Abstract
CXC chemokines, which induce angiogenesis, have glutamine-leucine-arginine amino acid residues (ELR motif) in the amino terminus and bind CXCR2 and the Duffy antigen chemokine-binding protein. Duffy, a seven transmembrane protein that binds CXC and CC chemokines, has not been shown to couple to trimeric G proteins or to transduce intracellular signals, although it is highly expressed on red blood cells, endothelial cells undergoing neovascularization, and neuronal cells. The binding of chemokines by Duffy could modulate chemokine responses positively or negatively. Positive regulation could come through the presentation of chemokine to functional receptors, and negative regulation could come through Duffy competition with functional chemokine receptors for chemokine binding, thus serving as a decoy receptor. To determine whether Duffy has a role in angiogenesis and/or maintenance of homeostasis, we developed transgenic mice expressing mDuffy under the control of the preproendothelin promoter/enhancer (PPEP), which directs expression of the transgene to the endothelium. Two PPEP-mDuffy-transgenic founders were identified, and expression of the transgene in the endothelium was verified by Northern blot, RT-PCR, and immunostaining of tissues. The phenotype of the mice carrying the transgene appeared normal by all visual parameters. However, careful comparison of transgenic and nontransgenic mice revealed two phenotypic differences: mDuffy-transgenic mice exhibited a diminished angiogenic response to MIP-2 in the corneal micropocket assay, and mDuffy-transgenic mice exhibited enhanced hepatocellular toxicity and necrosis as compared with nontransgenic littermates in response to overdose of acetaminophen (APAP; 400 mg/kg body weight). Morover, APAP treatment was lethal in 50% of the mDuffy-transgenic mice 24 h post challenge, and 100% of the nontransgenic litter-mates survived this treatment at the 24 h time point. Our data suggest that enhanced expression of mDuffy on endothelial cells can lead to impaired angiogenic response to chemokines and impaired maintenance of homeostasis in response to toxic stresses.
Keywords: hepatocellular toxicity, acetaminophen, chemotactic cytokines, MIP-2, CXCR2
INTRODUCTION
The Duffy antigen-binding protein was originally identified as a blood-group antigen required for invasion by the human malaria parasite, Plasmodium vivax, and related monkey malaria parasite, Plasmodium knowlesi [1–3]. It was demonstrated that the chemokine-binding protein expressed on red blood cells is identical to the Duffy antigen, and the new name, Duffy antigen receptor for chemokine (DARC), was given [4, 5]. Cells transfected with hDuffy bind to CXC chemokine family members, melanoma growth-stimulating activity (MGSA)/GRO, interleukin (IL)-8, and neutrophil-activating polypeptide-2 (NAP-2), and to CC chemokine family members, monocyte chemoattractant protein-1 (MCP-1) and regulated on activation, normal T expressed and secreted (RANTES), with similar high affinity [6]. In humans, this receptor is expressed on red blood cells, endothelial cells, and neuronal cells [5, 7–9]. Although Duffy does not couple to G proteins, it binds chemokine or P. vivax with high affinity, the binding event induces receptor internalization, and the binding of MGSA/GRO to Duffy blocks erythrocyte invasion by P. vivax [10, 11]. However, because Duffy is not coupled to trimeric G proteins, and ligand binding does not appear to evoke an intracellular signal [10], it was decided recently that Duffy should not be given receptor status and should be referred to as the Duffy antigen chemokine-binding protein [12].
Murine red blood cells exhibit a similar pattern of chemokine binding to that observed for human red blood cells, suggesting that the murine homolog of Duffy is also expressed in the mouse [13]. The murine ortholog of Duffy has been cloned, and the tissue expression of mDuffy appears to be nearly identical to that of hDuffy, exhibiting 63% identity at the amino acid level [14, 15]. The murine Duffy maps to chromosome 1 between Xmv41 and D1Mit166 [14, 15]. The genomic structure for mDuffy has been characterized, revealing two exons (100 and 1064 nucleotides in length, respectively), separated by a 461-bp intron [14, 15].
mDuffy is expressed during embryonic development between days 9.5 and 12 [15]. Duffy expression can be induced by cytokines through the GATA motif. To determine whether induction of Duffy expression might serve as a positive or negative modulator of homeostasis, we chose to develop transgenic mice expressing Duffy under the control of the preproendothelin promoter/enhancer (PPEP). This enhancer is shown to direct expression to the aorta, endothelial cells of large and small vessels, the brain, trachea, lung, and heart [16]. We show here that the observed phenotype of the animals at birth is fairly normal. However, transgene-positive mice exhibited a reduced angiogenic response to macrophage-inflammatory protein-2 (MIP-2) in the corneal micropocket assay, and hepatocytes around the central vein exhibited extensive vacuolization and lipid inclusions, without inflammatory infiltrate, elevated alanine amino transferase (ALT), or aspartate amino transferase (AST), two liver enzymes that indicate hepatocellular toxicity. In addition, we observed an abnormal chemical-induced stress response in the mDuffy-transgenic mice when overdosed with acetaminophen (APAP/tylenol). APAP-treated transgenic mice exhibited an early rise (8 h) in AST and ALT and a decline in serum MIP-2 levels compared with nontransgenic mice. Moreover, 50% of the transgenic mice died by 24 h, and nontransgenic APAP-treated mice exhibited 100% survival over the same period. Data are compatible with a role for mDuffy in maintenance of homeostasis, perhaps by sequestering chemokine from circulation, potentially reducing their effectiveness in mediating biological processes.
MATERIALS AND METHODS
Materials
Polyclonal antibodies to a peptide from the amino-terminus of mDuffy (NYFEDNYSYELSSDC-amide) conjugated to keyhole limpet hemocyanin (KLH) were developed in sheep in collaboration with Bionostics (Acton, MA) following standard protocols. Serum from immunized sheep was purified over a glutathione-S-transferase (GST)-mDuffy-affinity column, and the specificity of binding to the GST-mDuffy was confirmed against GST alone and by blocking immunostaining with the peptide to which the antibody was developed. C57Bl CXCR2−/+ mice were obtained from R. Terkletaub, VA Medical Center, San Diego, CA and bred to homozygosity.
mDuffy transgenics
Previously, we cloned the murine homologue of Duffy gene [15]. The 1.5 kb PstI/PvuII fragment of genomic mDuffy was ligated to the human growth hormone (hGH) polyadenylation fragment. This fragment was subsequently ligated to the vector containing the PPEP (5.9 kb), previously demonstrated to drive luciferase expression in transiently transfected endothelial cells. A fragment including the PPEP, mDuffy, and the hGH polyadenylation site was used for microinjection into fertilized ova from C57/BL6 black female mice, which were placed into the female B6D2 F1 mice (foster mothers). The microinjection was done by the Vanderbilt Cancer Center Transgenic Core (Nashville, TN). Germline transmission was confirmed by Southern blot and polymerase chain reaction (PCR) strategies as described below.
Genotyping of transgenic mice
The mice were weaned at 3 weeks of age. Newborns were genotyped at 2 weeks of age from DNA extracted from 1–1.5 cm of the tail. The tail cut was digested in 0.5 ml proteinase K buffer [50 mM Tris-HCl, pH 8.0/100 mM ethylenediaminetetraacetate (EDTA), pH 8.0/0.5% sodium dodecyl sulfate (SDS)], containing 25 μl proteinase K using an overnight incubation at 50°C. After three phenol/chloroform extractions, an equal volume of 100% ethanol containing 0.3 M sodium acetate (pH 6.0) was added, and genomic DNA was pulled out with a glass hook, followed by two 70% ethanol washes and one 100% ethanol wash. The DNA was air-dried and dissolved in milliQ water.
Screening of transgenic founders and gene-dosage determination by PCR and Southern blot
Three sets of transgene-specific PCR primers were designed at the junction of the PPEP and mDuffy (PPEP1/rev120, PPEP2/rev120) [5′-CCTGGATTGTCAGACGGC-3′/5′-GTCACTCGAGAGTTCATAGG-3′; 5′-TGCCTGTGGGTGACTAATC-3′/5′-GTCACTCGAGAGTTCATAGG-3′] and the junction of mDuffy and hGH polyadenylation fragment (mDuffy/hHGpA) [5′-GATGCAATGCTGAATGTGACAG3′/5′-TAATCCCAGCAATTTGGGAGGC-3′], respectively. Another set of endogenous mDuffy-specific primer sets was used to detect the presence of endogenous mDuffy [5′-GGCACTTATCTTGGAGCCAC-3′]. Typical PCR conditions were as follows: after 5 min of initial denaturation, 30 –35 cycles of denaturing at 94°C for 30 sec, annealing at 54°C for 30 sec, and extension at 72°C for 1 min, followed by 10 min of final extension at 72°C. PCR product (5 μl) was analyzed by agarose gel electrophoresis with ethidium bromide staining.
Southern blot
Genomic DNA from tailing (5 μg) was digested with selected restriction enzyme(s) in appropriate buffer(s). DNA fragments were separated in 0.7% agarose gel and transferred to HybondN+ membrane (Amersham-Pharmacia, Piscataway, NJ). A 470-bp mDuffy cDNA probe was labeled by 32P-dCTP using the Multi-Prime Labeling kit from Amersham-Pharmacia. The blot was washed extensively and exposed to X-film from Kodak or subjected to phosphoimage analysis. The transgene band differs from the endogenous mDuffy band. The gene dosage was determined by the intensity of the transgenic band as compared with the endogenous band.
