Abstract
Members of the nuclear factor-kappa beta (NF-κB) family maintain cellular homeostasis by enhancing the transcription of genes involved in inflammation, immune response, cell proliferation, and apoptosis. Melanoma tumor cells often express inflammatory mediators through enhanced activation of NF-κB. The NF-κB activation appears to result from the enhancer formation including NF-κB and lysine acetyl transferases such as p300, CREB (cyclic AMP-responsive element binding protein)-binding protein (CBP), and/or p300/CBP associating factor (PCAF). We observed that proteins expressed by Hs294T metastatic melanoma cells are highly acetylated compared with normal melanocytes, and dominant-negative PCAF reduced the basal and tumor necrosis factor-α-stimulated transcriptional activity of NF-κB. The promoter activity of NF-κB-regulated chemokines was also reduced by the expression of dominant-negative PCAF. The promoters of these chemokines contain a CCAAT displacement protein (CDP)-binding site near the NF-κB element. compared with vector-transduced cells, in CDP-transduced Hs294T cells: (i) over-expressed CDP bound efficiently to PCAF, (ii) tumor necrosis factor-α stimulated chemokine expression and NF-κB-mediated transcription were reduced, and (iii) the binding of CBP to Rel A was reduced. These data suggest that CDP inhibits cytokine-induced NF-κB-regulated chemokine transcription. This study contributes to our understanding of the role of CDP in an enhanceosome of NF-κB-mediated chemokine transcription in human melanoma cells.
Keywords: CCAAT displacement protein, chemokines, lysine acetyl transferase, melanoma, nuclear factor-kappa beta, transcription
Introduction
Melanoma is the most aggressive skin cancer and is notoriously resistant to current cancer therapies [1]. In normal skin, melanocytes synthesize melanin pigments and transfer them to surrounding keratinocytes, leading to skin pigmentation, which protects against solar ultraviolet radiation [2]. Melanoma lesions start as benign nevi, which progress to the radial growth phase, and then to the metastatic vertical growth phase [3]. Nuclear factor-kappa beta (NF-κB) activation has been proposed as a potential factor in melanoma tumor progression through enhanced expression of chemokines and resultant chemokine receptor-mediated signaling [4–6]. In human melanoma, a number of NF-κB-regulated chemokines are overexpressed: CXC ligand 8 [CXCL8 or interleukin-8 (IL-8)] [7], CXCL1 [Gro-α, or melanoma growth stimulatory activity (MSGA)] [8], and CCL5 [regulated on activation, normal T expressed, and secreted (RANTES)] [9,10]. These over-expressed NF-κB-regulated chemokines, including CXCL1–3 and CXCL8, enhance melanoma progression through autocrine and paracrine loops [5,11]. Indeed, over-expression of CXCL8 caused metastatic tumor growth in primary melanoma cells [7,12]. Over-expression of the murine homolog of CXCL1 in INK4a/ARF−/− immortalized melanocytes increased melanoma tumor incidence [13] and induced malignant progression of squamous cell carcinoma in nude mice [14].
All NF-κBs (Rel A/p65, Rel B, C-Rel, NF-κB1/p50 and NF-κB2/p52) contain a Rel homology domain that mediates dimerization, DNA binding, and IκB (inhibitor protein of NF-κB) binding [6,15,16]. IκB proteins, which associate with NF-κBs in the cytoplasm, are phosphorylated by the IκB kinase complex (IKK) and are subsequently degraded by the 26S proteasome [15,16]. Post-translational modifications generate active NF-κB complexes, especially the Rel A/p50 heterodimer, which represents the major activated form of NF-κB in many cell types [6,15,16]. Rel A is phosphorylated by a number of kinases during the phosphorylation and degradation of IκBs, and these events enhance the nuclear translocation of Rel A [17,18].
In the nucleus, Rel A is also known to be acetylated, leading to enhanced transcriptional activity [19]. Lysine acetyl transferases (LAT) such as CREB (cyclic AMP-responsive element binding protein)-binding protein (CBP), p300, and p300/CBP-association factor (PCAF) are also known as histone acetyl transferases, because they acetylate N-terminal tails of intrinsic histone molecules to enhance transcription [20,21]. In addition to modifying histones, these LAT proteins, however, have been shown to transfer acetyl groups to nonhistone proteins, including transcription factors such as NF-κB [22] and Stat 3 [23]. These activated transcription factors could in turn regulate the activation of cytokine/chemokine signaling. In melanoma cells, the downregulation of CBP and p300 results in growth inhibition, cyclin E downregulation, and activation of the senescence growth phase [24]. The mechanism by which acetylase activity associated with CBP and p300 affects transcription of the NF-κB-dependent chemokines is not clear.
The CCAAT motif is known as a binding site for the CCAAT enhancer binding protein (C/EBP), which is an activator of NF-κB-mediated transcription in many cells [25,26]. CCAAT displacement protein (CDP) also binds to DNA, competing with the binding of transcription activators such as C/EBP to the CCAAT box [27–29] and to the ATCGAT core site [30]. CDP is homologous to Drosophila cut, containing triplicate ‘cut repeats’ and a ‘cut homeodomain’, and is proposed to be a transcription repressor on promoters [31]. The carboxyl-terminal region of CDP is an active repression domain that interacts with histone deacetylase 1 in a binding-site-independent manner [32,33]. CDP indirectly reduces the transcription of the genes by repressing the transcription of other transcription activators such as C/EBP and c-Myc [34,35]. The gene locus of CDP is on 7q22 and has been considered to be a tumor suppressor gene, because many types of tumor show loss of heterozygosity on chromosome 7q. Though mutation analysis for loss of heterozygosity on 7q22 in 47 tumors failed to identify any somatic alteration in the CDP coding regions [36], amino-terminal-truncated CDP was found in human uterine leiomyomas [31] and in breast cancer cells [37], suggesting that aberrant expression of CDP is associated with the process of tumorigenesis.
We have previously shown that transient transfection of an antisense CDP expression vector increased CXCL1 promoter activity [38]. The mechanism for the affect of CDP on NF-κB-mediated chemokine transcription, however, has not been previously elucidated. We here provide experimental data that CDP inhibits NF-κB activation through the modulation of protein acetylation. These data contribute to our understanding of the regulation of chemokine transcription during melanoma tumor progression.
Materials and methods
Cell culture and reagents
Hs294T cells and human embryonic kidney (HEK) 293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum and 2 mmol/l L-glutamine. Normal human epidermal melanocytes (NHEM) were provided by the Tissue Culture Core of the Skin Disease Research Center at Vanderbilt University, and maintained in Medium-154 (Cascade Biologics, Portland, Oregon, USA) with 1% human melanocyte growth supplement (HMGS) (Cascade Biologics). The tumor necrosis factor-α (TNF-α) and trichostatin A (TSA) were purchased from PeproTech (Rocky Hill, New Jersey, USA) and Calbiochem (La Jolla, California, USA), respectively. All tissue culture reagents were from Life Technologies, Inc. (Rockville, Maryland, USA) unless otherwise specified.
Enzyme-linked immunosorbent assay
Cells were cultured in growth media to 80% confluency, washed twice with serum-free media, and incubated in serum-free media for 4 h with/without10 ng/ml TNF-α. The media were collected, centrifuged to eliminate cell debris, and subjected to enzyme-linked immunosorbent assay (ELISA) for CXCL1 and CXCL8 using Quantikine from R&D systems (Minneapolis, Minnesota, USA) and for CXCL2 using ELISA Development Kit (Leinco Technologies, St Louis, Missouri, USA). The ELISA values were normalized by cell number and represented in units of pg/104 cells or fold induction by TNF-α.
Plasmid constructs and protein expression
The expression vector pMX containing full length CDP cDNA (from + 29 to + 5100, acc no.: M74099) [39] was provided by Dr Alan Nepveu, McGill University. The CDP cDNA (5139 bp, from 76 bp upstream of the translation initiation site to just upstream of the polyadenylation site) was excised using Not I/Xho I enzymes from the vector, purified by agarose gel electrophoresis, and ligated into the retroviral expression vector pBMN-internal ribosomal entry sequence-gene for enhanced green fluorescent protein (pBMN-IRES-EGFP, provided by Dr Gary Nolan, Stanford University). The CDP insert was confirmed by enzyme digestion and sequencing using the forward primer: 5′-GACCTTACA-CAGTCCTGC-3′. The HEK293T retrovirus-packaging cells (2.5 × 106/100-mm dish) were transfected with 3 μg pBMN-CDP-IRES-EGFP or 3 μg pBMN-IRES-EGFP (control) using FUGENE 6 (Roche Molecular Biochemicals, Indianapolis, Indiana, USA). In each case, cells were cotransfected with 1 μg pHCMV-G (VSV-G) [40] and 3 μg pSV-Ψ-env−-MLV (pSV-pol/gag) [41] (provided by Dr Jane Burns, University of California, San Diego, USA). Virus-containing media were collected 48 h later and passed through a 45-μm filter (Pall Corporation, East Hills, New York, USA). Hs294T cells (105/60-mm dish) were incubated with virus-containing media for 2 h. After five passages, cells stably expressing EGFP were sorted by flow cytometry and expanded. EGFP expression was monitored with an inverted fluorescent microscope and cultures were maintained at a 100%-GFP expression level throughout all experiments.
