on mutant analysis approaches all these proteins have roles in nickel homeostasis, in urease expression, and in host colonization. The His-rich putative nickel storage proteins Hpn and Hpn-like play roles in nickel detoxification and may influence the levels of Ni-activated urease that can be achieved.
Keywords: Nickel enzyme, Hyp protein, Hydrogenase, Urease, Gastric colonization, Ulcer, Gastric cancer, Nickel storage, Nickel regualtion, Histidine-rich
Helicobacter pylori is a gram negative, motile, microaerophilic pathogen that colonizes the gastric mucosa of humans and causes chronic gastritis, peptic and duodenal ulcers, and even gastric cancers (Blaser 1990; Marshall and Warren 1984; Parsonnet et al. 1991; Parsonnet 1994; Parsonnet et al. 1994). The key aspect of its pathogenesis is its persistent nature (Israel and Peek 2006). The bacterium is characterized by its genetic diversity and its stringent adaptation to the human stomach, which includes ability to buffer the gastric acidity and adept motility attributes permitting transit of the stomach gastric contents or moving within the viscous mucous environment, respectively. Other important (virulence) characteristics are its ability to combat host-mediated oxidative stress and release of adhesions and cytotoxins that help maintain bacterial persistence (Mobley et al. 2001). Helicobacter pylori produces two final nickel sinks; they are the Ni-enzymes urease and hydrogenase (Evans et al. 1991; Maier et al. 1996). Much of the bacterium’s metal metabolism is apparently centered on expression and maturation of these two enzymes. Urease accounts for up to 10% of the total cellular protein made and is essential for colonization and virulence (Bauerfeind et al. 1997; Hu and Mobley 1990; Tsuda et al. 1994a; Tsuda et al. 1994b), while the hydrogen utilizing hydrogenase provides the bacterium a compact and high energy non-carbon substrate for respiratory based energy generation (Maier et al. 1996; Olson and Maier 2002). In addition to these nickel enzymes, H. pylori also possesses a NixA nickel specific permease (Bauerfeind et al. 1996), accessory proteins UreIEFGH and HypABCDEF (some of which bind nickel) required for maturation or activation of the apoenzymes for the two nickel enzymes (Benoit et al. 2004; Mehta et al. 2003a; Olson et al. 2001; Voland et al. 2003), a nickel dependent regulator, NikR (Contreras et al. 2003; van Vliet et al. 2002), a recently identified nickel efflux system (CznABC) (Stahler et al. 2006) and a histidine-rich heat shock protein, HspA, a GroES homologue (Kansau et al. 1996; Kansau and Labigne 1996). Here we address the roles and characteristics of these proteins, with emphasis on the roles of the accessory proteins directly required for Ni-enzyme maturation, and on the His-rich putative nickel storage proteins Hpn and Hpn-like. Helicobacter pylori NikR is the subject of another manuscript (A. van Vliet) in this issue and will therefore not be covered here.
NixA, the Nickel-specific permease
The nixA gene was first isolated in 1995 (as a gene enhancing urease activity) from a plasmid library composed of the H. pylori chromosome. It enhanced the Ni-enzyme activity when introduced into an E. coli strain carrying the urease structural genes (Mobley et al. 1995). Disruption of nixA in H. pylori led to a partial reduction of urease activity (up to 50% in some strains) (Bauerfeind et al. 1996; Nolan et al. 2002) which suggests the presence of additional nickel transporters in H. pylori yet to be characterized (Davis and Mobley 2005). Nevertheless, nixA mutants in the mouse-adapted strain SS1 failed to colonize the gastric mucosa, hence showing that NixA is required and that nickel uptake is of critical importance for the cell survival in the host (Nolan et al. 2002). NixA protein has been thoroughly characterized. It is an integral cytoplasmic membrane protein of approximately 37 kDa which belongs to the nickel-cobalt transporter family, found in a variety of microorganisms (Eitinger and Mandrand-Berthelot 2000; Saier et al. 1999). Members of this family usually have eight trans-membrane domains (Saier et al. 1999) and topology studies using lacZ and phoA gene fusions revealed this was indeed the case for H. pylori NixA (Fulkerson and Mobley 2000). Conserved Asp, Glu and His residues are located in the transmembrane domains and appear to be critical for nickel transport (Fulkerson et al. 1998); in addition, two essential domains have been characterized within helix II (GXXHAXDADH) and helix III (GX2FXXGHSSVV) of NixA (Fulkerson and Mobley 2000). Furthermore, other amino acids, including some with low in vitro affinity to nickel, have been shown to be also important for the function of NixA (Wolfram and Bauerfeind 2002). NixA has a high affinity for nickel, with an estimated Kd of 11.3 nM (Mobley et al. 1995), thus enabling H. pylori to efficiently scavenge the low concentrations of nickel found in the human body, which are estimated to be in the range of 2–11 nM (Sundermann 1993). Since high concentrations of nickel would be toxic to H. pylori, it is noteworthy that synthesis of NixA is repressed by nickel; this process is mediated by NikR (van Vliet et al. 2004; Wolfram et al. 2006).