Northern blot
Northern blot analyses were performed on total RNA extracted from organs of 5- to 6-week-old transgenic and nontransgenic mice. Total RNA was extracted with Ultraspec (Biotecx, Houston, TX) and quantitated by spectroscopy. RNA (20 μg) samples from each tissue [except for trachea (5 μg) and aorta (10 μg)] were electrophoresed through a 1.1% formaldehyde/agarose gel, transferred to Hybound N+ membrane (Amersham/Pharmacia), fixed by UV-cross-linking, prehybridized with Express-Hyb from Clontech (Palo Alto, CA), and hybridized in the same buffer containing the 1.1-kb full-length 32P-labeled mDuffy cDNA probe (9×107 cpm/ml) at 68°C for 2 h. The blot was subsequently washed four times at room temperature with 2 × saline sodium citrate (SSC)/0.05% SDS (10 min per wash) and twice at 50°C with 0.1 × SSC/0.1% SDS (30 min per wash) and then exposed to phosphoimage analysis (Molecular Dynamics, Sunnyvale, CA).
In situ hybridization
Deparaffinized sections were acid-treated to denature RNA at 25°C for 15 min in 0.2 M HCL, followed by 7.5 min of proteinase K (Sigma Chemical Co., St. Louis, MO) treatment (20 μg per ml) in 50 mM Tris, pH 7.5, 5 mM EDTA to permeabilize the cell. Sections were postfixed for 15 min in 4% paraformaldehyde. Samples were briefly acetylated and air-dried. The mDuffy probe was generated by subcloning a 474-bp EcoRV fragment into a pBluescript SK vector (Stratagene, San Diego, CA). The plasmid was linearized by BamHI or HindIII for antisense or sense riboprobe-labeling with T7 or T3 RNA polymerase (Amersham) and 35S-UTP. Hybridization was performed overnight at 50°C with 2 × SSC/chloride buffer/50% formamide for humidification using 200 μl hybridization mixture to treat the slides at a final probe concentration of 2 × 104 cpm per μl. The hybridization mixture contained 300 mM NaCl, 10 mM Tris-HCl, pH 7.4, 10 mM NaHPO4, pH 6.8, 5 mM EDTA, pH 8.0, 0.2% Ficoll 400, 0.2% polyvinyl pyrolidone, 50 mM dithiothreitol, 10% Dextran sulfate, 50% deionized formamide, and 20 mM β-mercaptoethanol. After hybridization at 55°C for 30 min each, slides were washed in a buffer containing 5 × SSC/chloride buffer, 20 mM Tris•HCl, pH 7.5, 2 mM EDTA, pH 8.0, at 37°C for 10 min. Slides were digested with RNase A (20 mg per μl) in the same buffer for 30 min. This procedure was then repeated. Slides were rinsed in water, air dried, dipped in Kodak NTB2 Emulsion, exposed for 2 weeks, and developed with D-19 (Kodak, Rochester, NY).
Reverse transcription (RT)-PCR analysis of tissue expression of mDuffy antigen and transgene Duffy antigen
Total RNA from tissue was extracted using the TRIZOL Reagent (Gibco/BRL, Grand Island, NY). Total RNA (10 –50 μG) was digested with 10 units of DNAseI for 30 min at 37°C. The mixture was then extracted with phenol/chloroform (3:1) to remove protein contamination and Dnase I from RNA. The clean RNA was collected by ethanol precipitation. RT-PCR analysis was used to detect the endogenous Duffy antigen and the transgene Duffy using the following primer sets: Duffy transgene PCR used the mPPEP primer [5′-CCTGGATTGTCAGACGGC-3′] and Rev 120 [5′-GTCACTCGAGAGTTCATAGG-3′]; endogenous Duffy PCR used the mDFA primer [5′-GGCACTTATCTTGGAGCCAC-3′] and the Rev 120 primer [5′-GTCACTCGAGAGTTCATAGG-3′]. The RT-PCR protocol was according the Access RT-PCR System (Promega, Madison, WI). First-strand cDNA synthesis is at 48°C for 45 min (RT), followed by 94°C for 2 min (RT inactivation and RNA/cDNA/primer denaturation), followed by 94°C for 30 sec (denaturation), 60°C for 1 min (annealing), 68°C for 2 min (extension), then 40 cycles at 68°C for 7 min (final extension), and finally cooled to 4°C (soak).
Western blot verification of mDuffy antibody specificity
Specificity of the antibody was determined by Western blot analysis of lysates from Escherichia coli expressing the mDuffy-GST fusion protein encoding the full amino terminus of mDuffy fused to the C-terminus of GST. Lysates were prepared for electrophoresis by solublization in SDS electrophoresis-loading buffer (0.125 M Tris-Cl, pH 6.8, 1% SDS, 2.5% β-mercaptoethanol, 10% glycerol, and 0.1% bromophenol blue) and electrophoresed on a 10% polyacrylamide gel with SDS and reducing agents. The samples were then transferred to nitrocellulose (Bio-Rad, Hercules, CA), blocked with a 5% solution of Carnation dry milk in Tris-buffered saline (TBS) with 0.05% TWEEN-20, incubated with a 1:500 dilution of sheep polyclonal antiserum to mDuffy in the same buffer at room temperature for 2 h, washed thrice in TBS with 0.05% TWEEN-20, incubated with a horseradish peroxidase-conjugated chicken antisheep antibody (1:5000) for 1 h at room temperature, and then washed three times with TBS prior to development with the enhanced chemiluminescence assay system (Amersham). Controls were lysates from E. coli expressing human CCR1-GST fusion protein. A second test of specificity of the mDuffy antiserum was immunodetection by dot blot analysis of the peptide, to which the antiserum was raised, coupled to bovine serum albumin (BSA). The mDuffy antiserum specifically bound the mDuffy peptide-BSA conjugate but not BSA alone. Based on these positive results, we affinity-purified the sheep mDuffy antiserum over a GST-mDuffy (amino terminus)-affinity column.
Histologic analysis
Organs from transgenic mice and wild-type (WT) mice were fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS), subjected to paraffin-embedded sectioning, followed by hematoxylin and eosin (H&E) staining prior to review. Sections were examined by two observers in a blinded fashion, without knowledge of the status of the mice. Immunostaining was performed using the standard Vectastain ABC methodology according to the Vectastain protocol, Vector Laboratories. PAS staining, Sudan Black, and Oil Red-O staining were according to published procedures using frozen sections [17]. To examine the expression of mDuffy in the brain, transgenic and nontransgenic mice were perfused through the dorsal aorta mice with saline followed by a 30 ml solution of 4% paraformaldehyde/PBS for 10 min. Brains were then dissected, postfixed in paraformaldehyde, embedded, and sectioned. Immunohistochemical staining was carried out using a 1:200 dilution of the mDuffy affinity-purified antibody for 1 h at 37°C and the Vectastain ABC kit to follow the binding of the sheep antibody to mDuffy. Sites of immunoreactivity were visualized using aminoethyl carbazole as a substrate for the peroxidase. Negative controls for the immunostaining included inclusion of a nonspecific immunoglobulin G (IgG) and/or elimination of the primary antibody, and the remainder of the Vectastain protocol was intact.
Corneal micropocket angiogenesis assay
Hydron pellets incorporating sucralfate with vehicle alone, basic fibroblast growth factor (bFGF; gift from Scios Inc., Sunnyvale, CA), recombinant (r)MIP-2 (gift of Elias Lolis, Yale University, New Haven, CT), and rMIP-2 in the presence or absence of affinity-purified antibody to murine CXCR2 (gift of R. M. S.) were prepared as described previously [18]. Pellets containing 160 ng MIP-2 or 90 ng bFGF were surgically implanted into corneal stromal micropockets created 1 mm medial to the lateral corneal limbus of C57BL male or female WT mice, those expressing the mDuffy transgene, or mice exhibiting a targeted deletion of the murine receptor for CXCR2 (CXCR2−/−). At day 5 after implantation, corneas were photographed at an incipient angle of 35–50° from the polar axis in the meridian containing the pellets using a Zeiss slip lamp. Quantitation was carried out in two ways: outgrowth of vessels from the limbus—response versus no response; and where there was an angiogenic response to the hydron pellet, the estimated length of vessel growth from the limbus was measured after photography.
APAP treatment
Male mice, 8 –16 weeks old, were used for the APAP treatment. Transgenic and WT mice (10/group) were fasted for 12 h prior to the intraperitoneal (i.p.) injection of APAP (400 mg/kg in PBS). PBS was injected in the control group. Mice were sacrificed at 8 h postinjection, the liver was removed and fixed in 4% paraformaldehyde/PBS for histology, and serum was separated for quantitation of ALT and AST as described below and for determination of chemokine levels by enzyme-linked immunosorbent assay (ELISA). This experiment was performed three times with three to six mice in each group for each repeat experiment. In a second set of experiments, mice (three to six per group) were observed for 24 h post-APAP-injection. For those that survived the 24-h time period, mice were sacrificed, serum was collected, and livers were processed for fixation as described above. Some of the mice in the second set of experiments were also injected i.p. with 10 μg MIP-2 at the time of the APAP treatment. In addition, mice not treated with APAP were injected with MIP-2, the mice were sacrificed, and MIP-2 levels in the serum and livers were determined 2 h postinjection.
Analysis of serum levels of APAP metabolites in Duffy transgenic and nontransgenic mice treated with APAP
Transgenic and nontransgenic mice received APAP (400 mg/kg) i.p., and blood was collected at 1, 2, and 4 h after APAP administration. The concentrations of APAP and its metabolites in plasma were detected by the high-pressure liquid chromatography (HPLC) method of Manautou et al. [19] with slight modification. The HPLC system is equipped with a Waters 501 pump, a manual injector, and 5 μm Hypersil C18 reverse-phase column (250×4.6mm). The mobile phase consisted of 1% glacial acetic acid, 12% methanol, and 87% water. The samples were run isocratically at a constant rate of 0.7 ml/min. APAP and its metabolites were detected using a Waters 740 detector at 254 nm. APAP and its metabolites in plasma samples were identified by comparing their retention times with those of authentic standards.