The expression vector pMX containing a carboxyl terminal CDP (from + 1605 to + 5100 nucleotide, acc. no.: M74099) in the antisense direction was provided by Dr Alan Nepveu of McGill University. This truncated CDP cDNA was excised using Not I/Xho I enzymes from the pMX vector, and ligated into the pcDNA3.1-His vector (Invitrogen, Carlsbad, California, USA) in the sense direction. The ligated plasmid was replicated in the bacteria strain BL21 (Stratagene, La Jolla, California, USA), and an expressed recombinant protein tagged with 6-histidine was isolated using the ProBound Purification System (Stratagene).
The expression vector pCi (Promega, Madison, Wisconsin, USA) containing the wild-type PCAF cDNA (PCAF-WT) (acc. no.: BC060823) [42] and PCAF-ΔLAT (deletion mutation in the domain for lysine acetyl transferase: amino acid position 609–618) were provided by Dr Tony Kouzarides of Cambridge University. PCAF-WT and PCAF-ΔLAT were excised using Eco RI/Not I enzymes from the vector. Purification of the plasmid, ligation into the retroviral expression vector, virus packaging, and infection of Hs294T cells were performed as stated above.
The restriction enzymes, T4-DNA ligase, and DNA polymerases were from New England BioLabs (Beverly, Massachusetts, USA) unless otherwise specified.
Immunoblot analysis and immunoprecipitation
Cells were washed twice with ice-cold phosphate-buffered saline and total cell lysates were isolated with a buffer containing 50 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% Na deoxycholate, 1 mmol/l ethylenediaminetetraacetic acid with 10 μl/ml protease inhibitor cocktail (P-8340, Sigma, St Louis, Missouri, USA) and 10 μl/ml phosphatase inhibitors (P-2850/P5726, Sigma). The protein lysates were then sonicated at power setting 4 for 10 s bursts using the Sonifier 250 (Banson, Golden, Colorado, USA). Protein extracts were quantitated using the Bicinchoninic acid Protein Assay reagent (Pierce, Rockford, Illinois, USA), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis as described previously [38]. Immunoreactive bands were visualized by a chemiluminescence reagent (Amersham Biosciences, Piscataway, New Jersey, USA) or by scanning the emitted infrared spectrum using the Odyssey System (LI-COR Biotechnology, Lincoln, Nebraska, USA). The cytoplasm/nucleus proteins were prepared with the same procedure as stated for the electrophoretic mobility shift assay (EMSA) below. Immunoprecipitations were performed after pre-clearing cell lysates with protein A-sepharose (Sigma) for 2 h at 4°C as described previously [43]. The antibodies (200 ng/ml) utilized were: CDP (sc-13024), Rel A (sc-109), CBP (sc-369) (Santa Cruz Biotechnology, Santa Cruz, California, USA); phospho-S536-Rel A, Acetylated lysine (Ac-K-103) (Cell Signaling, Beverly, Massachusetts, USA), and anti-Flag (Sigma).
Electrophoretic mobility shift assay
Cells were washed twice with ice-cold phosphate-buffered saline and collected in a cell suspension buffer containing 10 mmol/l Hepes (pH 7.9), 10 mmol/l NaCl, 1.5 mmol/l MgCl2, 0.5 mmol/l dithiothreitol, and 5 mmol/l β-mercapto-ethanol. To separate cytoplasm/nuclear proteins, cells were lysed in 1% Nonidet P-40 in the cell suspension buffer described above. The destruction of cell membranes and the presence of intact nuclei were observed by staining with 0.04% trypan blue. Cells were then centrifuged at 6000g and the supernatant was collected as the cytoplasm protein fraction. The pellet was washed with a nuclei suspension buffer containing 20 mmol/l Hepes (pH 7.9), 10 mmol/l NaCl, 1.5 mmol/l MgCl2, 0.5 mmol/l dithiothreitol, 5 mmol/l β-mercapto-ethanol, 0.2 mol/l ethylenediaminetetraacetic acid, and 1% Nonidet P-40. The nuclei were then lysed in 450 mmol/l NaCl hypertonic buffer in the nuclei suspension buffer described above. The protease inhibitors and phosphatase inhibitors were added to these suspension and lysis buffers as stated above. The nuclear protein (0.5 μg) was incubated with the following 32P end-labeled double-stranded oligonucleotide probes: NF-κB binding site, 5′-AGTTGAGGGGACTTTCCCAGG (Promega); CXCL1 promoter, 5′-GGGATCGATCTGGAACTCCGGGAATTTCCCTGGCCC (from −98 to −63); CXCL8 promoter, 5′-GCCATCAGTT GCAAATCGTGGAAT TTCCTCTGA (from −99 to −67). The procedures for the annealing of oligos to form double stranded probes, the labeling of probes, and the incubation of nuclear proteins with labeled probes were performed according to the protocol in the Gel Shift Assay System (Promega). Protein/oligo complexes were electrophoresed in a 6% native polyacrylamide gel, transferred to 3 MM chromatography paper (Whatmann, Clinton New Jersey, USA), and autoradiographed. The antibodies used for observing the super-shifted bands were Rel A (sc-109 ×), p50 (sc-114 ×), p52 (sc-298 ×), and CDP (sc-13024) (Santa Cruz Biotechnology).
Reporter gene assays
Cells were seeded in 24-well plates and transfected with 0.2 μg/well of one of the following luciferase reporter plasmid DNA containing (i) CXCL1 promoter [38,44], (ii) CXCL8 promoter [45], or (iii) NF-κB binding site (Clontech, Palo Alto, California, USA) using FUGENE 6 (Roche Molecular Biochemicals) according to the manufacturer’s protocol. Cells were cotransfected with 0.2 μg/well of one of the following transactivators: PCAF-WT, PCAF-ΔLAT, a CDP expression plasmid DNA, or an empty control plasmid DNA. Each well was also cotransfected with 0.02 μg/well of pNull-Renilla (promoter-less expression vector DNA of Renilla luciferase) (Promega) to normalize the transfection efficiency, as the ligand would not activate the promoter and the basal luciferase activity is high enough for the normalization of transfection efficiency. The luciferase activities were measured using the Dual Luciferase Assay System (Promega) according to the manufacturer’s protocol.
cDNA array analysis
Total RNAs were isolated with Trizol Reagent (Life Technologies), digested with RNase-free DNase (0.1 unit/1 μg RNA; Promega), and purified using a silica gel column (RNeasy Kit, Qiagen, Valencia, California, USA). Hs294T/vector-derived RNA was reverse-transcribed into cDNA with Cy5 labeling, and Hs294T/CDP-derived RNA was labeled with Cy3. The mixed cDNAs were hybridized to a human 30 000 oligo-array, and Cy3 and Cy5-labeled cDNAs were scanned at 532 and 635 nm, respectively, in GenePix Pro (Axon Instruments, Inc., Foster City, California, USA). The procedures for reverse transcription, RNA template digestion, and cDNA purification/hybridization to a human 30 000 oligo array were performed following protocols listed at www.array.vanderbilt.edu.
Results
Rel A-DNA binding and protein acetylation are highly activated in Hs294T metastatic melanoma cells compared with NHEM
The Hs294T cells were selected as an experimental system for human metastatic melanoma. Hs294T cells satisfy the parameters of malignancy, which include abnormal morphology, growth to high saturation density, lack of anchorage dependence, growth in semisolid media, colony formation on a contact-inhibited monolayer, and tumor formation in immunosuppressed mice with almost 100% efficiency [46]. Hs294T cells have a B-raf point transversion mutation (V600E, NM_004333) [47], which is associated with 66% of malignant melanoma lesions by stimulating downstream signaling molecules such as extracellular signal-regulated kinase 1 and 2 (ERK1/2) [47–49].