The CznA BC metal efflux pump
A novel efflux pump with metal binding capacity and that influences urease activity in H. pylori was recently described (Stahler et al. 2006). The unique feature of the metal export apparatus is CznC, a nickel binding efflux protein. Mutations individually in any of the czn A, B, or C genes rendered the pathogen ineffective in animal stomach colonization, underscoring the importance of metal homeostasis to virulence. While the structural organization of the Czn complex is probably like the Czc-type trans-envelope transporters of Ralstonia (and some other bacteria) that play key roles in conferring metal resistance (Nies 2003), the H. pylori efflux pump was shown to have a unique physiological role, that is modulation of urease activity (Stahler et al. 2006). Urease activity was significantly enhanced in the cznC and cznA mutants, and the CznC protein was assigned a role of particular importance to nickel export. Recombinant forms of both CznC and CznB could bind nickel. The individual cznA, cznB, and cznC mutants were all more sensitive to cadmium, nickel, and zinc than the parent strain. It is proposed that urease activity is modulated by nickel efflux via CznABC to reduce Ni-activated urease levels, and that efflux of other metals such as cadmium and zinc prevents urease inhibition. The model is strengthened by the observations that considerable apourease (devoid of nickel) is oftentimes observed in H. pylori, so that intracellular nickel levels would be expected to rapidly affect urease activity via enzyme activation/inactivation (Stingl and De Reuse 2005). An example of the importance of metal cation discrimination to H. pylori physiology is highlighted by the unique properties described for this Czn pump complex.
Accessory proteins for hydrogenase maturation
Helicobacter pylori contains all the Hyp accessory proteins necessary for maturation of NiFe hydrogenases. These are HypABCDEF. While the genes for these are clustered together in one operon in most H2 utilizing bacteria, they are instead separated into three different chromosomal locations on the H. pylori genome; hypA alone, hypBCD together, and hypEF together. In addition, the structural genes hydABCD are located at yet another area of the H. pylori chromosome. The most informative system for identifying the roles of these accessory proteins has been Escherichia coli, where the accessory proteins play roles in maturation of all three NiFe hydrogenases (the term hyp is derived from hydrogenase pleiotropic) and the currently-assigned roles are based primarily on the work of A.Böck and colleagues (Blokesch and Bock 2002; Blokesch et al. 2004a; Hube et al. 2002; Reissmann et al.2003). HypA (also known as HybF) is a Zn-containing protein (Atanassova and Zamble 2005)) but also binds stoichiometric amounts of nickel, and HypB is a GTPase (Blokesch et al. 2004a; Leach et al. 2005). The present model is that HypA serves as a nickel chaperone, and HypB as a regulator that thermodynamically controls the donation of the metal to the hydrogenase apoprotein or release of the nickel-free chaperone (Blokesch et al. 2004c; Leach et al. 2005; Reissmann et al. 2003). The present model for HypA/HypB function (Leach et al. 2005) involves a key metal binding site in HypB beyond the usual (Ni-binding) polyhistidine stretch. This high affinity site would serve as the nickel donor for hydrogenase. HypA would direct Ni(II)-bound HypB to the iron-loaded site of the hydrogenase large subunit, and facilitate the metal transfer. The model takes into account the weaker Ni-binding ability of HypA (compared to HypB), as important in facilitating the Ni-transfer step (Leach et al. 2005). The authors also suggested a feasible role for the low-affinity metal binding site of HypB; it may detect the nickel status of the enzyme in order to sense when to activate the GTPase. Such an activation step upon sensing proper nickel loading would be of benefit, as HypB’s have notoriously sluggish GTPase activities when tested in vitro.
Two other Hyp proteins, HypF and HypE are involved in the synthesis of the cyanide ligand of the Fe metal center of hydrogenase (Reissmann et al. 2003). For this role, HypF is a carbamoyl-transferase, carbamoylating the HypE protein using carbamoyphosphate as substrate. The thio-cyanate product comes from an ATP-dependent dehydratase activity of HypE (Blokesch et al. 2004b). The origin of the other diatomic ligand (carbon monoxide) attached to the Fe center is not known. A HypC-HypD complex is likely involved in accepting the Fe-cyanide ligand and transferring the fully liganded Fe to the large subunit of hydrogenase. These maturation steps involve dynamic complexes between these accessory proteins and sometimes between these accessory proteins and the hydrogenase subunits (Blokesch et al. 2004a; Blokesch et al. 2004b). There is no reason to believe that the hydrogenase maturation steps performed by these proteins are different in H. pylori. As for mutants studied in other H2 oxidizing bacteria, mutant strains in each of the H. pylori hyp genes yielded strains with a hydrogenase deficiency phenotype (Benoit et al. 2004; Mehta et al. 2003a; Olson et al. 2001). Similarly, the deficiency could be connected to nickel, as nickel supplementation to the medium of mutant strains partially restored the hydrogenase activity.