ALT and AST levels in sera were determined by kinetic method using the Sigma Diagnostics kits. Results from the kits were compared with results obtained from the Chemistry Laboratory of the Vanderbilt Clinic. Comparable results were obtained with both assays, so subsequent assays were performed manually using the Sigma Diagnostics kits for ALT and AST.
ELISA assays for chemokine-level determination
MIP-2, MCP-1 [murine homologue of MCP-1 (JE)], KC (probable murine orthology of human CXCL3), and RANTES levels in diluted mouse sera and liver lysates were measured by ELISA using the Quantikine ELISA kit for these murine chemokines from R&D Systems (Minneapolis, MN). Snap frozen liver from APAP-treated and -nontreated mice was homogenized in 50 mM Tris•HCl, pH 8.0/25 mM NaCl, containing complete proteinase inhibitor (Boehringer Mannheim, Indianapolis, IN). After two rounds of centrifugation (15,000 rpm) for 30 min at 4°C, the supernatant was used for ELISA.
Analysis of serum lipid levels
Blood was collected from mice at 2 months of age and processed after clotting to collect serum. Aliquots (20 μl) were used for analysis of triglycerides and cholesterol using standard protocols [20].
RESULTS
Development of mDuffy-transgenic mice
The mDuffy-transgene construct placing the entire mDuffy cDNA under the control of 5.9 kb of the PPEP was developed using the protocols described in Materials and Methods (Fig. 1). Using standard microinjection and transgenic protocols, two male founders expressing the mDuffy transgene were generated on the C57 Black background (#35 and #43; Fig. 1, B–D). The gross phenotype of the founder mice and their offspring was visually normal. There was no evidence for gross motor abnormalities or behavioral problems.
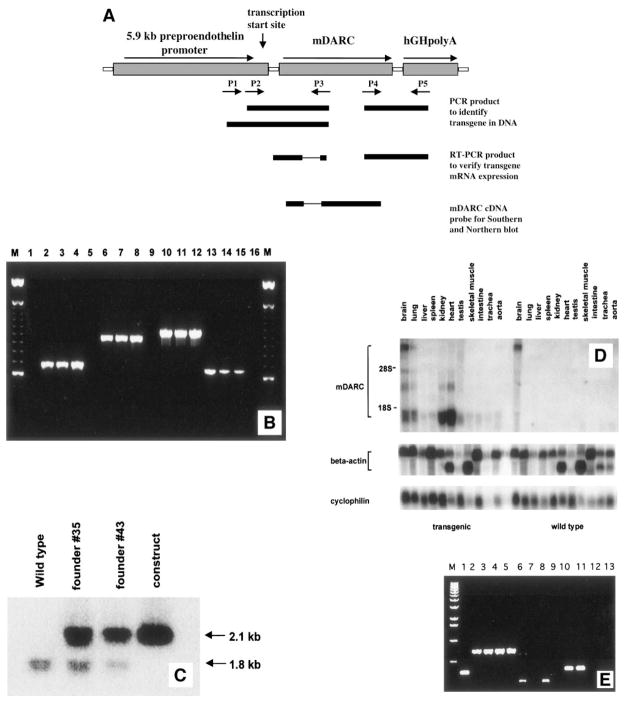
Fig. 1.
(A) Construction of the mDuffy transgene. The PPEP directs expression to endothelial cells, aorta, brain, trachea, lung, and heart. The transgene included 5.9 kb of the PPEP to direct expression of a 1.5-kb fragment of genomic mDuffy sequence encoding the mDuffy first intron and entire coding sequence of the mDuffy protein. The hGH polyadenylation sequence was ligated to the 1.5-kb mDuffy genomic fragment. A fragment comprised of the PPEP, mDuffy, and hGH polyadenylation site was used for microinjection for development of C57Bl transgenic mice as described in Materials and Methods. (B) PCR strategy for transgene screening. Three sets of transgene-specific primer sets were designed to detect the transgene. Two sets (P1/P3 and P2/P3) are located in the PPEP-mDuffy junction, and the third one (P4/P5) is located in the mDuffy-hGH polyA junction. Genomic DNA (50 ng) was used as template, and an endogenous-specific primer set was also used to monitor the quality of DNA samples. DNA from lanes 1, 5, 9, and 13 is from WT mice; lanes 2, 6, 10, and 14, from founder #35; lanes 3, 7, 11, and 15, from founder #43; lanes 4, 8, 12, and 16, from the transgene-plasmid construct; and lane M, the 100-bp DNA ladder. The PCR product represented in lanes 2– 4 is approximately 700 bp; lanes 6 – 8, 950 bp; lanes 10 –12, 1050 bp; and lanes 13–15, 650 bp. (C) Typical southern blot analysis of mDuffy transgene. Genomic DNA (5 μg) was digested with PstI and subjected to Southern blot analysis using the mDuffy 470-bp cDNA fragment as a probe. The lower 1.8-kb band represents the endogenous mDuffy gene, and the 2.1-kb band represents the transgene. By normalizing transgene dosage to the endogenous mDuffy band, we determined that the transgene copy number was equivalent in founder #35 and founder #43. (D) Northern analysis of RNA from transgenic and WT mice. Total RNA was extracted from tissues from mDuffy-transgenic and WT mice. Northern blots were performed as described in Materials and Methods, probing with the mDuffy cDNA probe, β-actin, and cyclophilin (IB15). Note the strong expression of mDuffy in kidney, heart, brain, and lung in transgenic mice as compared with WT mice. (E) PCR analysis of murine Duffy antigen and transgene Duffy antigen expression in transgenic and WT mice. Total RNA was extracted from tissues from mDuffy-transgenic and WT mice. RT-PCR analysis was performed as described in Materials and Methods, and for a control, β-actin mRNA was detected by RT-PCR. The size of the PCR products was determined based on comparison with a λ DNA 1-kb ladder. Lane 1, RT-PCR product generated from the positive control RNA from the kit (323 bp); lanes 2–5, actin RT-PCR (~750 bp) product from brain and kidney of mDuffy-transgenic mice (lanes 2 and 3) and from brain and kidney from WT mice (lanes 4 and 5); lanes 6 –9, endogenous mDuffy RT-PCR product (~250 bp) of brain and kidney from mDuffy-transgenic mice (lanes 6 and 7) and brain and kidney from WT mice (lanes 8 and 9); lanes 10 –13, transgene RT-PCR product of brain and kidney from mDuffy-transgenic mice (lanes 10 and 11) and brain and kidney from WT mice (lanes 12 and 13).
Expression of the mDuffy transgene
Northern blot analysis of tissues confirmed high levels of expression of mDuffy in the brain, lung, kidney, and heart of transgenic mice compared with nontransgenic littermates (Fig. 1D). There was also a low level of expression of mDuffy in liver, spleen, testis, skeletal muscle, intestine, and trachea in transgenic mice, which could be better visualized with a longer exposure of the Northern blot. In WT mice, mDuffy expression is also visualized by prolonged exposure of the Northern, although it is markedly lower than in transgenic mice. Earlier studies using the PPEP did not show that this promoter directs transgene expression to kidney. Although mDuffy has been shown previously to be expressed in the kidney, Northern analysis of mRNA from the kidney of nontransgenic mice did not reveal detectable Duffy mRNA in the kidney. To determine whether the Duffy mRNA in the kidney was derived from transcription of the endogenous mDARC gene or the transgene, RT-PCR analysis was performed using primers designed to distinguish between these two sources of mRNA. Results show that the source of Duffy mRNA in the kidney is the transgene (Fig. 1E).
Immunostaining for expression of mDuffy protein using a sheep polyclonal antibody developed by Bionostics revealed that the blood vessels, trachea, and brain of transgenic mice exhibited significantly more immunoreactive mDuffy than in nontransgenic littermates. These results are compatible with previous studies that the PPEP directs the highest level of expression to the endothelium (aorta), followed by the brain, trachea, lung, and heart [21]. In situ hybridization studies of brains revealed much greater density of silver grains over the Purkinje cells of the cerebellum of transgenic compared with nontransgenic mice (unpublished results). Transgenic and nontransgenic mice showed some faint immunoreactivity in the cerebral cortex, most often localized to pyramidal neurons. The corpus collosum also exhibited immunoreactivity for mDuffy in the transgenic mice. Purkinje and granule cells of the cerebellar cortex exhibit immunoreactivity for mDuffy in transgenic and nontransgenic animals, and the transgenic mice exhibit stronger mDuffy immunoreactivity (Fig. 2, A and B). The molecular layer of the cerebellum of transgenic and nontransgenic mice showed faint immunostaining, possibly indicative of dendritic (Purkinje cells) or axonal immunoreactivity (Fig. 2, A and B). The biological role for mDuffy expression in brain is unclear at this time. IL-8 and MGSA/GRO have been shown to modulate Purkinje cells [22] and neurotransmitter release by cerebellar neurons [23]. One possible influence of expression of mDuffy is that there could be enhanced sequestration of brain chemokines. Recently, Andjelkovic et al. [24] have visualized the binding sites for two Duffy ligands, MIP-1α and MCP-1, on the microvessels of the brain. There is no evidence to date that an intracellular signal is initiated when ligand binds to Duffy.