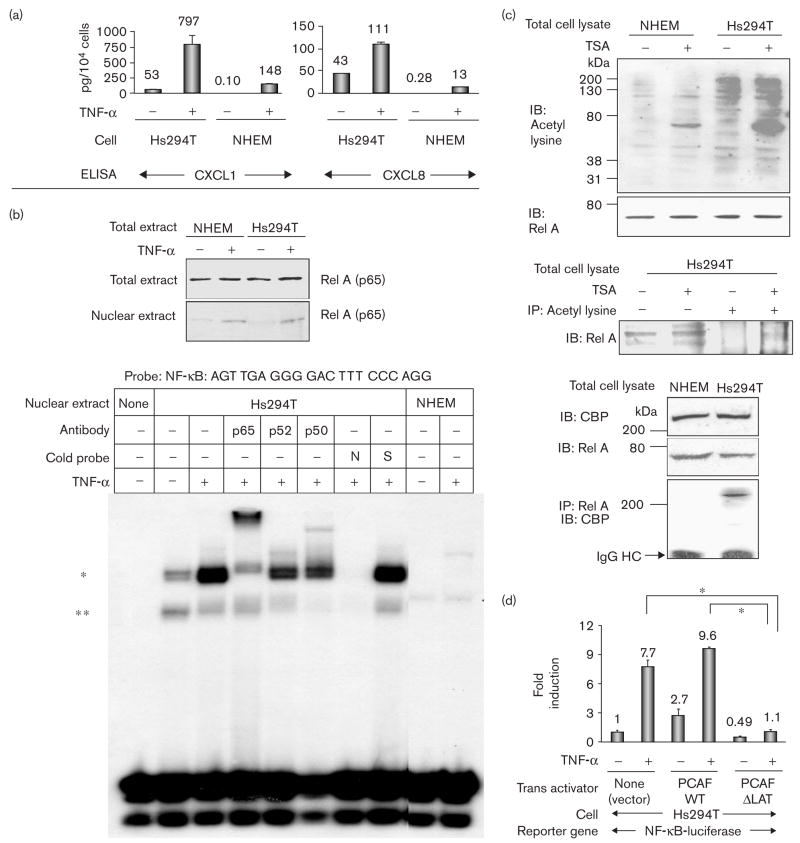
To verify the downstream events of the activated NF-κB in Hs294T cells, culture media were subjected to ELISA for CXCL1 and CXCL8 as representative of NF-κB-regulated gene transcription. Compared with NHEM, Hs294T cells (104 cells) secreted a significantly higher amount of CXCL1 (53 pg/104 cells) and CXCL8 (43 pg/104 cells) even without TNF-α stimulation. With TNF-α stimulation, secretion of these chemokines was significantly amplified in Hs294T cells (Fig. 1a).
Fig. 1.
Rel A-DNA binding and acetylases are highly activated in Hs294T metastatic melanoma cells compared with normal human epidermal melanocytes (NHEM). (a) Hs294T cells and NHEM were incubated in normal growth media, washed twice with serum-free medium (SFM), and then incubated with/without tumor necrosis factor-α (TNF-α) (10 ng/ml) in SFM for 4 h. Media were collected, centrifuged to eliminate cell debris, and aliquots were subjected to enzyme-linked immunosorbent assay (ELISA) to determine the presence of CXCL1 and CXCL8. The ELISA was repeated thrice and the values were normalized in the unit of pg/104 cells. (b) Top: Hs294T cells and NHEM were incubated in normal growth media, washed twice with SFM, incubated with/without TNF-α (10 ng/ml) in SFM for 1 h, and lysed to obtain total and cytoplasm/nuclear fractionated proteins. Proteins (40 μg) were subjected to immunoblot analysis with an αRel A antibody. Bottom: nuclear proteins (0.5 μg) were subjected to electrophoretic mobility shift assay (EMSA) with the probe of Nuclear factor kappa beta (NF-κB) (promega) as indicated. (*) EMSA band that significantly increased with TNF-α and was shifted by coincubation with the αRel A antibody; (**) EMSA band that was mainly shifted by coincubation with the αP50 antibody; N: 50-fold excess of unlabeled NF-κB consensus oligonucleotide; S: 50-fold excess of unlabeled Sp-1 consensus oligonucleotide. (c) Top: the total cell lysates from Hs294T and NHEM with/without 500 nmol/l trichostatin A (TSA) were subjected to immunoblot analysis with α-acetyl-lysine and αRel A antibodies. IB: immunoblot analysis. Middle: the total cell lysates from Hs294T with/without 500 nmol/l TSA were subjected to immunoblot analysis (lanes 1, 2). The total cell lysates from Hs294T with/without 500 nmol/l TSA were also subjected to immunoprecipitation with α acetyl-lysine antibody followed by immunoblot analysis with αRel A antibodiy (lane 3, 4). Bottom: the total cell lysate from Hs294T and NHEM were subjected to immunoblot analysis with αCBP and αRel A antibodies. Cell lysates were also immunoprecipitated with αRel A antibody, and the immunoprecipitated complexes were subjected to immunoblot analysis with αCBP antibody. IP: immunoprecipitation; IgG HC: immunoglobulin G heavy chain. (d) Cells (4 × 105 cells) were seeded in 24-well plates and transfected with 0.2 μg/well of NF-κB-luciferase reporter plasmid and 0.02 μg/well of pNull-Renilla luciferase reporter plasmid in a complete growth medium. Cells were also co-transfected with 0.2 μg/well of one of the following trans-activators: (i) PCAF-WT, (ii) PCAF-ΔLAT or (iii) pcDNA3 empty control vector. Cells were incubated in SFM for 16 h and then further incubated with/without TNF-α (10 ng/ml) for 4 h. NF-κB-luciferase reporter activity was divided by Renilla luciferase activity to normalize the transfection efficiency. Standard deviations of the mean fold induction were calculated from the relative luciferase activity from the three wells. The assays were done thrice. *P < 0.002 (Student’s t-test).
To analyze whether the activated NF-κB-mediated chemokine production is due to the nuclear translocation of NF-κB, the cytoplasm/nuclei fractionated proteins from Hs294T cells and NHEM were subjected to immunoblot analysis with α-Rel A antibody (Fig. 1b, top). The amount of nuclear Rel A with/without TNF-α stimulation was about the same in both NHEM and Hs294T cells (Fig. 1b, top). The DNA binding of NF-κBs was then analyzed by EMSA using nuclear extracts from Hs294T cells and NHEM with the NF-κB binding probe sequence (Promega, AGTTGAGGGGACTTTCCCAGG). Two bands (noted as * and **) were observed using Hs294T nuclear protein without TNF-α stimulation, but these bands were not clearly detected in NHEM (Fig. 1b, bottom). With TNF-α stimulation, the upper band (*) significantly increased and shifted with coincubation with the αRel A antibody. The lower band (**), however, decreased slightly in the presence of the αRel A antibody, and shifted with coincubation with the anti-p50 antibody (Fig. 1b, bottom). These two EMSA bands were also partially shifted by the anti-P52 antibody (Fig. 1b, bottom). These EMSA data show that DNA binding of Rel A and P50 in Hs294T cells were activated to a higher degree than in NHEM.
To explore why NF-κB/DNA interaction is activated to a greater extent in Hs294T cells compared with NHEM, acetylase activity in these cells was examined. Total cell lysates from Hs294T cells and NHEM were subjected to immunoblot analysis with α-acetylated lysine antibody. Hs294T melanoma cells are more highly acetylated than proteins from NHEM, and the frequency of acetylated proteins significantly increased upon treatment with the deacetylase inhibitor TSA (Fig. 1c, top). The acetylation of Rel A is detected in Hs294T cells with TSA treatment (Fig. 1c, middle). In Hs294T cells, immunoprecipitation analysis revealed an interaction between Rel A and CBP, but this interaction was not detected in NHEM (Fig. 1c, bottom).
To examine the activation of NF-κB by acetylases in Hs294T cells, cells were transiently cotransfected an NF-κB-luciferase reporter gene with acetylases expression vectors containing (i) CBP, (ii) p300, or (iii) p300/CBP association factor (PCAF). The transient expression of wild-type PCAF (PCAF-WT) in Hs294T cells increased basal and TNF-α-stimulated NF-κB-luciferase reporter gene activity. The expression of PCAF mutated in the lysine acetylase domain (PCAF-ΔLAT) reduced basal and TNF-α-stimulated NF-κB-luciferase reporter gene activity (Fig. 1d).