Accessory proteins for urease maturation
The gene products of the ureIEFGH operon, located downstream of the ureAB urease structural genes, are required for the production of active urease. Indeed, coexpression of the ureI, ureE, ureF, ureG and ureH genes was shown to be required along with structural genes ureA and ureB to yield fully active urease in E. coli (Cussac et al. 1992). More recently, Voland and coworkers have shown that H. pylori ureE, ureF, ureG and ureH mutants are severely deficient in urease activity (Voland et al. 2003). Among these four genes, the roles of ureE and ureG have drawn the most attention. While addition of excess nickel can restore urease activity in hypA or hypB mutants (see below), this is not the case for either the ureE or the ureG mutant (Benoit, Mehta Maier, unpublished data) which indicates that these genes are absolutely required for urease activity under any circumstance. However the UreE and the UreG protein do not appear to be involved in hydrogenase maturation, since mutants in either gene have hydrogenase activities comparable to those of wild type cells (Benoit and Maier 2003; Mehta et al. 2003a).
Helicobacter pylori UreE (19.4 kDa) is found as a dimer in solution and equilibrium dialysis studies show each dimer is able to bind only 1 Ni2+ ion (Benoit and Maier 2003). The protein contains one conserved His residue (His102), shown to be involved in nickel binding in other well studied UreE proteins, including Klebsiella aerogenes UreE (residue His96) (Colpas et al. 1999; Song et al. 2001) or Bacillus pasteurii UreE (residue His100) (Stola et al. 2006). By adding several His residues (His6-tag or His-rich tail of K. aerogenes UreE) to the C-terminus of the H. pylori UreE, it is possible to artificially increase the nickel binding capacity of the protein (as confirmed in vitro with purified proteins); introduction of any one of a number of engineered versions into the chromosome of mutants deficient in nickel sequestration led to an increase of urease activity. It was concluded that there is a correlation between the nickel-sequestering ability of UreE and the nickel activation steps yielding urease activity in H. pylori (Benoit and Maier 2003).
UreG (22 kDa) is thought to play a role in the urease maturation process as a GTPase (Mehta et al. 2003a); GTP hydrolysis is apparently required for a nickel transfer or a protein-protein interaction/release step. Although purified UreG showed negligible GTPase activity, the protein contains a conserved nucleotide-binding domain (GSGKT) known as a “P-loop” motif, and site-directed mutation in this conserved domain (Lys14 to Ala14) abolished the urease activity in H. pylori (Mehta et al. 2003a). The assignment of UreG as a GTPase required for the urease activation, as well as the critical role played by the P-loop motif have been reported in K. aerogenes (Moncrief and Hausinger 1997) or B. pasteurii (Zambelli et al. 2005).
Although UreF (28.6 kDa) and UreH (29.7 kDa) have not yet been characterized in H. pylori, their role as chaperone proteins can be anticipated by observation of their homologs in K. aerogenes. In this bacterium, UreD (homologous to H. pylori UreH) binds to apourease, thus inducing a conformational change required before the next steps (Park et al. 1994), which are (i) insertion of UreF to make a UreD-UreF apourease complex (Moncrief and Hausinger 1996) (ii) addition of UreG to yield a UreD-UreF-UreG apourease complex (Park and Hausinger 1995; Soriano and Hausinger 1999) and (iii) involvement of UreE with this complex (Soriano et al. 2000). This is in good agreement with a recent study from Park and coworkers, which suggests that the products of plasmid-borne H. pylori ureFGH genes expressed in E. coli cooperate together before interacting with UreE and UreI (Park et al. 2005).
UreI (21.7 kDa) is a proton-gated inner membrane protein involved in urea transport as well as acid resistance (Rektorschek et al. 2000; Skouloubris et al. 1998). UreI possesses six membrane-spanning segments (Weeks et al. 2000) and conserved histidine residues, including His123, are essential for acid activation of the channel (Bury-Mone et al. 2001; Weeks et al. 2000; Weeks and Sachs 2001). The UreI protein is required for survival of the bacterium in the mouse or the gerbil gastric mucosa (Skouloubris et al. 1998) (Mollenhauer-Rektorschek et al. 2002).
Direct interactions among H. pylori urease accessory proteins have been reported, including UreF-UreH and UreG-UreE interactions, as well as interactions between the urease catalytic subunits UreA/B and UreI (Voland et al. 2003).