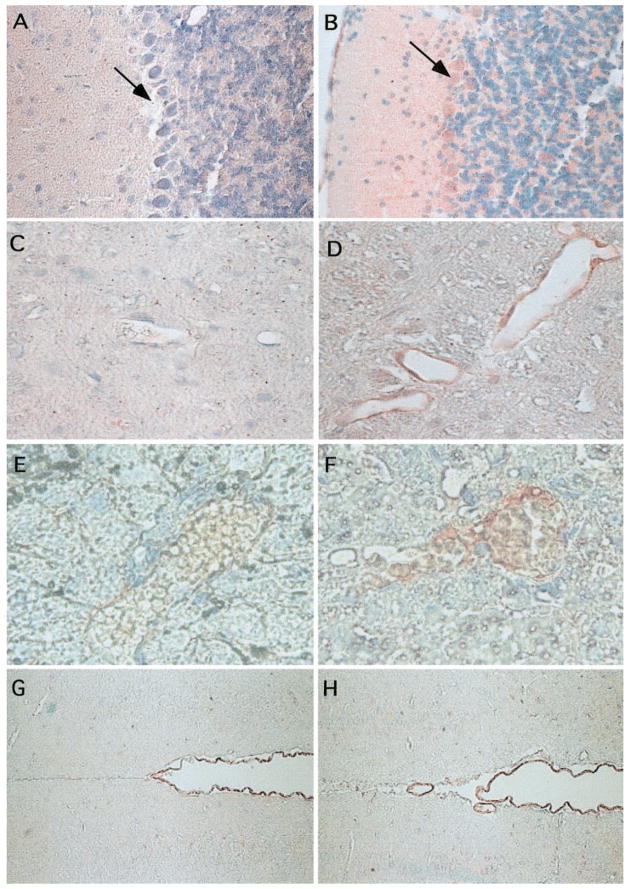
Fig. 2.
Localization of mDuffy mRNA and protein by immunohistochemistry. Brains and livers of transgenic and nontransgenic mice were fixed, embedded, and stained for immunoreactive mDuffy using the protocols described in Materials and Methods. Expression of the mDuffy protein in brain was confirmed by immunostaining using the mDuffy polyclonal antibody. Note the high level of expression of mDuffy (as visualized by the red staining) in the Purkinje cells as well as in the granular and molecular layers of the cerebellum in transgenic mice (B), compared with nontransgenic mice (A; original magnification, 100×). Note the expression of mDuffy in endothelial cells lining the blood vessels of the brain and APAP-treated liver in transgenic mice (D and F; original magnification, 50× and 125×, respectively), compared with that of nontransgenic mice (C and E; original magnification, 50× and 125×, respectively). The thin walls of the vessels suggest these are veins/venules. In contrast, the pia lining of the ventricles of the brain shows strong immunoreactivity for mDuffy in nontransgenic (G) and mDuffy-transgenic mice (H; original magnification, 50×).
Immunohistochemical staining for mDuffy was also concentrated along the endothelial cells of the intimal lining of the aorta (not shown) and in the endothelial cells lining some of the the blood vessels in the brain (Fig. 2, C and D), in the blood vessels of the liver (Fig. 2, E and F), and in the eye (see Fig. 4). In the trachea, immunoreactivity was associated with the epithelial cells (not shown). In the brain, immunoreactivity was also associated with cells of the pia lining the ventricles in the transgenic and nontransgenic mice (Fig. 2, G and H).
Fig. 4.
Immunolocalization of mDuffy and CXCR2 in the eye of transgenic mice treated with MIP-2. Immunoreactive CXCR2 (red staining) was observed in the endothelial cells of capillaries and blood vessels near the limbus, apparently involved in the angiogenic response to MIP-2 in transgenic mice (A) and WT mice (B), compared with control sections stained with a nonspecific IgG (C; original magnification, 125×). In transgenic mice, mDuffy immunoreactivity was stronger in the endothelial cells of the choroid, in the cells of the neural retina, particularly the rods and cones, in the cell processes comprising the outer plexiform layer, and the inner plexiform layer (D), compared with WT mice (E) and control treated with nonspecific IgG (F; original magnification, 50×). The blood vessels of the iris stain positively for immunoreactive mDuffy in transgenic mice (G) and to a lesser extent, in WT mice (H), and control tissues treated with a nonspecific isotype-matched IgG exhibit no immunoreactivity (I; original magnification, 125×). Arrows indicate positive staining for CXCR2 (A and B) and Duffy (D, E, G, and H).
Corneal micropocket assay for angiogenesis
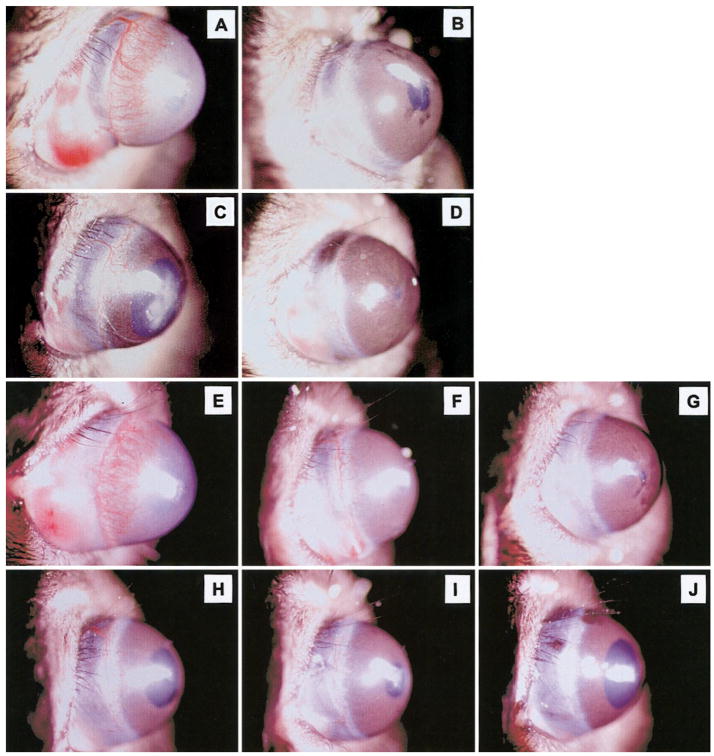
It has been suggested previously that Duffy may play a role as a regulator of angiogenesis. To determine whether overexpression of mDuffy in endothelial cells altered angiogenic response to the angiogenic chemokine, MIP-2, the corneal micropocket angiogenesis assay was performed. Hydron pellets containing 160 ng MIP-2, 90 ng bFGF, or vehicle alone were placed into the corneal pockets of anesthetized transgenic or nontransgenic mice of equal age using standard protocols [18]. Mice were followed over a time course of 5 days. On day 5, the outgrowth of blood vessels from the limbus of the eye was examined microscopically and photographed. Responses were evaluated as positive or negative based on whether the growth of blood vessels from the limbus was greater than observed with vehicle alone. The length of the vessels extending from the limbus could be compared and measured morphometrically after photography. In some experiments, neutralizing antibody against CXCR2 was added to pellets containing MIP-2, as indicated in the figure legend. Controls included bFGF and vehicle only (Fig. 3, A–J).
Fig. 3.
Angiogenic response to MIP-2 in nontransgenic (WT) mice and transgenic (mDuffy) mice. Corneal micropocket assays were carried out as described in Materials and Methods. For nontransgenic mice (A–D), a brisk angiogenic response was observed in response to bFGF (90 ng) in nontransgenic mice (A), compared with the vehicle control (B). The response to MIP-2 (160 ng) was clearly visible (C) but reduced as compared with bFGF (A). The angiogenic response to MIP-2 was totally suppressed by including blocking antibody to murine CXCR2 in the hydron pellet (D). Pictures shown are representative of three experiments. For mDuffy-transgenic mice (E–J), a brisk angiogenic response was observed in response to bFGF (90 ng; E), compared with the vehicle control (F), which was equivalent to that in the nontransgenic mice (compare A and B with E and F). The response to MIP-2 (160 ng) was markedly reduced in transgenic mice (H–J), compared with nontransgenic mice (C). Antibody to murine CXCR2 eliminated the small angiogenic response to MIP-2 in nontransgenic mice (D) and in the mDuffy-transgenic mice (G). The pictures shown are from one experiment representative of three independent experiments.
No alteration in the angiogenic response to bFGF was noted in the transgenic mice (TG) as compared with nontransgenic mice (WT; Fig. 3A, WT mice; 3E, TG mice). Approximately two-thirds of the mDuffy-transgenic mice implanted with the MIP-2 hydron pellet did not produce an angiogenic response to MIP-2, and the other third showed a marked reduction in the angiogenic response to MIP-2 (Fig. 3, H–J), compared with the response in nontransgenic mice (Fig. 3C). In WT mice, the response to MIP-2 was not as vigorous as the response to bFGF in these assays (compare Fig. 3A with 3C). The antibody to mCXCR2 blocked the angiogenic response to MIP-2 in the nontransgenic mice (Fig. 3H) and in the transgenic mice (Fig. 3G). Moreover, MIP-2 hydron pellets imbedded into CXCR2−/− mice did not produce an angiogenic response (unpublished results). Altogether, these data demonstrate that the receptor for MIP-2, which mediates the angiogenic response, is CXCR2. Moreover, overexpression of mDuffy in endothelial cells of mDuffy-transgenic mice, impairs or retards the angiogenic response to MIP-2.