Putative CDP sites are near the NF-κB sites in the promoters of chemokines for CXCR1 and CXCR2
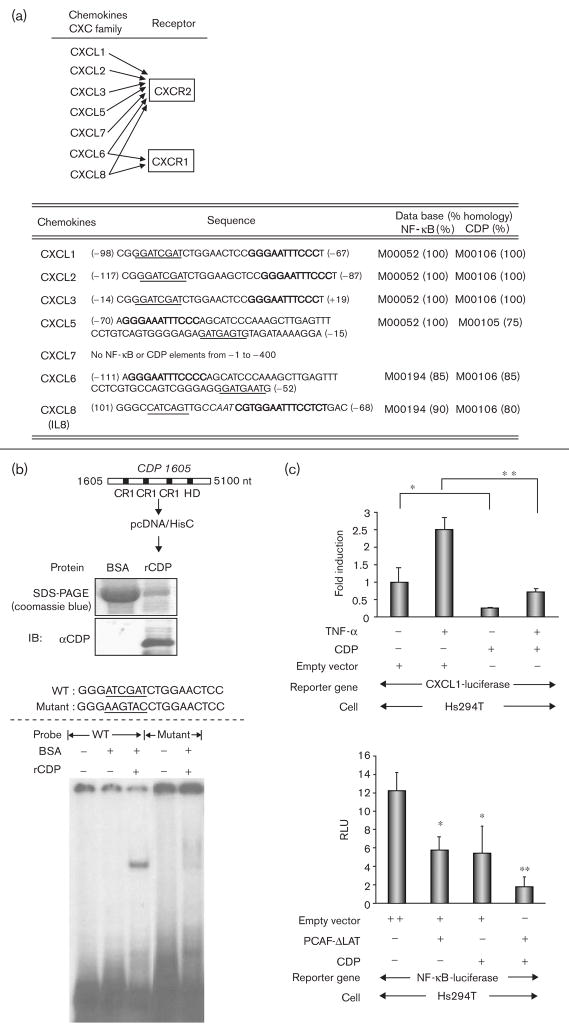
Chemokines, CXCL1 and CXCL8, are abundantly produced and secreted in Hs294T metastatic melanoma cells (Fig. 1a), and the receptors of these chemokines are known to be CXCR1 and CXCR2 (Fig. 2a, top). Enhanced expression of these chemokines has been reported to promote melanoma progression and to be associated with constitutive activation of NF-κB [11]. According to the TFSEARCH (www.cbrc.jp), the promoters of chemokines for CXCR1 and CXCR2 contain the C(A/G)AT motif near the NF-κB binding sites (Fig. 2a, bottom), which are predicted binding sites for the C/EBP and/or CDP. Previously, we reported that the transient transfection of full length CDP reduced the expression of a luciferase reporter gene with a 355 bp CXCL1 promoter (from −301 to + 54) [38]. We here verified by EMSA that the isolated CDP protein binds to the 19 bp DNA sequence GGGATCGATCTGGAACTCC (Fig. 2a, bottom), including the CDP binding motif C(A/G)T for CXCL1–3. Recombinant CDP encoding DNA binding domains, three cut Repeats (CR1–3), and one homeodomain were purified (Fig. 2b, top). This recombinant protein was capable of binding to the 19 mer probe, but did not bind to a probe of identical length with a substitution mutation in the CDP biding motif (Fig. 2b, bottom). We also verified that transient transfection of a CDP expression vector reduced the expression of the CXCL1-luciferase reporter gene in Hs294T cells treated or not treated with TNF-α (Fig. 2c, top). Furthermore, NF-κB-mediated gene transcription was reduced by about 50% by the transfection of the CDP-expression vector, as well as by the transfection of PCAF-ΔLAT expression vector. Moreover, the cotransfection of CDP and PCAF-ΔLAT further reduced the basal NF-κB-mediated transcription by 85% (Fig. 2c, bottom).
Fig. 2.
Putative CCAAT displacement protein (CDP) sites are near the nuclear factor-kappa beta (NF-κB) sites in the promoters of chemokines for CXCR1 and CXCR2. (a) Top: CXC ligands for CXCR1 and CXCR2. Bottom: the sequences of the putative NF-κB and CDP binding sites in the promoters of the CXC ligands. The promoter ID numbers and the percentages of homologies are followed by the TFSEARCH (www.cbrc.jp). Block letters: NF-κB binding sequences; underlined letters: CDP binding sequences; Italic letters: CCAAT sequence. (b) Top: purified DNA-binding domain of CDP. CDP cDNA (from + 1605 to + 5100, acc. no.: M74099) containing DNA binding domains (CR1-3 and homeodomain) was subcloned into the expression vector pcDNA/HisC. The expressed CDP recombinant protein (5 μg) was stained with coomassie blue after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subjected to immunoblot analysis with the αCDP antibody. CR, cut repeat; HD, homeodomain; nt, nucleotide. Bottom: electrophoretic mobility shift assay (EMSA) using a representative CDP binding sequence as indicated. The binding of the CDP recombinant protein was analyzed in EMSA using the CDP binding sequence from the CXCL1–3 promoters. (c) Top: cells were seeded in 24-well plates and the transfection of the reporter constructs were the same as stated in Fig. 1e. Cells were cotransfected with 0.2 μg/well of CDP expression plasmid or control empty plasmid. The following day, cells were incubated in serum-free medium (SFM) for 16 h and further incubated with/without TNF-α (10 ng/ml) for 4 h in SFM. The normalization of the transfection efficiency, and data analysis were the same as stated in Fig. 1e. The assays were done thrice. *P < 0.05; **P < 0.01 (Student’s t-test). Bottom: cells were cotransfected with following transactivators: (i) 0.2 μg/well of empty vector, (ii) 0.1 μg/well of empty vector + 0.1 μg/well of CDP expression plasmid, (iii) 0.1 μg/well of empty vector + 0.1 μg/well of PCAF-ΔLAT, and (iv) 0.1 μg/well of CDP expression plasmid + 0.1 μg/well of PCAF-ΔLAT. The measurement of luciferase activities and the data analysis are the same as above. *P < 0.05; **P < 0.01 (Student’s t-test).
In CDP-transduced melanoma cells, NF-κB-mediated chemokine transcription is suppressed
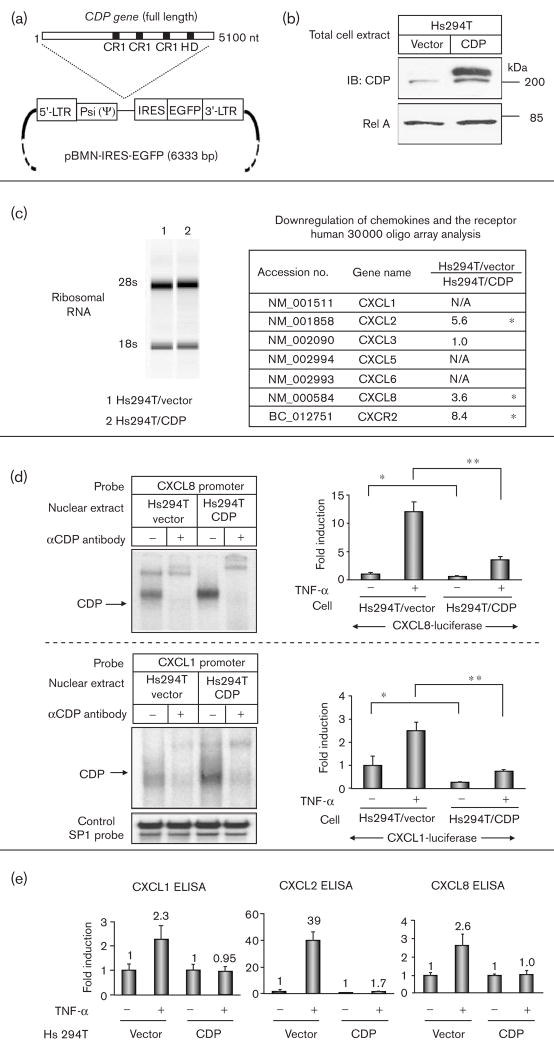
To analyse the function of CDP in the transcription of the NF-κB-regulated chemokines, we transduced the full-length CDP gene into Hs294T metastatic melanoma cells using a retroviral expression vector to create a polyclonal stable cell line over-expressing CDP (Fig. 3a). In Western blot analysis, a low level of endogenous CDP (200 kDa) was detected in vector-transduced Hs294T cells (Hs294T/vector), whereas a high level of expression was verified in CDP-transduced Hs294T cells (Hs294T/CDP) (Fig. 3b).