Accessory proteins common to both Ni-enzyme maturation systems
Helicobacter pylori has all the known accessory proteins needed for the individual maturation of hydrogenase and urease. Nevertheless, when mutant strains in each of the hydrogenase genes were assayed for urease activity an unexpected phenotype was obtained for two mutants, hypA or hypB (see table 1). These two strains were almost devoid of urease activity, and full urease activity could be achieved by supplementing the cultures with 5 µM Ni (Olson et al. 2001) The mutant strains synthesized normal levels of urease apoproteins, but the urease pool was lacking in nickel content (Olson et al. 2001). Also, polar affects of the mutation were ruled out as the cause of this phenotype as chromosomal complementation of the hypA strain (to create strain hypA:kanHA) with the wild type version of the gene restored both hydrogenase and urease activities (see table 1); complementation of the hypB strain was unnecessary as the next open reading frame was hypC, and a hypC mutant was normal in urease activity. It seems that H. pylori is unique in that the two Hyp proteins are involved in maturation of both Ni-enzymes. Gene targeted mutant strains in HypA and HypB in which key residues were targeted (i.e. the Ni-binding His2 for HypA, and the GTP binding Lys59 of HypB) resulted in the conclusion that these functions (Ni binding and GTPase, respectively) are involved in the Ni-dependent maturation of both hydrogenase and urease (Mehta et al. 2003a,b). H. pylori HypA and HypB interact with each other in a 1:1 molar ratio based on cross-linking and immunoblotting approaches, and this interaction did not depend on added nickel or GTP. H. pylori HypA and HypB must have the unique ability to recognize Ni-dependent maturation machinery for two different Ni-enzymes, and these two Ni-enzymes have completely different nickel centers. It therefore seems likely that HypA/B would interact with other accessory proteins at relatively early steps in the sequential Ni-enzyme maturation process. Preliminary studies involving mixing of pure H. pylori Ure accessory proteins and HypA/B, then capturing the intimate contacts by crosslinking have indicated that HypA can recognize accessory proteins from the heterologous system (Benoit, Mehta, Maier, unpublished data). Most likely, the HypA/B complex is able to donate/mobilize Ni to a UreE/G complex to facilitate Ni donation ultimately to urease.
Table 1.
Ni-enzyme activities of Helicobacter pylori accessory protein mutants
| Strain | Hydrogenase activitya | Urease activityb |
|---|---|---|
| Wild type | 1.4 ± 0.3 | 64 ± 23 |
| HypA | <0.01 | 0.3 ± 0.1 |
| HypB | <0.01 | 1.6 ± 0.5 |
| HypC | <0.01 | 62 ± 19 |
| HypD | <0.01 | 71 ± 11 |
| HypE | <0.01 | 76 ± 17 |
| HypF | <0.01 | 78 ± 14 |
| hypA:kanHA | 1.2 ± 0.3 | 62 ± 8 |
nmol H2 oxid per min per 108 cells
µmole urea hydrolyzed per min per mg protein
Urease activity of hypA and hypB strains was fully restored by adding 5 µM nickel. Data is from Olson et al. 2001, Mehta et al. 2003a and Benoit et al. 2004
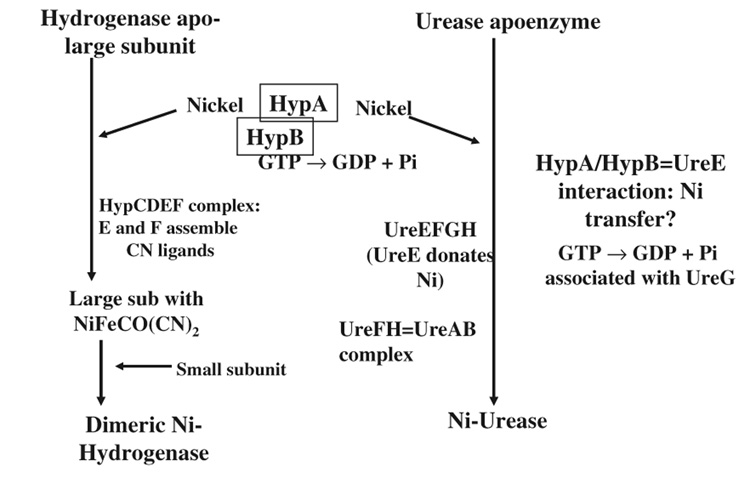
The Helicobacter pylori Ni-enzyme maturation summary is shown in Fig. 1. It relies heavily on the best-studied systems E. coli (for hydrogenase) and K. aerogenes (for urease) for function of many components, but also incorporates the unique roles of hypA/B, and their preliminarily-defined interactions with UreE. The reason for the additional complexity in maturation components (use HypA/B when all the other Ure accessory components should suffice) required for H. pylori urease maturation may be related to the high nickel demand of the gastric pathogen for abundant urease expression. While K. aerogenes UreE can bind 5–6 nickel ions per dimer, the H. pylori UreE dimer can bind only one (see table 2). At the same time the Ni-enzyme (urease) that depends on UreE for nickel is present as only about 0.1% of the total cell protein (K. aerogenes) compared to up to 10% (H. pylori) (see table 2 legend). It seems likely that H. pylori may have incorporated an already needed (for hydrogenase) Ni-sequestering protein to aid its nickel sequestering capacity for urease maturation, as its UreE is relatively poor in total Ni-binding capacity. In support of this, it was observed that introduction of engineered versions of UreE containing additional His residues into H. pylori hypA or hypB strains resulted in 5–10 fold greater urease activities than the mutant strains (see accessory proteins for urease maturation section above).