For some experiments, the eye was removed, fixed, embedded, sectioned, and stained for immunoreactive mDuffy and immunoreactive CXCR2. Results indicated that mDuffy and CXCR2 were expressed in the endothelial cells. Immunoreactive CXCR2 was observed in the endothelial cells of capillaries and blood vessels near the limbus, apparently involved in the angiogenic response to MIP-2 in transgenic (Fig. 4A) and WT mice (Fig. 4B), compared with control tissues stained with nonspecific IgG alone (Fig. 4C). In transgenic mice, mDuffy immunoreactivity was stronger in the endothelial cells of the choroid, in the cells of the neural retina, particularly the rods and cones, and in the cell processes comprising the outer plexiform layer and the inner plexiform layer (Fig. 4D), compared with WT mice (Fig. 4E) and nonspecific IgG antibody controls (Fig. 4F). The blood vessels of the iris stained positively for immunoreactive mDuffy in transgenic mice (Fig. 4G) and to a lesser extent in WT mice (Fig. 4H), compared with control tissues treated with a nonspecific IgG (Fig. 4I).
Histology on organ tissues
Organs from transgenic and nontransgenic littermates were fixed in paraformaldehyde, embedded in paraffin, sectioned, mounted on slides, and stained with H&E. The histology of the tissues from all the organs was within the limit of “normal”, with one exception—the liver. Analysis of the tissue slides revealed that the liver tissue of the transgenic mice, but not the nontransgenic littermates, exhibited extensive vacuolization and lipid deposition around the central vein, without evidence of leukocyte infiltration (unpublished results). Sudan Black-staining of the livers of transgenic mice revealed extensive lipid deposition in some mice, and for others, the lipid deposition was not as extensive (unpublished results). Liver from nontransgenic mice did not exhibit lipid deposition detectable by Sudan Black-staining. Other tissues appeared to be normal. The livers of male and female mice generated from both founders exhibited histological abnormalities.
Serum lipid levels
Serum levels of cholesterol and triglycerides were determined to be within the normal range for transgenic and nontransgenic animals. However, the cholesterol levels for the transgenic mice were approximately 44% elevated as compared with control (131 vs. 91), and the triglyceride levels were comparable for the two sets of mice (mean of 247 for mDuffy transgenics and mean of 230 for the nontransgenic mice). The levels of cholesterol and triglycerides were not particularly high in transgenic or nontransgenic mice, probably because of the low-fat content of the mouse chow diet. Because the cholesterol levels are not strikingly elevated in transgenic mice, we presume that the lipid deposition in the liver is not the result of systemic abnormalities in the handling of lipids in the transgenic mice but rather results from lipid production by hepatocytes that are not healthy. This is a common problem in hepatic diseases, which lead to hepatocellular necrosis.
Challenge with APAP
To determine whether the hepatocellular necrosis observed in transgenic mice was magnified with APAP challenge, 400 mg APAP/kg of body weight was injected into the peritoneum of 10 mDuffy-transgenic mice and 10 nontransgenic littermates. The mice were followed over a 24-h period for survival, at which time surviving mice were euthanized. Upon death, the mice were dissected, and organs were fixed, embedded, sectioned, and stained with H&E. APAP is metabolized by the liver, and overdose of APAP results in death as a result of hepatocellular toxicity. The toxicity of APAP has been shown to be mediated through N-acetyl-p-benzoquinone imine (NAPQI), which is produced through the action of the P450 system. Initially, this is neutralized by glutathione but exerts a direct effect on hepatic proteins when this glutathione sink is consumed. It has been proposed that an inflammatory mechanism also contributes to the hepatic damage of APAP. Increases in hepatic myeloperoxidase content and protective effects of antineutrophil sera have suggested the involvement of acute inflammatory agents [25]. Although elevations in tumor necrosis factor-α and IL-1-α implicated these cytokines as proinflammatory mediators after APAP challenge, mice nullizygous for the genes encoding these cytokines had manifestations of APAP toxicity similar to those in WT mice [25].
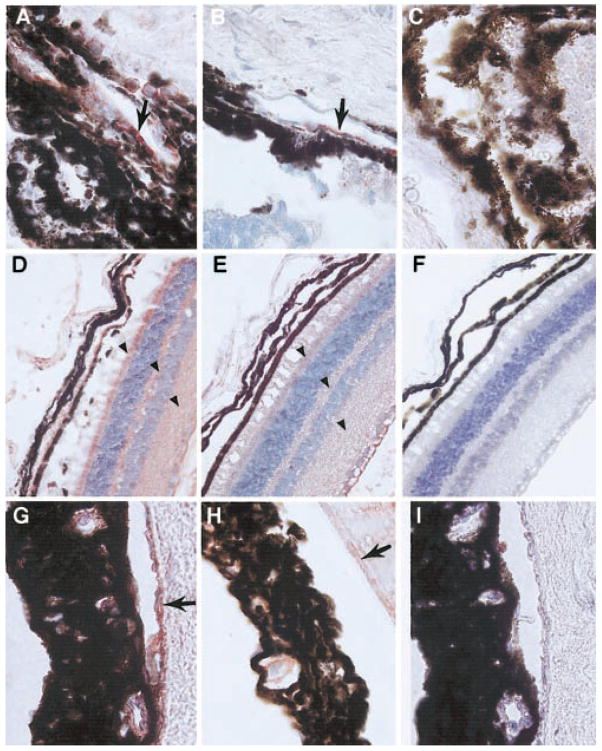
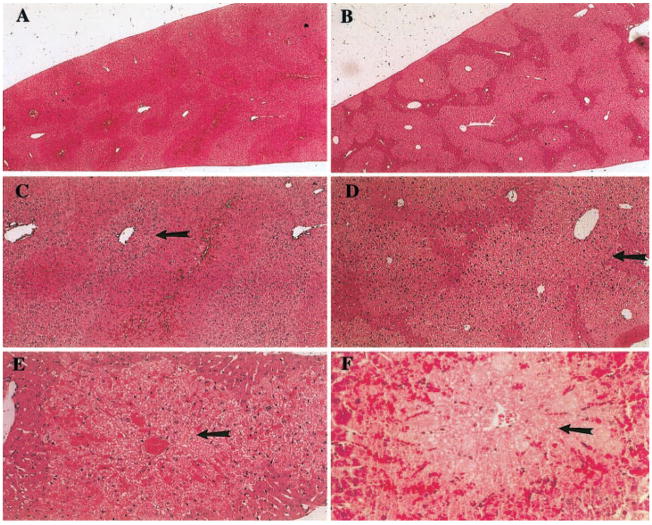
The livers of the APAP-treated mDuffy-transgenic and nontransgenic mice were visibly damaged. Histologic examination of sections of liver revealed a range of lesions with a spectrum of pathological manifestations, most prominently zonal hepatocellular necrosis accompanied by hyperemia, which extended to the development of hemorrhagic pools (Fig. 5, A–F). The necrosis, which was generally extensive, was perivenular in location and extended into the mid-zonal region. Focal regions of necrosis were typically bound by a rim of intact hepatocytes (Fig. 5, A–D). There was overlap of pathologic features between the WT and transgenic mice, and in many instances, members of these groups could not be distinguished solely on the basis of the nature of the lesion. However, one characteristic, the presence of perivenular coagulative necrosis in the absence of significant hyperemia or hemorrhage, appeared be a feature of the hepatic damage present in mice carrying the mDuffy transgene, thus helping to distinguish between the pathology of these groups (Fig. 5, E and F). Some transgenic animals had extensive necrosis that was disproportionate to the small amount of hyperemia. In the transgenic animals, the hyperemia was typically in the portal or mid-zonal segments of the hepatic lobule. In contrast, the hyperemia frequently spanned the hepatic lobule in control animals, and the appearance of distended hepatic sinusoids and the development of pools of erythrocytes were in severely affected animals. There was no evidence of inflammatory infiltrate in transgenic or nontransgenic mice treated with APAP.
Fig. 5.
APAP challenge causes greater hepatocellular damage in mDuffy-transgenic mice than in nontransgenic mice. Response to APAP challenge (400 mg/kg) of 8- to 16-week-old WT and mDuffy-transgenic mice. Eight hours after APAP challenge, livers were removed and fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with H&E. Histology revealed that hepatocellular damage was concentrated around the central vein with extensive lipid deposition and vacuolarization. This occurred in the absence of leukocyte infiltration, and the histology was compatible with the pathology observed in conjunction with necrosis. In the left-hand panel (A), observe the pattern of damage occurring in WT mice (original magnification, 5×), with lipid deposition around the central vein (C and E; original magnification, 12.5× and 25×, respectively). Red blood cells were deposited throughout the hepatic islands around the central veins where the damage occurred. In the right-hand panel (B), observe the more extensive damage that occurred in the mDuffy-transgenic mice (original magnification, 5×), the striking deposition of lipid, and the extensive vacuolarization of the hepatocytes (D and F; original magnification, 12.5× and 25×, respectively). This was also observed in the absence of leukocytic infiltration and was compatible with necrosis around the central vein. The mean necrotic index for the mDuffy transgenics was >3 and for the WT animals was <2.
To determine whether transgenic mice metabolized APAP differently than nontransgenic mice, we examined the plasma of the challenged mice at 1, 2, and 4 h post-APAP treatment. Results show that the only time point showing differences in APAP metabolism was the 1-h time point, where we observed that the nontransgenic mice produced more of the A-sulfate metabolite (220±5 nmol/ml vs. 130±28 nmol/ml) and retained a higher level of the nonmetabolized APAP (1100±50 nmol/ml vs. 694±137 nmol/ml) than did the Duffy-transgenic mice (Table 1). However, by 2 and 4 h, there were no significant differences between transgenic and nontransgenic mice in the plasma levels of any of the APAP metabolites. The APAP-reduced glutathione (GSH) conjugate is used as an indirect index of NAPQI formation (A-GSH in Table 1). At the 1-h time point, but not the other time points, we observed a significantly higher level of the A-GSH metabolite in the transgenic mice as compared with nontransgenic mice (20.26±8.17 vs. 2.15±2.47). Thus, we can assume that the formation of NAPQI is elevated in transgenic mice at the 1-h time point but not at the 2- and 4-h time points. Thus, we cannot rule out the possibility that some early variability with regard to ability to metabolize APAP could have minor effects on the overall degree of hepatic damage suffered after APAP challenge.