Fig. 3.
In CCAAT displacement protein (CDP)-transduced metastatic melanoma cells, nuclear factor-kappa beta (NF-κB)-mediated chemokine transcription is suppressed. (a) Map of the retroviral vector, pBMN-IRES-EGFP, for the stable expression of full-length CDP. LTR, long terminal repeat; Psi(Ψ), consensus sequence for viral packaging; IRES, internal ribosomal entry sequence; EGFP, gene for enhanced green fluorescent protein; CR, cut repeat; HD, homeodomain; nt, nucleotide. (b) Total cell lysates from CDP-transduced and vector-transduced control Hs294T were subjected to SDS-PAGE (50 μg/lane) followed by an immunoblot analysis with αCDP and αRel A antibodies. (c) Left: Total RNA was isolated from Hs294T/vector and Hs294T/CDP, treated with DNase, and electrophoresed in the eukaryote total RNA Nano-DE114000902 to measure RNA integrity value. 28S and 18S, ribosomal RNA. Right: the list of differentially expressed genes identified by microarray analysis. Hs294T/vector-derived RNA was reverse-transcribed into cDNA with Cy5 labeling, and Hs294T/CDP-derived RNA was labeled with Cy3. The mixed cDNAs were hybridized to a human 30 000 oligo-array, and Cy5/Cy3 fluorescent ratio was analyzed with GeneSpring software. The chemokines and the receptors in Fig. 2a were analyzed from array data, and differentially expressed genes with over a three-fold expression were listed as significantly differentially expressed and noted as (*). Over-saturated or undetected fluorescent intensity was listed as N/A. (d) Top: nuclear proteins (0.5 μg) from Hs294T/vector and Hs294T/CDP cells were subjected to electrophoretic shift assay (EMSA) with the IL-8 probe (from −99 to −67). Cells were seeded in 24-well plates (4 × 105 cells/well) and transfected with (i) 0.2 μg/well of a luciferase reporter plasmid DNA with the CXCL8 promoter sequence (from133 to + 44) and 0.02 μg/well of pNull-Renilla luciferase reporter plasmid DNA (Promega) to normalize variations in transfection efficiency. Cells were incubated in serum-free medium (SFM) for 16 h and further incubated with/without tumor necrosis factor (TNF-α) (10 ng/ml) for 4 h in SFM. Standard deviations of the mean fold induction were calculated from the relative luciferase activity from the three wells. The assays were performed thrice. *P < 0.02; **P < 0.002 (Student’s t-test). Bottom: nuclear proteins (0.5 μg) from Hs294T/vector and Hs294T/CDP cells were subjected to EMSA with the CXCL1 promoter sequence probe (from −98 to −63) and SP1 control probe (Promega). CXCL1 promoter activity was measured using CXCL1 promoter sequence (from −301 to + 54) and SP1 control probe (Promega). The EMSA and luciferase reporter gene assays were performed thrice. *P < 0.02; **P < 0.002 (Student’s t-test). (e) Hs294T/vector and Hs294T/CDP cells were incubated in normal growth media, washed twice with SFM, and then incubated with/without TNF-α (10 ng/ml) in SFM for 4 h. Media were collected, centrifuged to eliminate cell debris, and aliquots were subjected to enzyme-linked immunosorbent assay (ELISA) to determine the presence of CXCL1, CXCL2, and CXCL8. The ELISA values were normalized in the unit of pg/105 cells and presented as fold induction by TNF-α stimulation.
To compare the expression of NF-κB-targeted gene expression between Hs294T/CDP and Hs294T/vector expressing cells, we isolated total RNA from these cells (Fig. 3c, left) and analyzed the reverse-transcribed cDNAs using a 30 000 oligo-array. Genes exhibiting a three-fold difference in the Cy5/Cy3 intensity ratio were evaluated as differentially expressed. Among these differentially expressed genes, chemokines and the receptors shown in Fig. 2a were listed (Fig. 3c, right).
The differential gene transcription between Hs294T/CDP and Hs294T/vector was verified by EMSA and luciferase reporter gene analysis. The nuclear protein from Hs294T/CDP cells exhibited increased binding to a sequence of the CXCL8 promoter (33mer, from −99 to −67, 5′-GCCATCAGTTGCAAATCGTGGAATTTCCTCTGA) in EMSA, and the coincubation of the αCDP antibody shifted the EMSA band (Fig. 3d, top). In CDP-transduced Hs294T cells, transcription activity of the CXCL8 promoter (177 bp, from −133 to + 44) [45] was decreased in a luciferase reporter gene assay (Fig. 3d, top). CDP also bound to a sequence common to the CXCL1–3 promoters (36mer, 5′-GGGATCGATCTGGAACTCCGGGAATTTCCCTGGCCC) in EMSA, and decreased luciferase reporter gene activity with the CXCL1 promoter (355 bp, from −301 to + 54) (Fig. 3d, bottom).
Under the experimental conditions shown here, we did not obtain clear NF-κB binding patterns on the EMSA probes from the 36mer CXCL1 promoter or the 33mer CXCL8 promoter, both of which include one copy of an NF-κB binding element and CDP binding element. These EMSA probes containing the putative CDP binding site with NF-κB element seem to be optimized for CDP binding. In contrast, the EMSA probe containing only the NF-κB binding site (AGTTGAGGGGACT TTCCCAGG, Promega) seems to be optimized for NF-κB binding.
The differential induction of chemokine expression by TNF-α in Hs294T/CDP cells and Hs294T/vector cells was then verified by ELISA (Fig. 3e). Although TNF-α induced the expression of CXCL1, 2 and 8 in vector-transduced Hs294T control cells, in Hs294T/CDP cells, TNF-α did not significantly induce the expression of these chemokines (Fig. 3e).
CDP reduces NF-κB-mediated chemokine transcription in metastatic melanoma cells
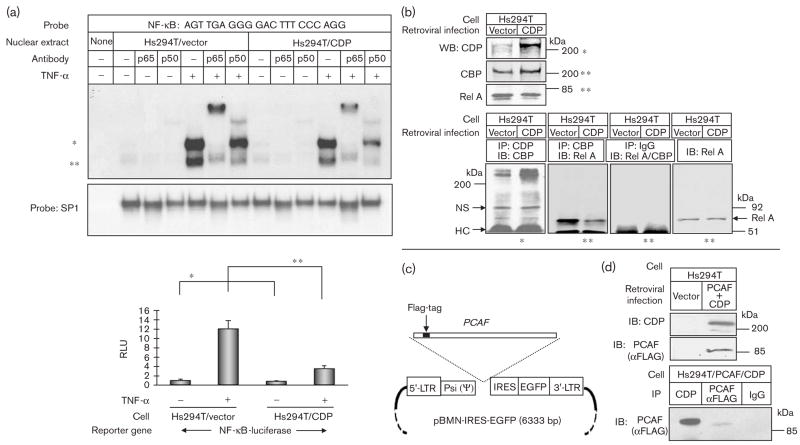
When we compared CDP-transduced Hs294T cells with vector-transduced control cells by EMSA using an NF-κB element probe, we observed that nuclear protein from TNF-α-stimulated CDP-transduced cells exhibited decreased binding of RelA/p50 subunits of NF-κB (Fig. 4a, top). We also observed that NF-κB-luciferase reporter gene activity was reduced in basal and TNF-α-stimulated Hs294T/CDP cells compared with the Hs294T/vector cells (Fig. 4a, bottom).
Fig. 4.