Fig. 1.
Helicobacter Ni-Enzyme Maturation
Table 2.
Comparisons of UreE from two sources
| Bacterium | Ni-binding residues | Ni-binding abilities (per homodimeter) | Urease madea |
|---|---|---|---|
| K. aerogenes | H-96, H-110, H-112 | 5–6 | 0.1% |
| H. pylori | H-102 | 1 | 10% |
percent of total cell protein, for Ka from R. Hausinger (pers. commun); for Hp, from Bauer and Mobley (1997).
The Ni-binding chaperone-homologue HspA
An interesting H. pylori heat-shock protein in the GroES family was characterized (Kansau et al. 1996; Kansau and Labigne 1996). The N-terminal portion is homologous to other GroES proteins, but HspA uniquely contains 27 additional residues at the His-rich C-terminus that are involved in nickel binding. The C-terminus is rich in His residues. Among a series of divalent cations tested, HspA showed the greatest specificity and affinity for nickel; it bound about two nickel ions per HspA molecule and with a Kd of 1.8 µM. Another Ni-binding domain but with lower metal affinity was identified by use of nickel levels above 30 µM. With its two identified and distinct functional domains, HspA is proposed to play a role as a nickel carrier (domain B), and as a classical GroES chaperonin (the A domain). Among many clinical isolates tested, the A domain is highly conserved, whereas the B domain encompasses two variant type sequences. The different domains elicit distinct host immunological responses (Kansau et al. 1996). HspA may play a role in urease maturation as expression of HspA in E. coli cells containing urease-encoding genes resulted in a fourfold increase in urease activity. It would be interesting to assess the urease expression with truncated (domain-specific) or site-changed versions of the HspA.
Hpn and Hpn-like proteins
An H. pylori protein rich in histidine residues (47% histidine residues, TIGR annotation HP1427, see Fig. 2) was identified almost 10 years ago. It was named ‘Hpn’ because it was first identified in H. p ylori and had affinity for nickel ions (Gilbert et al. 1995). Hpn is also present in the ferret and cat gastric Helicobacter pathogens, but it is absent from the mouse (liver) pathogen H. hepaticus. H. pylori mutants lacking hpn were more sensitive to nickel and bismuth than the parent strain, but a nixA strain was not more sensitive; this indicates that Hpn may sequester metals that accumulate internally via a passive equilibrium mechanism (from a high external metals environment) (Mobley et al. 1999). As nixA itself is regulated by nickel levels (through NikR) , it would seem to be of benefit to H. pylori to have Hpn proteins available to detoxify rapid fluxes in metal levels. Recombinant Hpn was recently purified, characterized and analyzed for metal content; it exists mainly as a range of multimeric forms in solution. In the presence of Ni2+ and imidazole, a high molecular weight aggregate ( > 500 kDa) of Hpn is observed, but with addition of DTT, the amount of the high MW form decreases greatly and the predominant form is a 136 kDa species. This size corresponds to a 20-mer, and it was suggested that this is the physiologically relevant form in vivo (Ge et al.2006). Still, the 136 kDa form is apparently in equilibrium with the other multimers; subjection of the pure (from gel filtration chromatography) 136 kDa form to gel filtration again yields not only the 136 kDa form but the other multimers observed in the original sample as well. Each Hpn monomer of 7 kDa reversibly binds 5 nickel ions at pH of 7.4. Although it is presumed the bulk of the nickel sequestering occurs via the multiple imidazole groups, Ni2+ -bound Hpn was very low in free thiolates, indicating the four Cys residues also take part in Ni-binding. The moderate Kd of 7.1 µM for nickel means the protein could facilitate nickel transfer to H. pylori Ni-chaperones with lower Kd’s, such as HypA (Kd of 1.3 µM ) or HspA (Kd of 1.8 µM). Such an intimate interaction and perhaps Ni transfer steps could be investigated by cross-linking approaches.
Fig. 2.
Histidine rich proteins of H. pylori
Nickel can be released from the Hpn protein by decreasing the pH or by adding nickel chelating agents such as EDTA (Ge et al. 2006). The former observation may be highly relevant to H. pylori nickel physiology and Ni-enzyme expression, as the bacterium can survive a wide pH range and can alter its pH environment. Also, if Hpn serves a (nickel) storage role it would be expected to use a facile mechanism for release of stored nickel. Nickel levels in E. coli BL21 cells expressing H. pylori Hpn from an inducible plasmid was higher than in uninduced cells or those lacking the (pET-hpn) plasmid (Ge et al. 2006). The level of nickel in the H. pylori strain 26695 cytoplasm was observed to be slightly higher than in an hpn deletion mutant. All the above is consistent with a proposed Ni-storage and detoxification role for the His-rich protein. Additionally, Hpn represents 2% of the total protein of H. pylori 26695, so it would be expected to play a large role in the cells nickel budget and Ni-homeostasis. No differences in the urease activities in the hpn mutant compared to the wild-type were reported in the two published studies in which hpn mutants were described (Ge et al. 2006; Gilbert et al. 1995).