TABLE 1.
APAP Metabolites in Plasma of Transgenic and Nontransgenic Mice
| Treatment | A-Sulfate nmol/ml | A-Glu nmol/ml | APAP nmol/ml | A-Cysteine nmol/ml | A-Mercap nmol/ml | A-GSH nmol/ml |
|---|---|---|---|---|---|---|
| mDuffy (−) 1 h | 220.55 ± 5.46a | 667.54 ± 74.29 | 1100 ± 50.15a | 35.66 ± 12.05 | 35.62 ± 16.04 | 2.15 ± 2.47a |
| mDuffy (+) 1 h | 130.17 ± 28.40 | 546.96 ± 77.97 | 694.49 ± 137.32 | 52.69 ± 23.44 | 14.66 ± 6.16 | 20.26 ± 8.17 |
| mDuffy (−) 2 h | 83.49 ± 13.44 | 327.73 ± 138.40 | 276.78 ± 144.24 | 64.90 ± 15.41 | 33.27 ± 7.44 | 25.45 ± 13.53 |
| mDuffy (+) 2 h | 107.75 ± 62.17 | 487.26 ± 412.34 | 373.99 ± 200.93 | 51.34 ± 29.12 | 25.02 ± 13.09 | 27.69 ± 7.43 |
| mDuffy (−) 4 h | 79.34 ± 44.12 | 234.92 ± 215.87 | 117.06 ± 121.78 | 39.55 ± 29.33 | 27.28 ± 17.81 | 28.19 ± 34.23 |
| mDuffy (+) 4 h | 33.19 ± 21.69 | 71.69 ± 82.14 | 30.62 ± 12.30 | 9.02 ± 5.66 | 12.18 ± 6.06 | 0.91 ± 1.64 |
APAP (400 mg/kg) was administered i.p., and blood samples were taken at 1, 2, and 4 h after APAP administration. Values represent mean ± SD (n = 2–4).
Significantly different from mDuffy (+) at 1 h, as determined by two-tailed t-test for independent samples (P < 0.05).
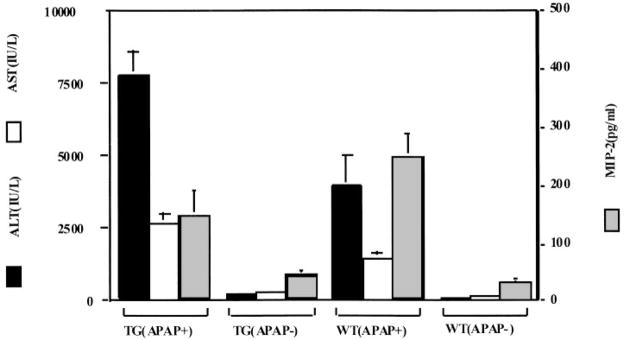
A more quantitative measure of hepatocellular damage after APAP challenge is obtained by monitoring the rise in ALT and AST. Analysis of ALT and AST in transgenic and nontransgenic mice 8 h after treatment with APAP (Fig. 6) revealed a higher elevation of ALT and AST in transgenic mice [7750±851 IU/L ALT and 2697±314 IU/L AST for mDuffy-transgenic mice, compared with 3965±1068 IU/L ALT and 1480±217 IU/L AST for nontransgenic mice (P<.02 for ALT; P<.01 for AST; two-sample t-test)]. Significant differences in ALT and AST levels in the untreated transgenic versus nontransgenic mice were not observed [237±29 IU/L ALT and 195±35 IU/L AST in transgenic mice vs. 222±49 IU/L ALT and 211 IU/L AST in WT mice (P>0.6 for ALT and AST; two-sample t-test)] (Fig. 6). These data suggest that the enhanced liver damage occurring in the mDuffy-transgenic mice occurred in coincidence with increases in the ALT and AST levels. This was true for progeny from both founders.
Fig. 6.
APAP alters the levels of ALT and AST in the serum of transgenic mice more than nontransgenic mice. Treatment of mDuffy-transgenic mice and WT mice with APAP for 8 h induced an increase in the level of ALT and AST, two liver enzymes that indicate hepatocellular damage. The induction of ALT and AST was greater in the transgenic mice than in the nontransgenic mice. Prior to APAP challenge, the levels of these enzymes were quite low in mDuffy-transgenic and nontransgenic mice. Serum MIP-2 levels were not induced to the same level in the mDuffy-transgenic mice as in the nontransgenic mice, suggesting that expression of the mDuffy transgene may have sequestered a significant amount of the MIP-2.
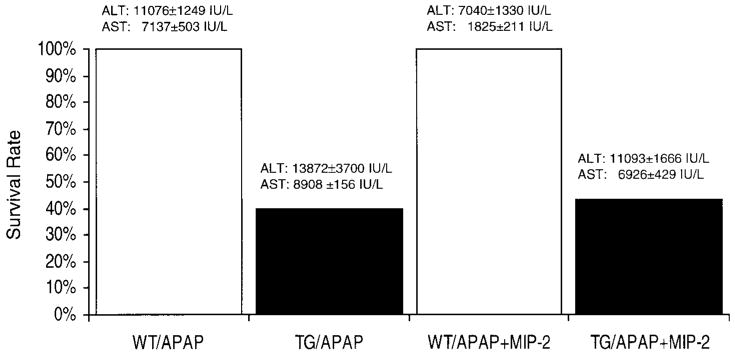
The serum MIP-2 levels increased significantly in the APAP-challenged WT mice by 8-h and 24-h posttreatment (Fig. 6 and Table 2, respectively). An increase in the level of two other chemokines that bind to mDuffy, MCP-1(JE), and KC was also observed (Table 2). APAP-challenged transgenic mice had mean serum MIP-2 levels that were not significantly different from that observed in nontransgenic APAP-challenged mice 8 h post-APAP (Fig. 6; P<.05; two tailed t-test). At the 24-h time point, transgenic mice that survived the challenge with APAP did not exhibit significant differences in serum MCP-1(JE), MIP-2, or KC compared with APAP-challenged nontransgenic littermates, and liver tissue levels of KC and MIP-2, but not JE, were reduced in transgenic mice (Table 2). Fifty percent of the mDuffy-transgenic mice died within 24 h after APAP challenge, and all the WT mice survived (Fig. 7). To test the hypothesis that reduced survival of mDuffy-transgenic mice was directly related to the early reduction in MIP-2, an attempt was made to rescue the mDuffy-transgenic phenotype upon APAP challenge with i.p. injection of 10 μg MIP-2 at the time of APAP challenge. This MIP-2 injection was accompanied by a decline in the ALT level in the APAP-challenged nontransgenic mice, which was not significant at the P < 0.05 level in the two-tailed test. In contrast, differences in AST levels in MIP-2-treated transgenic versus nontransgenic mice were significantly reduced at the P < 0.01 level based on the two-tailed t-test. Similar results were observed when intravenous (i.v.) MIP-2 (10 μg) injection was done (unpublished results). There was also some improvement in the histology of the liver tissue (unpublished results). However, there was no significant enhancement of survival in the MIP-2-treated APAP-challenged transgenic mice. Attempts to use higher concentrations of MIP-2 were aborted because of induction of harmful side effects. These data suggest that the mechanism for enhanced hepatocellular damage in mDARC transgenic mice in response to APAP challenge may be more complex than simple sequestration of MIP-2. Alternatively, the improvement in response to MIP-2 after APAP challenge may not be sufficient to reverse the extensive damage induced by APAP overdose.
TABLE 2.
Effect of APAP Challenge on ALT and AST Serum Levels, Chemokine Levels in Serum and Liver in Transgenic and Nontransgenic Mice with or without MIP-2 Treatment
| Serum enzyme levels (IU/L) |
Chemokine levels in serum (pg/ml) |
Chemokine levels in liver (pg/mg) |
||||||
|---|---|---|---|---|---|---|---|---|
| Treatment | ALT (IU/L) | AST (IU/L) | MIP-2 (pg/ml) | KC (pg/ml) | JE (pg/ml) | MIP-2 (pg/mg) | KC (pg/mg) | JE (pg/mg) |
| WT | 361 ± 32 | 72 ± 2 | 17.7 ± 4.7 | 134 ± 32 | 92 ± 28 | 1.3 ± 0.1 | 3.6 ± 1.1 | 0.1 ± 0.2 |
| TG | 298 ± 44 | 83 ± 8 | 17.7 ± 3.4 | 257 ± 61 | 148 ± 75 | 3.9 ± 1.4 | 6.3 ± 1.3 | 1.7 ± 1.6 |
| WT + APAP* | 11,076 ± 1249 | 7137 ± 503 | 296 ± 235 | 651 ± 67 | 605 ± 415 | 201 ± 124 | 103 ± 74 | 25 ± 16 |
| TG + APAP* | 13,872 ± 3700 | 8908 ± 156 | 144 ± 117 | 820 ± 159 | 708 ± 308 | 125 ± 138 | 43 ± 31 | 29 ± 1 |
| WT + MIP-2 + APAP* | 7040 ± 1330 | 1825 ± 211 | 223 ± 50 | 424 ± 94 | 343 ± 157 | 86 ± 47 | 16 ± 3 | 20 ± 11 |
| TG + MIP-2 + APAP* | 11,093 ± 1666 | 6926 ± 429 | 205 ± 79 | 481 ± 197 | 590 ± 212 | 17 ± 6 | 20 ± 5 | 25 ± 6 |
Values for ALT, AST, MIP-2, KC, and JE in APAP-challenged mice were obtained 24 h after APAP challenge.