Rel A-DNA binding is reduced in CCAAT displacement protein (CDP)-transduced Hs294T cells, and over-expressed CDP interacted with lysine acetyl transferase, CBP/PCAF. (a) Top: nuclear proteins (0.5 μg) from Hs294T/vector cells and Hs294T/CDP cells were subjected to electrophoretic mobility shift assay (EMSA) with the nuclear factor-kappa beta (NF-κB) probe as indicated and SP1 control probe with the same procedure as stated in Fig. 1b. Bottom: NF-κB promoter activity was measured in Hs294T/vector cells and Hs294T/CDP cells using an NF-κB-luciferase reporter gene with the same procedure as stated in Fig. 3d. *P < 0.02; **P < 0.002 (Student’s t-test). (b) Nuclear cell lysates (noted as *) from Hs294T/vector and Hs294T/CDP were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (50 μg/lane) with αCDP antibody. Total cell lysate (noted as **) from Hs294T/vector and Hs294T/CDP were subjected to SDS-PAGE (50 μg/lane) with αCBP and αRel A antibodies. Nuclear cell lysates were immunoprecipitated with αCDP antibody, followed by immunoblot analysis with αCBP antibodies. The total cell lysates were immunoprecipitated with αCBP antibody, followed by immunoblot analysis with the αRel A antibody. As a control, normal IgG was used for immunoprecipitation, and αCBP/αRel A antibodies were used for immunoblot analysis. IgG HC, immunoglobulin G heavy chain; NS, nonspecific. (c) Map of the retroviral vector pBMN-IRES-EGFP, which stably expresses PCAF-WT. The vector information is the same as stated in Fig. 3a. (d) The Hs294T cells were infected with (i) two retroviruses encoding CDP and PCAF-WT and (ii) an empty vector. The total cell lysates were subjected to immunoblot analysis with the αFLAG antibody to detect transduced PCAF or CDP. Protein lysates were also immunoprecipitated with either αCDP or αFlag antibodies, and then subjected to the immunoblot analysis with the αFlag antibody to detect PCAF.
Previously, Li et al. [50] also showed that CDP binds to CBP and PCAF. We observed that, in CDP-transduced Hs294T cells, over-expressed CDP bound efficiently to CBP, as determined by immunoprecipitation of cell lysates with an αCDP antibody followed by immunoblot analysis with an αCBP antibody (Fig. 4b). This was not observed in vector-transduced control cells. Moreover, as compared with vector-transduced control Hs294T cells, in Hs294T/CDP cells, the binding of CBP with Rel A is reduced when cell lysates were immunoprecipitated with an αCBP antibody followed by immunoblot analysis with a Rel A antibody (Fig. 4b). Under the experimental conditions shown here, cell lysates from the nuclear fraction were used for the detection of CDP in the Western blot analysis (noted as *). Total cell lysate prepared with RIPA buffer, however, was used for the efficient detection of CBP in the Western blot analysis (noted as **). The total cell lysate was also used for the detection of Rel A (noted as **), because under conditions employed here, Rel A was not efficiently detected in the nuclear fraction by Western blot analysis without TNF-α treatment.
To analyze the hypothesis that CDP affects CBP/PCAF-mediated NF-κB transcription, we over-expressed both CDP and PCAF in human embryonic kidney (HEK) 293T cells, and verified the association between CDP and PCAF-WT by immunoprecipitation analysis with an αCDP antibody followed by immunoblot analysis using an αFLAG antibody to detect PCAF-WT and PCAF-ΔLAT (data not shown). We then stably co-transduced CDP and PCAF in Hs294T cells using a retroviral vector (Fig. 4c) and verified the association between the two proteins in Hs294T cells by immunoprecipitation and Western blot analysis (Fig. 4d).
Discussion
Over-expression of chemokines such as CXCL1 and CXCL8 (as shown in Fig. 1a) in melanocytes is known to enhance melanoma tumor progression [5]. These chemokines are transcribed through the NF-κB canonical activation pathway, where (i) Rel A is phosphorylated and translocated into the nucleus and (ii) homo/heterodimerized Rel A binds to DNA to activate transcription. Our data show that the level of Rel A expression and the level of Rel A nuclear translocation before and after TNF-α stimulation are approximately equivalent in Hs294T metastatic melanoma cells and normal melanocytes based upon Western blot analysis of nuclear preparations. The DNA-binding of Rel A based upon EMSA, however, was found to differ significantly in melanoma cells as compared with normal melanocytes.
In the nucleus, Rel A is also known to be acetylated, which leads to enhanced transcriptional activity [19]. Acetylated Rel A binds weakly to IκBα and deacetylated Rel A interacts strongly with IκBα, which leads to an IκBα-dependent nuclear export [19]. In the nucleus, IKKγ is known to associate with CBP, an event that leads to the interference of the acetylase activity of CBP for Rel A [51]. Thus, the reversible acetylation of Rel A serves as a dynamic control point for intranuclear NF-κB action [22]. Indeed, in Hs294T melanoma cells, compared with NHEMs, a number of proteins are highly acetylated. Expression of the dominant-negative PCAF acetylase decreased NF-κB luciferase reporter activity in Hs294T cells. These observations may indicate that PCAF and the other lysine acetylases constitutively enhance NF-κB-regulated transcription in Hs294T cells, possibly through stabilization of NF-κB activation. In melanoma cells, the expression of dominant-negative p300 resulted in growth inhibition through the downregulation of cyclin E in melanoma cells [24]. Moreover, constitutive acetylation by CBP is associated with tumor progression through the overexpression of HOX genes [52]. These earlier reports indicate that deregulation of lysine acetyl transferase activity can facilitate tumor progression, and are in agreement with our findings.
We previously reported that the CDP binding site is located upstream of the NF-κB binding site in the CXCL1 promoter [38]. As we expected, the promoters for CXCR1 and CXCR2 (receptors for CXCL1, 2, 3, 8) contain putative CDP binding motifs [CCAAT and GATC(G/A) motifs] around the NF-κB binding sequences as shown in Fig. 2a. This led to the hypothesis that the binding of CDP to DNA may reduce not only CXCL1 transcription but also the transcription of other NF-κB regulated genes. Indeed, the expression of CDP and PCAF-ΔLAT both individually reduced the NF-κB-derived luciferase reporter gene activity. In addition, these molecules act cooperatively to further reduce NF-κB-mediated transcription. These data indicate that protein acetylases are a positive regulator of NF-κB-mediated transcription and CDP simultaneously can negatively regulate this transcription, potentially through the recruitment of histone deacetylases.
In CDP-transduced Hs294T melanoma cells, the decreased expression of chemokines (CXCL2 and CXCL8) and their receptor, CXCR2, was detected in human oligo array analysis. The downregulation of CXCL1 was not detected in the array analysis owing to the inappropriate fluorescent intensity, however, decreased transcriptional activity of the CXCL1 reporter gene and the CXCL8 reporter gene was observed in CDP-transduced Hs294T cells. In ELISA, we showed that TNF-α significantly activates the expression of CXCL1, 2, and 8 in vector-transduced cells, but not in CDP transduced cells. Interestingly, ELISA results indicate that the basal expression level of these chemokines is higher in CDP-transduced cells, compared with vector-transduced cells, even though TNFα-induced expression of these chemokines was markedly reduced in CDP-transfected cells. This may indicate that, in CDP transduced cells, the endogenous expression of these chemokines is affected differently by CDP than is the TNF-α induced expression of these chemokines, perhaps indirectly through enhanced translation or stabilization of chemokine.
Human melanoma cells are reported to express CXCR1 and CXCR2. These chemokine receptors play a role in the chemokine-mediated signaling pathways in melanoma [53,54]. The constitutive expression of CXCR1 and CXCR2 leads to a CXCL8-mediated metastatic phenotype in human malignant melanoma cells [54]. Expression of CXCL8 also leads to a CXCL8-dependent proliferation and angiogenesis in human melanomas [55–57]. Expression of CXCL1 leads to the IKK activation [43]. Over-expression of the murine homolog of CXCL1 in INK4a/ARF−/− immortalized melanocytes increased melanoma tumor incidence [13] and induced malignant progression of squamous cell carcinoma in nude mice [14]. These data suggest that G-protein-coupled receptors, especially CXCR1 and CXCR2, may facilitate to melanoma tumor progression through an autocrine system, which may in turn lead to the constitutive activation of NF-κB in melanoma cells.
In CDP-transduced Hs294T melanoma cells, both NF-κB/DNA association and NF-κB-mediated transcription are reduced in comparison to vector-transduced Hs294T cells. These data indicate that the expression of CDP reduces the NF-κB-mediated transcription. In CDP-transduced Hs294T melanoma cells, the binding of CBP to Rel A is reduced, whereas the overexpressed CDP binds efficiently to CBP in comparison to vector-transduced Hs294T cells. Moreover, over-expression of CDP enables the detection of the binding of CDP with PCAF as well as with CBP. This increased binding of CDP to CBP/PCAF may lead to the sequestration of CBP/PCAF from Rel A, which may cause the binding between DNA and Rel A to become unstable. Previously, Li et al. [50] showed that PCAF/CBP binds to and acetylates CDP, leading to the inhibited binding of CDP to DNA. The lysine acetylases seem to enhance NF-κB-mediated transcription by increasing the DNA binding of RelA/p50 NF-κB and simultaneously decreasing the DNA binding of CDP [50]. Here, we show that the coexpression of CDP and PCAF-ΔLAT efficiently leads to a decrease in NF-κB activity.