Another H. pylori His-rich protein is annotated as histidine and glutamine rich protein (HP1432, see Fig 2). The N-terminal sequence (46 residues) of HP1432 shows 56% identity to Hpn and the protein was thus termed Hpn-like. It has not been purified, but has been preliminarily studied by a targeted mutagenesis approach (Seshadri and Maier, abstract D21, 105th general meeting ASM, 2005). Sequence analysis indicates that 25% of the amino acids are histidine residues including a stretch of six consecutive histidine residues while 30 of the 72 amino acid residues are glutamine residues (42%) (Fig. 2). The His residues can predictably be assigned metal binding roles, but the role of the glutamines is a mystery. Additionally, Hpn-like has an Met-Ala-His motif at the N-terminus which is similar to Asp-Ala-His motif involved in nickel binding in human serum albumin (Callan and Sunderman 1973; Gilbert et al. 1995; Tabata and Sarkar 1992).
Hpn-like is upregulated at pH 5.0 (compared to pH 7.0) by the two-component system ArsRS (HP166–HP165) (Bury-Mone et al. 2004; Pflock et al. 2004; Pflock et al. 2006). Both Hpn and Hpn-like are activated in the presence of nickel by the nickel sensor NikR (Contreras et al. 2003).The regulation studies thus far suggest that the Hpn proteins would be expressed at a time when nickel detoxification and/or nickel storage would be an advantage. To further explore the roles of these (hpn and hpn-like) genes, individual gene-disruption mutant strains were created, and a double mutant lacking genes for both these histidine rich proteins was also studied (Seshadri, Benoit, Maier, unpublished data, ASM Abstract 2005). Previous results showed that an hpn mutant is more sensitive to nickel than is wild-type (Mobley et al. 1999). Recent preliminary studies (S. Seshadri and R. Maier, abstract D21, 105th general meeting ASM, 2005) indicate the Hpn-like protein also may play a role in nickel detoxification. While Hpn is a predominant protein in H. pylori, we have no information on the relative amount of Hpn-like that is synthesized.
Only a minor percentage of H. pylori urease is Ni-activated (Stingl and De Reuse, 2005) and that nickel can be a factor sometimes limiting urease activity rather than activity being limited by the amount of urease protein (van Vliet et al. 2001). It was observed that addition of just 1 µM nickel to the growth medium increased the urease activities but did not cause an increase in urease expression levels (van Vliet et al. 2002) and disrupting H. pylori genes encoding a metal efflux pump (cznABC) led to an increase in urease activity (Stahler et al. 2006), presumably by making cytosolic nickel available.
H. pylori apparently synthesizes more urease apo-enzyme than it can (Ni) activate in some lab conditions, but such low nickel levels would also be expected in the host environment. In some low Ni conditions the Hpn and Hpn-like mutant strains contain significantly more urease activity than the parent strain (Seshadri and Maier, abstract D21, 105th general meeting ASM, 2005). It seems likely that these Hpn proteins compete with Ni-dependent urease maturation enzymes in nickel deficient conditions. This may not be a negative situation for H. pylori even though it relies on active urease to transit the stomach; the successful gastric pathogen would be expected to accomplish transit rapidly and spend the vast majority of its life residing in the mucin layer or adjacent to gastric epithelia in a highly nickel-restricted environment. As such the bacterium would need to conserve its scarce nickel reserves for H2 oxidation (Maier et al. 1996) or for later stage urease production (Tanahashi et al. 2000). In addition, synthesis of apo-urease (lacking metal, but still extruded from the cell) may have advantages to H pylori in vivo existence. For example, non-ureolytic (inactive) urease has been shown to play roles in pathogenesis and (for H. pylori) to significantly alter inflammatory responses of gastric cells (Suzuki et al. 2002; Tanahashi et al. 2000). It is clear we have much to learn about the roles of H. pylori’s nickel metabolism that allows the bacterium overwhelming success in colonizing and persisting in the human stomach.
Acknowledgements
These authors’ work was supported by NIH grant number DK06285201.