Fig. 7.
Effect of APAP challenge with or without MIP-2 injection on survival, ALT, and AST serum levels in transgenic and nontransgenic mice. Transgenic and nontransgenic mice were treated with APAP overdose (400 μ/kg) with or without i.p. injection with 10 μg MIP-2; 24 h later, the surviving mice were euthanized, and serum levels of ALT and AST were determined as described in Materials and Methods. Three to six mice from each group were studied.
DISCUSSION
It has been shown previously in humans that Duffy expression is not limited to erythrocytes but is also expressed on endothelial cells and certain neuronal cells [26]. Based on the endothelial cell expression of mDuffy, it has been postulated that this chemokine receptor might play some positive role in regulating angiogenesis or development of the vascular system. However, we found no evidence for this in our transgenic model. Our data show that overexpression of mDuffy resulted in reduced angiogenic responses to MIP-2. Moreover, because CXCR2−/− mice did not exhibit an angiogenic response to MIP-2, and antibody to murine CXCR2 completely suppressed the angiogenic response to MIP-2, it appears that CXCR2 is the receptor that mediates positive angiogenic responses to MIP-2. Expression of mDuffy may provide a negative throttle for this response by sequestering MIP-2. Middleton et al. [27] have postulated that the combined interaction of Duffy and heparan sulfate molecules on the surface of endothelial cells is involved in mediating the interaction of chemokines with endothelial cells. Our data suggest that this mediation could also serve to repress response to chemokines.
The unchallenged PPEP-mDuffy-transgenic mice were phenotypically normal except for baseline differences in liver histopathology, but no other significant histological abnormalities were observed. These transgenic mice had hepatic lipid deposition that ranged from mild to extensive fatty changes that spanned the lobule. Small foci of necrosis were observed but were not a prominent feature of the changes, and there was no evidence of cholestasis and fibrosis. At this time, we do not know the mechanism for this baseline liver abnormality. Hepatic toxicity was observed in WT and transgenic animals following treatment with APAP, consistent with the dose administered. Histopathologic examination revealed characteristic changes, which included hemorrhagic necrosis having a perivenular distribution. These hemorrhagic lesions showed a spectrum of evolution in most animals, ranging from lobules affected by hyperemia to those containing geographic areas of necrosis and hemorrhagic lakes. The zonal region of necrosis was striking in the transgenic mice and lacked evidence of hemorrhage in several of these animals, although this was a constant feature in the WT mice. The lack of a significant inflammatory infiltrate in the treated mice is consistent with the direct action on hepatocytes of the P450-generated APAP biometabolite. Although histologic examination suggested that necrosis appeared to be more prominent in the livers from the transgenic mice, this approach is semiquantitative at best, and the relative levels of hepatic damage can best be judged from the elevations in ALT and AST in serum.
APAP challenge also resulted in enhanced detection of the mDuffy protein in blood vessels of APAP-challenged transgenic mice in the experiments described here (Fig. 2, C–F), which could possibly occur through APAP induction of the PPEP [25, 28 –33]. However, in this model, we propose that it is likely that APAP challenge causes hepatocellular damage, and as a result, there are increases in ALT and AST enzyme levels in the serum. Because the enzymatic markers for hepatotoxicity, ALT and AST, are not elevated in the transgenic mice as compared with nontransgenic littermates prior to APAP challenge but are elevated after APAP challenge, this increase in ALT and AST in transgenic mice after APAP challenge is probably related to the constant and increased expression of mDARC in the transgenic mice. In summary, expression of the PPEP-mDuffy transgene appears to make the mice more susceptible to APAP-mediated hepatotoxicity.
We observed differences in the metabolism of APAP between transgenic and nontransgenic mice during the first hour after APAP challenge. MDARC-transgenic mice exhibited reduced appearance of the A-sulfate metabolite but increased concentrations of the A-GSH metabolite in the plasma at 1 h post-APAP challenge. However, by 2 h post-APAP challenge, there were no significant differences in the levels of the APAP metabolites in the plasma of transgenic and nontransgenic mice. When all of these data on the metabolite levels are considered in total, we cannot rule out the possibilty that early differences in APAP metabolism can contribute to the phenotypic differences in APAP-challenged transgenic versus nontransgenic mice.
In our study, APAP challenge increased the serum levels of MIP-2 as early as 8 h after challenge, and MIP-2, KC, and JE were induced by 24 h after challenge in transgenic and nontransgenic mice. Normally, the expression of mDuffy on red blood cells is thought to sequester excess chemokine [34], however the additional expression of mDuffy in the endothelium may trap a significant amount of MIP-2. It is not clear at this time why MIP-2 and KC, but not JE, are reduced in the liver of transgenic mice, because mDuffy binds all three of these chemokines. One could postulate that in transgenic mice, the early reduction in MIP-2 (8 h) results in a loss of the protective effect of MIP-2 during APAP treatment, ultimately resulting in an increase in the level of necrosis in the livers of transgenic mice as compared with nontransgenic littermates. Indeed, the histology of the APAP-treated livers from transgenic and WT mice showed a different pattern of necrotic damage (unpublished results), and more important, 50% of the mDuffy-transgenic mice died within 24 h after APAP challenge, and WT mice survived.
Administration of MIP-2 by i.v. injection of MIP-2 or by injection of adenovirus expressing MIP-2 has been shown to rescue APAP-induced hepatocellular toxicity in CD1 mice [35]. The mechanism for this protection was shown to be through induction of hepatocyte proliferation [35] or reduction in the APAP-induced decline in hepatocyte proliferation, and this could occur even when MIP-2 was administered as late as 10 h after APAP [36]. Earlier studies have demonstrated that MIP-2 will facilitate liver regeneration by inducing hepatocyte proliferation [37]. We also observed that in vitro, MIP-2 treatment slightly enhanced the MTT metabolism of APAP-challenged hepatocytes (unpublished results). We observe that ALT and AST levels are higher in mDuffy-transgenic mice than in nontransgenic mice 8 h post-APAP challenge. By 24 h posttreatment, these differences are much less apparent in the population of mice that survived APAP challenge, although the AST and ALT levels in the transgenic mice that died would be predictably much higher. We observed that MIP-2 injection lowered AST levels in APAP-challenged C57Bl-transgenic mice, but this did not enhance survival of the transgenic mice at the 24-h time point. There appear to be strain differences between the C57Bl and CD1 mice in the level of induction of MIP-2 and KC in response to APAP challenge [35]. Because administration of MIP-2 at the time of APAP challenge did not rescue the enhanced sensitivity of the mDuffy-transgenic mice to APAP challenge, our data suggest that overexpression of mDuffy can have effects on homeostasis that go beyond simply serving as a sink for chemokine ligand. Indeed, 50% of the the mDuffy-transgenic mice were dead within 10 h after APAP challenge, with or without administration of MIP-2, suggesting that Duffy is involved as a negative modulator of homeostasis. However, the mDuffy knockout mice had a normal appearance [38, 39], and although one study did not find any phenotypic abnormalities in the face of many environmental challenges [38], the other study showed that challenge with lipopolysaccharide resulted in a significant increase in inflammatory infiltrates in lung and liver in the mice null for Duffy [39]. The latter study suggested that by serving as a sink for chemokines, Duffy may modulate the intensity of the inflammatory response. Those results are complimentary to our own, where we observe that overexpression of Duffy in the endothelium results in enhanced toxicity in response to APAP challenge and reduced angiogenic response to MIP-2 in the corneal micropocket assay.
Acknowledgments
This work was funded by grants from the Department of Veterans Affairs—a Merit Award and a Career Scientist Award to A. R., by a grant from the TJ Martell Foundation and National Institutes of Health (DK38517) to T. O. D., and by NIH CA87879 and HL66027 grants to R. M. S. We are indebted to Jeanette Norden for help in the analysis of the immunoreactivity for mDuffy in the mouse brain, to Amy Pruitt for help in editing this manuscript, to Kay Washington for a second opinion on the liver histology/pathology, to Sergio Fascio for the serum lipid analysis, to Tong Tang for technical assistance with the design and construction of the PPEP-mDuffy-expression construct, and to Tom Quartemous for providing the PPEP DNA. The endotoxin-free, purified rMIP-2 was the kind gift of Elias Lolis. The transgenic mice were developed in the Vanderbilt-Ingram Cancer Center Transgenic Core (CA68485). The histology processing was performed by the Morphology Core of the NIH-funded Skin Disease Research Center at Vanderbilt University School of Medicine (AR41943).