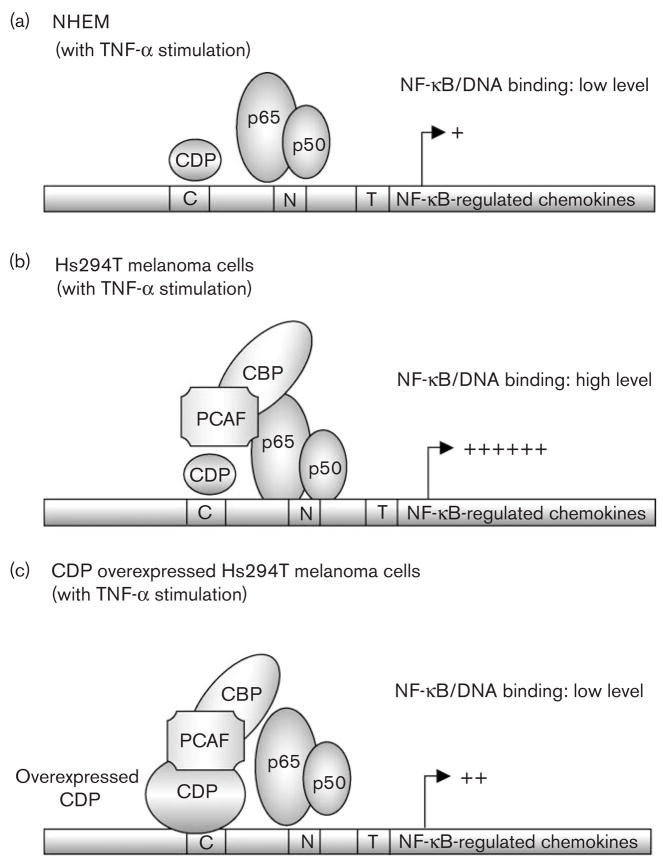
According to our observation, we proposed a hypothetical model whereby CDP overexpression leads to the inhibition of NF-κB activation. In NHEM normal melanocytes, Rel A/p50 dimers translocate to the nucleus after TNF-α stimulation. The DNA-binding activity of Rel A/p50 dimers, however, may be destabilized due to insufficient acetylation. This destabilization likely results in reduced transcription activity (Fig. 5a). In Hs294T cells, Rel A/p50 dimers bind to the NF-κB consensus sequences, and this binding is stabilized by increased acetylation of RelA, resulting in significantly enhanced transcription (Fig. 5b). Likewise, in CDP-transduced Hs294Tcells, the interaction of the over-expressed CDP with CBP/PCAF is increased, which may lead to the sequestration of CBP/PCAF from Rel A. This in turn may lead to the destabilization of DNA/Rel A binding, as well as to the reduction of transcriptional activity (Fig. 5c). In addition, previous studies suggest that overall transcription is activated by CBP/p300/PCAF-mediated acetylation of histones [38], and CDP recruits histone deacetylases, leading to the inhibition of the transcription [38]. These data together suggest that CDP is a regulator of NF-κB-mediated chemokine/cytokine transcription in melanoma cells.
Fig. 5.
Hypothetical model: CCAAT displacement protein (CDP) interacts with lysine acetyl transferase, CBP/PCAF and interferes with the activation of nuclear factor-kappa beta (NF-κB). (a) Model of NF-κB-regulated transcription in normal human epidermal melanocytes (NHEM): NF-κB proteins (Rel A/p50 dimers) translocate into the nucleus after tumor necrosis factor α (TNF-α) stimulation. The binding of NF-κB proteins to DNA, however, is reduced in NHEM as compared with Hs294T cells. This may be due to the lack of interaction between NF-κB and the protein acetylase CBP/PCAF complex. (b) Model of NF-κB-regulated transcription in Hs294T melanoma cells: NF-κB proteins (Rel A/p50 dimers) translocate into the nucleus after TNF-α stimulation. NF-κB proteins bind to its consensus sequences in the nucleus, and this binding is possibly stabilized by the protein acetylase complex CBP/PCAF. (c) Model of NF-κB-regulated transcription in Hs294T melanoma cells with over-expressed CDP: NF-κB proteins (Rel A/p50 dimers) translocate into the nucleus after TNF-α stimulation. Over-expressed CDP stably binds to DNA and interacts with CBP/PCAF, which may lead to the sequestration of CBP/PCAF from Rel A and to the unstable DNA binding of Rel A/p50 complex.
This study contributes to our understanding of the mechanism for NF-κB constitutive activation in human melanoma cells. We observed that enhanced acetylation of NF-κB in melanoma cells is associated with enhanced binding of NF-κB protein to their DNA promoter element. Moreover, we showed that binding of CDP to CBP/PCAF, leads to the inhibition of the activated NF-κB in human melanoma cells. We predict that the positioning of the CDP binding motif C(G/A)AT and C/EBP binding motif CCAATadjacent to NF-κB binding element occurs in many NF-κB-regulated gene promoters. We show here that CDP reduces not only CXCL1/CXCL8 promoter activity, but also reduces NF-κB-mediated gene transcription in melanoma cells. We anticipate that the NF-κB-mediated transcription may be a useful target in the treatment of melanoma. Thus, raising CDP expression might provide a new window into lowering NF-κB-mediated transcription and blocking melanoma tumor growth.
Acknowledgments
We would like to acknowledge the Vanderbilt Microarray Core Laboratory for the oligo-array analysis (supported by CA68425), Dr Naofumi Mukaida of Kanazawa University, Japan, for the IL-8-luciferase reporter gene construct, Dr Tony Kouzarides of Cambridge University for pCi/PCAF plasmid constructs, and Dr Alan Nepveu of McGill University for the pMX/CDP plasmid constructs. This work was supported by grants from the Department of Veterans Affairs (Merit Award and Senior Career Scientist Award to A.R.) and the National Institutes of Health (CA56704 and CA116021 to A.R.).