References
- Atanassova A, Zamble DB. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind P, Garner RM, Mobley LT. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind P, Garner R, Dunn BE, et al. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Mehta N, Wang G, et al. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb Pathog. 2004;36:153–157. doi: 10.1016/j.micpath.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Böck A. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J Mol Biol. 2002;324:287–296. doi: 10.1016/s0022-2836(02)01070-7. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Albracht SP, Matzanke BF, et al. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of[NiFe]-hydrogenases. J Mol Biol. 2004a;344:155–167. doi: 10.1016/j.jmb.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Paschos A, Bauer A, et al. Analysis of the transcarbamoylation-dehydration reaction catalyzed by the hydrogenase maturation proteins HypF and hypE. Eur J Biochem. 2004b;271:3428–3436. doi: 10.1111/j.1432-1033.2004.04280.x. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Rohrmoser M, Rode S, et al. HybF, a zinc-containing protein involved in NiFe hydrogenase maturation. J Bacteriol. 2004c;186:2603–2611. doi: 10.1128/JB.186.9.2603-2611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Mone S, Skouloubris S, Labigne A, et al. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol Microbiol. 2001;42:1021–1034. doi: 10.1046/j.1365-2958.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- Bury-Mone S, Thiberge JM, Contreras M, et al. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- Callan WM, Sunderman FW., Jr Species variations in binding of 63 Ni(II) by serum albumin. Res Commun Chem Pathol Pharmacol. 1973;5:459–472. [PubMed] [Google Scholar]
- Colpas GJ, Brayman TG, Ming LJ, et al. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry. 1999;38:4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]
- Contreras M, Thiberge JM, Mandrand-Berthelot MA, et al. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol Microbiol. 2003;49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- Cussac V, Ferrero RL, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GS, Mobley HL. Contribution of dppA to urease activity in Helicobacter pylori 26695. Helicobacter. 2005;10:416–423. doi: 10.1111/j.1523-5378.2005.00348.x. [DOI] [PubMed] [Google Scholar]
- Eitinger T, Mandrand-Berthelot MA. Nickel transport systems in microorganisms. Arch Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- Evans DJ, Jr, Evans DG, Kirkpatrick SS, et al. Characterization of the Helicobacter pylori urease and purification of its subunits. Microb Pathog. 1991;10:15–26. doi: 10.1016/0882-4010(91)90062-f. [DOI] [PubMed] [Google Scholar]
- Fulkerson JF, Jr, Garner RM, Mobley HL. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J Biol Chem. 1998;273:235–241. doi: 10.1074/jbc.273.1.235. [DOI] [PubMed] [Google Scholar]
- Fulkerson JF, Jr, Mobley HL. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J Bacteriol. 2000;182:1722–1730. doi: 10.1128/jb.182.6.1722-1730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Watt RM, Sun X, et al. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem J. 2006;393:285–293. doi: 10.1042/BJ20051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JV, Ramakrishna J, Sunderman FW, Jr, et al. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect Immun. 1995;63:2682–2688. doi: 10.1128/iai.63.7.2682-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LT, Mobley HL. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube M, Blokesch M, Böck A. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J Bacteriol. 2002;184:3879–3885. doi: 10.1128/JB.184.14.3879-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DA, Peek RM., Jr The role of persistence in Helicobacter pylori pathogenesis. Curr Opin Gastro-enterol. 2006;22:3–7. doi: 10.1097/01.mog.0000194790.51714.f0. [DOI] [PubMed] [Google Scholar]
- Kansau I, Guillain F, Thiberge JM, et al. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA) Mol Microbiol. 1996;22:1013–1023. doi: 10.1046/j.1365-2958.1996.01536.x. [DOI] [PubMed] [Google Scholar]
- Kansau I, Labigne A. Heat shock proteins of Helicobacter pylori. Aliment Pharmacol Ther. 1996;10 Suppl 1:51–56. doi: 10.1046/j.1365-2036.1996.22164005.x. [DOI] [PubMed] [Google Scholar]
- Leach MR, Sandal S, Sun H, et al. Metal binding activity of the Escherichia coli hydrogenase maturation factor HypB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Fu C, Gilbert J, et al. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003a;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins HypB and UreG in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathogen. 2003b;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Garner RM, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Garner RM, Chippendale GR, et al. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter. 1999;4:162–169. doi: 10.1046/j.1523-5378.1999.99286.x. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Mendz GL, Hazell SL. Helicobacter pylori: physiology and genetics. ASM Press; 2001. [PubMed] [Google Scholar]
- Mollenhauer-Rektorschek M, Hanauer G, Sachs G, et al. Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res Microbiol. 2002;153:659–666. doi: 10.1016/s0923-2508(02)01380-3. [DOI] [PubMed] [Google Scholar]