References
- 1.Chaudhuri A, Polyakova J, Zbrzezna V, Williams K, Gulati S, Pogo AO. Cloning of glycoprotein D cDNA, which encodes the major subunit of the Duffy blood group system and the receptor for the Plasmodium vivax malaria parasite. Proc Natl Acad Sci USA. 1993;90:10793–10797. doi: 10.1073/pnas.90.22.10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LH, Good MF, Milon G. Malaria pathogenesis. Science. 1994;264:1878 –1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 3.Miller LJ, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 4.Hadley TJ, Lu ZH, Wasniowska K, Martin AW, Peiper SC, Hesselgesser J, Horuk R. Postcapillary venule endothelial cells in kidney express a multispecific chemokine receptor that is structurally and functionally identical to the erythroid isoform, which is the Duffy blood group antigen. J Clin Investig. 1994;94:985–991. doi: 10.1172/JCI117465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiper SC, Wang Z, Neote K, Martin AW, Showell HJ, Conklyn MJ, Ogborne K, Hadley TJ, Lu ZH, Hesselgesser J, et al. The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. J Exp Med. 1995;181:1311–1317. doi: 10.1084/jem.181.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri A, Zbrzezna V, Polyakova J, Pogo AO, Hesselgesser J, Horuk R. Expression of the Duffy antigen in K562 cells. J Biol Chem. 1994;269:7835–7838. [PubMed] [Google Scholar]
- 7.Chaudhuri A, Nielsen S, Elkjaer ML, Zbrzezna V, Fang F, Pogo AO. Detection of Duffy antigen in the plasma membranes and caveolae of vascular endothelial and epithelial cells of non-erythroid organs. Blood. 1997;89:701–712. [PubMed] [Google Scholar]
- 8.Hesselgesser J, Horuk R. Chemokine and chemokine receptor expression in the central nervous system. J Neurovirol. 1999;1:13–26. doi: 10.3109/13550289909029741. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen S, Chaudhuri A, Pogo AO. Which are the nonerythroid cells that constitutively express the duffy antigen? Blood. 1997;90:3231–3232. [PubMed] [Google Scholar]
- 10.Neote K, Mak JY, Kolakowski LF, Jr, Schall C. Functional and biochemical analysis of the cloned Duffy antigen: identity with the red blood cell chemokine receptor. Blood. 1994;84:44 –52. [PubMed] [Google Scholar]
- 11.Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 12.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. XXII Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 13.Szabo MC, Loo KS, Zlotnik A, Schall TJ. Chemokine class differences in binding to the Duffy antigen-erythrocyte chemokine receptor. J Biol Chem. 1995;270:25348 –25351. doi: 10.1074/jbc.270.43.25348. [DOI] [PubMed] [Google Scholar]
- 14.Luo H, Chaudhuri A, Johnson KR, Neote K, Zbrzezna V, He Y, Pogo AO. Cloning, characterization, and mapping of a murine promiscuous chemokine receptor gene: homolog of the human Duffy gene. Genome Res. 1997;7:932–941. doi: 10.1101/gr.7.9.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang T, Owen JD, Du J, Walker CL, Richmond A. Molecular cloning and characterization of a mouse gene with homology to the Duffy-antigen receptor for chemokines. DNA Seq. 1998;9:129 –143. doi: 10.3109/10425179809072188. [DOI] [PubMed] [Google Scholar]
- 16.Bu X, Quertermous V. Identification of an endothelial cell-specific regulatory region in the murine endothelin-1 gene. J Biol Chem. 1997;272:32613–32622. doi: 10.1074/jbc.272.51.32613. [DOI] [PubMed] [Google Scholar]
- 17.Chang LW. A Color Atlas and Manual for Applied Histochemistry. Springfield, IL: Thomas; 1979. pp. 34–39. [Google Scholar]
- 18.Kenyon BM, Voest EE, Chen CC, Flynn E, Folkman J, D’Amato RJ. Model of angiogenesis in the mouse cornea. Investig Ophthalmol Vis Sci. 1996;37:1625–1632. [PubMed] [Google Scholar]
- 19.Manautou JE, Tvit A, Hoivik DJ, Khairallah EA, Cohen SD. Protection by clofibrate against acetaminophen hepatotoxicity in male CD-1 mice is associated with an early increase in biliary concentration of acetaminophen-glutathione adducts. Toxicol Appl Pharmacol. 1996;140:30 –38. doi: 10.1006/taap.1996.0194. [DOI] [PubMed] [Google Scholar]
- 20.Linton MF, Hasty AH, Babaev VR, Fazio S. Hepatic apo E expression is required for remnant lipoprotein clearance in the absence of the low density lipoprotein receptor. J Clin Investig. 1999;101:1726 –1736. doi: 10.1172/JCI2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harats D, Kurihara H, Belloni P, Oakley H, Ziober A, Ackley D, Cain G, Kurihara Y, Lawn R, Sigal E. Targeting gene expression to the vascular wall in transgenic mice using the murine preproendothelin-1 promoter. J Clin Investig. 1996;95:1335–1344. doi: 10.1172/JCI117784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giovannelli A, Limatola C, Ragozzino D, Mileo AM, Ruggieri A, Ciotti MT, Mercanti D, Santoni A, Eusebi F. CXC chemokines interleukin-8 (IL-8) and growth-related gene product alpha (GRO alpha) modulate Purkinje neuron activity in mouse cerebellum. J Neuroimmunol. 1998;92:122–132. doi: 10.1016/s0165-5728(98)00192-1. [DOI] [PubMed] [Google Scholar]
- 23.Ragozzino D, Giovannelli A, Mileo AN, Limatola C, Santoni A, Eusebi F. Modulation of the neurotransmitter release in rat cerebellar neurons by GRO beta. Neuroreport. 1998;9:3601–3606. doi: 10.1097/00001756-199811160-00011. [DOI] [PubMed] [Google Scholar]
- 24.Andjelkovic AV, Spencer DD, Pachter JS. Visualization of chemokine binding sites on human brain microvessels. J Cell Biol. 1999;145:403– 412. doi: 10.1083/jcb.145.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blazka ME, Wilmer JL, Holladay SD, Wilson RE, Luster MI. Role of proinflammatory cytokines in acetaminophen hepatotoxicity. Toxicol Appl Pharmacol. 1995;133:43–52. doi: 10.1006/taap.1995.1125. [DOI] [PubMed] [Google Scholar]
- 26.Horuk R, Martin AW, Wang Z, Schweitzer L, Gerassimides A, Guo H, Lu Z, Hesselgesser J, Perez HD, Kim J, Parker J, Hadley TJ, Peiper SC. Expression of chemokine receptors by subsets of neurons in the central nervous system. J Immunol. 1997;158:2882–2890. [PubMed] [Google Scholar]
- 27.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, Rot A. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 1997;91:385–395. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 28.Lee ME, Temizer DH, Clifford JA, Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J Biol Chem. 1991;266:16188 –16192. [PubMed] [Google Scholar]
- 29.Lee ME, Bloch KD, Clifford JA, Quertermous T. Functional analysis of the endothelin-1 gene promoter. Evidence for an endothelial cell-specific cis-acting sequence. J Biol Chem. 1990;265:10446 –10500. [PubMed] [Google Scholar]
- 30.Blazka ME, Bruccoleri A, Simeonova PP, Germolec DR, Pennypacker KR, Luster V. Acetaminophen-induced hepatotoxicity is associated with early changes in AP-1 DNA binding activity. Res Commun Mol Pathol Pharmacol. 1996;92:259 –273. [PubMed] [Google Scholar]
- 31.Hu J, Discher DJ, Bishopric NH, Webster KA. Hypoxia regulates expression of the endothelin-1 gene through a proximal hypoxia-inducible factor-1 binding site on the antisense strand. Biochem Biophys Res Commun. 1998;245:894 – 899. doi: 10.1006/bbrc.1998.8543. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Chen H, Wang D. Distribution and significance of endothelin-1 and endothelin receptor A mRNA in liver after endotoxemia. Zhonghua Yixue Zazhi. 1997;77:171–174. [PubMed] [Google Scholar]
- 33.Marsden PA, Brenner BM. Transcriptional regulation of the endothelin-1 gene by TNF-alpha. Am J Physiol. 1992;262:C854 –C861. doi: 10.1152/ajpcell.1992.262.4.C854. [DOI] [PubMed] [Google Scholar]
- 34.Horuk R, Wang ZX, Peiper SC, Hesselgesser J. Identification and characterization of a promiscuous chemokine-binding protein in a human erythroleukemic cell line. J Biol Chem. 1994;269:17730 –17733. [PubMed] [Google Scholar]
- 35.Hogaboam CM, Simpson KJ, Chensue SW, Steinhauser ML, Lukacs NW, Gauldie J, Strieter RM, Kunkel SL. Macrophage inflammatory protein-2 gene therapy attenuates adenovirus- and acetaminophen-mediated hepatic injury. Gene Ther. 1999;6:573–584. doi: 10.1038/sj.gt.3300858. [DOI] [PubMed] [Google Scholar]
- 36.Hogaboam CM, Bone-Larson CL, Steinhauser ML, Lukacs NW, Colletti LM, Simpson KJ, Strieter RM, Kunkel V. Novel CXCR-dependent liver regenerative qualities of ELR-containing CXC chemokines. FASEB J. 1999;13:1565–1574. doi: 10.1096/fasebj.13.12.1565. [DOI] [PubMed] [Google Scholar]
- 37.Colletti LM, Green M, Burdick MD, Kunkel SL, Strieter RM. Proliferative effects of CXC chemokines in rat hepatocytes in vitro and in vivo. Shock. 1998;10:248 –257. doi: 10.1097/00024382-199810000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Luo H, Chaudhuri A, Zbrezezna B, He Y, Pogo AO. Deletion of the murine Duffy gene (Dfy) reveals that the Duffy receptor is functionally redundant. Mol Cell Biol. 2000;20:3097–3101. doi: 10.1128/mcb.20.9.3097-3101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawson TC, Lentsch AB, Wang Z, Cowhig JE, Rot A, Maeda N, Peiper SC. Exaggerated response to endotoxin in mice lacking the Duffy antigen/receptor for chemokines (DARC) Blood. 2000;96:1681–1684. [PubMed] [Google Scholar]