References
- 1.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay D, Timchenko N, Suwa T, Hornsby PJ, Campisi J, Medrano EE. The human melanocyte: a model system to study the complexity of cellular aging and transformation in non-fibroblastic cells. Exp Gerontol. 2001;36:1265. doi: 10.1016/s0531-5565(01)00098-5. [DOI] [PubMed] [Google Scholar]
- 3.Nyormoi O, Bar-Eli M. Transcriptional regulation of metastasis-related genes in human melanoma. Clin Exp Metastasis. 2003;20:251. doi: 10.1023/a:1022991302172. [DOI] [PubMed] [Google Scholar]
- 4.Huang S, DeGuzman A, Bucana CD, Fidler IJ. Level of interleukin-8 expression by metastatic human melanoma cells directly correlates with constitutive NF-kappaB activity. Cytokines Cell Mol Ther. 2000;6:9. doi: 10.1080/13684730050515868. [DOI] [PubMed] [Google Scholar]
- 5.Ueda Y, Richmond A. NF-kappa B activation in melanoma. Pigment Cell Res. 2006;19:112. doi: 10.1111/j.1600-0749.2006.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richmond A. NF-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol. 2002;2:664. doi: 10.1038/nri887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh RK, Gutman M, Radinsky R, Bucana CD, Fidler IJ. Expression of interleukin 8 correlates with the metastatic potential of human melanoma cells in nude mice. Cancer Res. 1994;54:3242. [PubMed] [Google Scholar]
- 8.Richmond A, Lawson DH, Nixon DW, Chawla RK. Characterization of autostimulatory and transforming growth factors from human melanoma cells. Cancer Res. 1985;45 (12 Pt 1):6390. [PubMed] [Google Scholar]
- 9.Mrowietz U, Schwenk U, Maune S, Bartels J, Kupper M, Fichtner I, et al. The chemokine RANTES is secreted by human melanoma cells and is associated with enhanced tumour formation in nude mice. Br J Cancer. 1999;79:1025. doi: 10.1038/sj.bjc.6690164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottazzi B, Walter S, Govoni D, Colotta F, Mantovani A. Monocyte chemotactic cytokine gene transfer modulates macrophage infiltration, growth, and susceptibility to IL-2 therapy of a murine melanoma. J Immunol. 1992;148:1280. [PubMed] [Google Scholar]
- 11.Owen JD, Strieter R, Burdick M, Haghnegahdar H, Nanney L, Shattuck-Brandt R, Richmond A. Enhanced tumor-forming capacity for immortalized melanocytes expressing melanma growth stimulatory activity/growth-regulated cytokine beta and gamma proteins. Int J Cancer. 1997;73:94. doi: 10.1002/(sici)1097-0215(19970926)73:1<94::aid-ijc15>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Schaider H, Oka M, Bogenrieder T, Nesbit M, Satyamoorthy K, Berking C, et al. Differential response of primary and metastatic melanomas to neutrophils attracted by IL-8. Int J Cancer. 2003;103:335. doi: 10.1002/ijc.10775. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Luan J, Yu Y, Li C, DePinho RA, Chin L, Richmond A. Induction of melanoma in murine macrophage inflammatory protein 2 transgenic mice heterozygous for inhibitor of kinase/alternate reading frame. Cancer Res. 2001;61:8150. [PubMed] [Google Scholar]
- 14.Loukinova E, Dong G, Enamorado-Ayalya I, Thomas GR, Chen Z, Schreiber H, Van Waes C. Growth regulated oncogene-alpha expression by murine squamous cell carcinoma promotes tumor growth, metastasis, leukocyte infiltration and angiogenesis by a host CXC receptor-2 dependent mechanism. Oncogene. 2000;19:3477. doi: 10.1038/sj.onc.1203687. [DOI] [PubMed] [Google Scholar]
- 15.Dixit V, Mak TW. NF-kappaB signaling. Many roads lead to madrid. Cell. 2002;111:615. doi: 10.1016/s0092-8674(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 17.Naumann M, Scheidereit C. Activation of NF-kappa B in vivo is regulated by multiple phosphorylations. Embo J. 1994;13:4597. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 20.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 21.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 22.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J Mol Med. 2003;81:549. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea JJ, Kanno Y, Chen X, Levy DE. Cell signaling. Stat acetylation: a key facet of cytokine signaling? Science. 2005;307:217. doi: 10.1126/science.1108164. [DOI] [PubMed] [Google Scholar]
- 24.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62:6231. [PubMed] [Google Scholar]
- 25.Soloff MS, Cook DL, Jr, Jeng YJ, Anderson GD. In situ analysis of interleukin-1-induced transcription of cox-2 and il-8 in cultured human myometrial cells. Endocrinology. 2004;145:1248. doi: 10.1210/en.2003-1310. [DOI] [PubMed] [Google Scholar]
- 26.Stein B, Baldwin AS., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol Cell Biol. 1993;13:7191. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antes TJ, Chen J, Cooper AD, Levy-Wilson B. The nuclear matrix protein CDP represses hepatic transcription of the human cholesterol-7alpha hydroxylase gene. J Biol Chem. 2000;275:26649. doi: 10.1074/jbc.M002852200. [DOI] [PubMed] [Google Scholar]
- 28.Khanna-Gupta A, Zibello T, Kolla S, Neufeld EJ, Berliner N. CCAAT displacement protein (CDP/cut) recognizes a silencer element within the lactoferrin gene promoter. Blood. 1997;90:2784. [PubMed] [Google Scholar]
- 29.Wu GD, Lai EJ, Huang N, Wen X. Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. The role of Oct-1 as a transcriptional repressor. J Biol Chem. 1997;272:2396. [PubMed] [Google Scholar]
- 30.Boudreau F, Rings EH, Swain GP, Sinclair AM, Suh ER, Silberg DG, et al. A novel colonic repressor element regulates intestinal gene expression by interacting with Cux/CDP. Mol Cell Biol. 2002;22:5467. doi: 10.1128/MCB.22.15.5467-5478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon NS, Rong Zeng W, Premdas P, Santaguida M, Berube G, Nepveu A. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int J Cancer. 2002;100:429. doi: 10.1002/ijc.10510. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, et al. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 33.Mailly F, Berube G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dufort D, Nepveu A. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N. C/EBP epsilon mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut) Proc Natl Acad Sci U S A. 2001;98:8000. doi: 10.1073/pnas.141229598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neville PJ, Thomas N, Campbell IG. Loss of heterozygosity at 7q22 and mutation analysis of the CDP gene in human epithelial ovarian tumors. Int J Cancer. 2001;91:345. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1050>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 37.Goulet B, Watson P, Poirier M, Leduy L, Berube G, Meterissian S, et al. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625. [PubMed] [Google Scholar]
- 38.Nirodi C, Hart J, Dhawan P, Moon NS, Nepveu A, Richmond A. The role of CDP in the negative regulation of CXCL1 gene expression. J Biol Chem. 2001;276:26122. doi: 10.1074/jbc.M102872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 40.Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci U S A. 1994;91:9564. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau NR, Littman DR. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Richmond A. Constitutive IkappaB kinase activity correlates with nuclear factor-kappaB activation in human melanoma cells. Cancer Res. 2001;61:4901. [PubMed] [Google Scholar]
- 44.Iwata K, Tomita K, Sano H, Fujii Y, Yamasaki A, Shimizu E. Trichostatin A, a histone deacetylase inhibitor, down-regulates interleukin-12 transcription in SV-40-transformed lung epithelial cells. Cell Immunol. 2002;218:26. doi: 10.1016/s0008-8749(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa Y, Mukaida N, Kuno K, Rice N, Okamoto S, Matsushima K. Establishment of lipopolysaccharide-dependent nuclear factor kappa B activation in a cell-free system. J Biol Chem. 1995;270:4158. [PubMed] [Google Scholar]
- 46.Creasey AA, Smith HS, Hackett AJ, Fukuyama K, Epstein WL, Madin SH. Biological properties of human melanoma cells in culture. In Vitro. 1979;15:342. doi: 10.1007/BF02616140. [DOI] [PubMed] [Google Scholar]
- 47.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 48.Ikenoue T, Hikiba Y, Kanai F, Aragaki J, Tanaka Y, Imamura J, et al. Different effects of point mutations within the B-Raf glycine-rich loop in colorectal tumors on mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase and nuclear factor kappaB pathway and cellular transformation. Cancer Res. 2004;64:3428. doi: 10.1158/0008-5472.CAN-03-3591. [DOI] [PubMed] [Google Scholar]
- 49.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Aufiero B, Schiltz RL, Walsh MJ. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc Natl Acad Sci U S A. 2000;97:7166. doi: 10.1073/pnas.130028697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma UN, Yamamoto Y, Prajapati S, Gaynor RB. Nuclear role of I kappa B Kinase-gamma/NF-kappa B essential modulator (IKK gamma/NEMO) in NF-kappa B-dependent gene expression. J Biol Chem. 2004;279:3509. doi: 10.1074/jbc.M309300200. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 53.Luan J, Shattuck-Brandt R, Haghnegahdar H, Owen JD, Strieter R, Burdick M, et al. Mechanism and biological significance of constitutive expression of MGSA/GRO chemokines in malignant melanoma tumor progression. J Leukoc Biol. 1997;62:588. doi: 10.1002/jlb.62.5.588. [DOI] [PubMed] [Google Scholar]
- 54.Varney ML, Li A, Dave BJ, Bucana CD, Johansson SL, Singh RK. Expression of CXCR1 and CXCR2 receptors in malignant melanoma with different metastatic potential and their role in interleukin-8 (CXCL-8)-mediated modulation of metastatic phenotype. Clin Exp Metastasis. 2003;20:723. doi: 10.1023/b:clin.0000006814.48627.bd. [DOI] [PubMed] [Google Scholar]
- 55.Bobrovnikova-Marjon EV, Marjon PL, Barbash O, Vander Jagt DL, Abcouwer SF. Expression of angiogenic factors vascular endothelial growth factor and interleukin-8/CXCL8 is highly responsive to ambient glutamine availability: role of nuclear factor-kappaB and activating protein-1. Cancer Res. 2004;64:4858. doi: 10.1158/0008-5472.CAN-04-0682. [DOI] [PubMed] [Google Scholar]
- 56.Leslie MC, Bar-Eli M. Regulation of gene expression in melanoma: new approaches for treatment. J Cell Biochem. 2005;94:25. doi: 10.1002/jcb.20296. [DOI] [PubMed] [Google Scholar]
- 57.Singh RK, Varney ML, Bucana CD, Johansson SL. Expression of interleukin-8 in primary and metastatic malignant melanoma of the skin. Melanoma Res. 1999;9:383. doi: 10.1097/00008390-199908000-00007. [DOI] [PubMed] [Google Scholar]