- Moncrief MB, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief MB, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- Nolan KJ, McGee DJ, Mitchell HM, et al. In vivo behavior of a Helicobacter pylori SS1 nixA mutant with reduced urease activity. Infect Immun. 2002;70:685–691. doi: 10.1128/iai.70.2.685-691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- Park IS, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci U S A. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IS, Hausinger RP. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J Bacteriol. 1995;177:1947–1951. doi: 10.1128/jb.177.8.1947-1951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JU, Song JY, Kwon YC, et al. Effect of the urease accessory genes on activation of the Helicobacter pylori urease apoprotein. Mol Cells. 2005;20:371–377. [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Parsonnet J. Gastric adenocarcinoma and Helicobacter pylori infection. West J Med. 1994;161:60. [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J, Hansen S, Rodriguez L, et al. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- Pflock M, Dietz P, Schar J, et al. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett. 2004;234:51–61. doi: 10.1016/j.femsle.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Pflock M, Finsterer N, Joseph B, et al. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol. 2006;188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S, Hochleitner E, Wang H, et al. Taming of a poison: biosynthesis of the NiFe-hydrogenase cyanide ligands. Science. 2003;299:1067–1070. doi: 10.1126/science.1080972. [DOI] [PubMed] [Google Scholar]
- Rektorschek M, Buhmann A, Weeks D, et al. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol Microbiol. 2000;36:141–152. doi: 10.1046/j.1365-2958.2000.01835.x. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Eng BH, Fard S, et al. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- Skouloubris S, Thiberge JM, Labigne A, et al. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HK, Mulrooney SB, Huber R, et al. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J Biol Chem. 2001;276:49359–49364. doi: 10.1074/jbc.M108619200. [DOI] [PubMed] [Google Scholar]
- Soriano A, Hausinger RP. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc Natl Acad Sci U S A. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano A, Colpas GJ, Hausinger RP. UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry. 2000;39:12435–12440. doi: 10.1021/bi001296o. [DOI] [PubMed] [Google Scholar]
- Stahler FN, Odenbreit S, Haas R, et al. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun. 2006;74:3845–3852. doi: 10.1128/IAI.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, De Reuse H. Staying alive overdosed: how does Helicobacter pylori control urease activity? Int J Med Microbiol. 2005;295:307–315. doi: 10.1016/j.ijmm.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Stola M, Musiani F, Mangani S, et al. The nickel site of Bacillus pasteurii UreE, a urease metallo-chaperone, as revealed by metal-binding studies and X-ray absorption spectroscopy. Biochemistry. 2006;45:6495–6509. doi: 10.1021/bi0601003. [DOI] [PubMed] [Google Scholar]
- Sundermann FW., Jr Biological monitoring of nickel in humans. Scand J Work Environ Health. 1993;19:34–38. [PubMed] [Google Scholar]
- Suzuki H, Masaoka T, Miyazawa M, et al. Gastric mucosal response to Helicobacter pylori. Keio J Med. 2002;51 Supple 2:40–44. doi: 10.2302/kjm.51.supplement2_40. [DOI] [PubMed] [Google Scholar]
- Tabata M, Sarkar B. Specific nickel(II)-transfer process between the native sequence peptide representing the nickel(II)-transport site of human serum albumin and L-histidine. J Inorg Biochem. 1992;45:93–104. doi: 10.1016/0162-0134(92)80003-e. [DOI] [PubMed] [Google Scholar]
- Tanahashi T, Kita M, Kodama T, et al. Cytokine expression and production by purified Helicobacter pylori urease in human gastric epithelial cells. Infect Immun. 2000;68:664–671. doi: 10.1128/iai.68.2.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Karita M, Mizote T, et al. Essential role of Helicobacter pylori urease in gastric colonization: definite proof using a urease-negative mutant constructed by gene replacement. Eur J Gastroenterol Hepatol. 1994a;6 Supple 1:S49–S52. [PubMed] [Google Scholar]
- Tsuda M, Karita M, Morshed MG, et al. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994b;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AH, Kuipers EJ, Waidner B, et al. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect Immun. 2001;69:4891–4897. doi: 10.1128/IAI.69.8.4891-4897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AH, Poppelaars SW, Davies BJ, et al. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect Immun. 2002;70:2846–2852. doi: 10.1128/IAI.70.6.2846-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AH, Ernst FD, Kusters JG. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 2004;12:489–494. doi: 10.1016/j.tim.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Voland P, Weeks DL, Marcus EA, et al. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Eskandari S, Scott DR, et al. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Sachs G. Sites of pH regulation of the urea channel of Helicobacter pylori. Mol Microbiol. 2001;40:1249–1259. doi: 10.1046/j.1365-2958.2001.02466.x. [DOI] [PubMed] [Google Scholar]
- Wolfram L, Bauerfeind P. Conserved low-affinity nickel-binding amino acids are essential for the function of the nickel permease NixA of Helicobacter pylori. J Bacteriol. 2002;184:1438–1443. doi: 10.1128/JB.184.5.1438-1443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacteriol J, Haas E, Bauerfeind P. Nickel represses the synthesis of the nickel permease NixA of Helicobacter pylori. J Bacteriol. 2006;188:1245–1250. doi: 10.1128/JB.188.4.1245-1250.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli B, Stola M, Musiani F, et al. UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+ J Biol Chem. 2005;280:4684–4695. doi: 10.1074/jbc.M408483200. [DOI] [PubMed] [Google Scholar]