Abstract
The continuous production of the CXC ligand 1 (CXCL1) chemokine by melanoma cells is a major effector of tumor growth. We have previously shown that the constitutive expression of this chemokine is dependent upon transcription factors nuclear factor-kappa B (NF-κB), stimulating protein-1 (SP1), high-mobility group-I/Y (HMGI/Y), CAAT displacement protein (CDP) and poly(ADPribose) polymerase-1 (PARP-1). In this study, we demonstrate for the first time the mechanism of transcriptional regulation of CXCL1 through PARP-1 in melanoma cells. In its inactive state, PARP-1 binds to the CXCL1 promoter in a sequence-specific manner and prevents binding of NF-κB (p65/p50) to its element. However, activation of the PARP-1 enzymatic activity enhances CXCL1 expression, owing to the loss of PARP- 1 binding to the CXCL1 promoter, accompanied by enhanced binding of p65 to the promoter. The delineation of the role of NF-κB-interacting factors in the putative CXCL1 enhanceosome will provide key information in developing strategies to block constitutive expression of this and other chemokines in cancer and to develop targeted therapy.
Keywords: PARP-1, CXCL1, NF-κB, transcription, melanoma
Introduction
Melanoma is the most aggressive form of skin cancer and is highly resistant to conventional chemotherapy. A growing body of evidence suggests that melanoma tumor cells acquire the ability to attenuate apoptotic signals by activating transcription of antiapoptotic and growth-promoting factors through constitutive upregulation of nuclear factor-kappa B (NF-κB). The NF-κB transcription factor represents a central and important component in the transcription of many chemokines. Interestingly, elevated expression of angiogenic chemokines has been observed in many tumor cell types, implicating a role for chemokines in neoplasia (Olbina et al., 1996; Richards et al., 1997; Takamori et al., 2000; Li et al., 2001; Haqq et al., 2005). Constitutive expression of the angiogenic chemokine, CXC ligand 1 (CXCL1), has been shown to transform immortalized melanocytes (Balentien et al., 1991), and high levels of endogenous CXCL1 and CXCL8 have been detected in melanoma (Schadendorf et al., 1994, 1996; Owen et al., 1997; Haghnegahdar et al., 2000; Li et al., 2001; Yang and Richmond, 2001). Studies in our laboratory have shown that the level of CXCL1 chemokine is elevated in many melanoma cell lines, and that this elevation in chemokine expression may be due to deregulation in the transcription of CXCL1 (Haghnegahdar et al., 2000).
Transcription of CXCL1 requires a 306 bp minimal promoter containing the following five cis elements: TATA box (−25 to −30), NF-κB binding site (−67 to 77), AT-rich high mobility group-I/Y (HMGI/Y) binding element within the NF-κB site, an immediate upstream region (IUR) (−78 to −93) and a GC-rich stimulating protein-1 (SP1) binding site (−117 to −128) (Wood et al., 1995). Within the CXCL1 promoter, the IUR element is a 20-bp sequence that is located immediately upstream of the NF-κB site. The IUR is thought to regulate the transcription of CXCL1, both positively and negatively. It contains the sequence TCGATC, which binds the positive modulator, poly-(ADP-ribose) polymerase-1 (PARP-1) (Nirodi et al., 2001). Although PARP-1 was identified as a putative positive regulator of CXCL1 transcription, the mechanism of PARP-1 regulation of CXCL1 gene expression is yet unknown.
The mammalian PARP-1, the major isoform of the PARP family, is comprised of 1014 amino acids (114 kDa) and is continuously expressed in eucaryotes. It has a 46 kDa DNA-binding domain at the N terminus containing the DEVD sequence, which is the target of caspase-3 during apoptosis. When cleaved by caspase-3, PARP-1 is inactivated, resulting in the formation of two proteolytic fragments of PARP-1, a 29 kDa amino terminus and an 85 kDa carboxyl terminus (Alvarez- Gonzalez et al., 1999; Smulson et al., 2000). A 54 kDa domain of PARP-1 located in the carboxyl terminus represents the β-nicotinamide adenine dinucleotide (NAD+)-binding domain. Between the DNA-binding domain and the NAD+-binding domain is a 22 kDa ‘automodification domain’, which facilitates the homoand/or heterodimerization of PARP-1 with other proteins (Alvarez-Gonzalez et al., 1999).
The catalytic activity of PARP-1 is stimulated 500- fold by non-covalent contact of the DNA-binding domain with DNA strand breaks and results in the transfer of successive units of the adenosine diphosphate (ADP)-ribose moiety from NAD+ to itself and other nuclear protein acceptors such as topoisomerase I and II, histones, HMG proteins, p53 and NF-κB (Meisterernst et al., 1997; Hassa and Hottiger, 1999; Smulson et al., 2000; Burkle, 2001). Studies with PARP-1- deficient cells and animals have revealed diverse functions of PARP-1, including roles in anti-recombination and genomic instability, DNA replication, regulation of telomere function and transcriptional regulation (Simbulan-Rosenthal et al., 1999; Smulson et al., 2000; Cayuela et al., 2001). Although no consensus DNA binding sequence for PARP-1 has been established, the ability of PARP-1 to bind DNA in a sequence-specific manner was demonstrated by Huang et al. (2004), where PARP-1 preferentially bound to DNA oligonucleotides containing the 5′-TGTTG-3′ nucleotide sequence motif (Simbulan-Rosenthal et al., 2003). PARP-1 has also been shown to activate the human T-cell leukemia virus type I Tax-mediated transcription in murine lymphocytic leukemia cells through sequence-specific binding to the Tax-responsive element TTGACGACAA (Zhang et al., 2002).
In this study, the role of PARP-1 in transcription within the context of the CXCL1 promoter has been characterized. We show for the first time that PARP-1 may regulate CXCL1 gene expression both negatively and positively. The presence of enzymatically inactive PARP-1 inhibits transcriptional activation of CXCL1, whereas activation of PARP-1 enzymatic activity is an inducer of the transcription. Furthermore, our study demonstrates that PARP-1 is overexpressed in melanoma cell lines, and this overexpression correlates with higher PARP-1 activity. We propose that the constitutive activation of NF-κB converged with elevated PARP-1 activity in melanoma results in elevated CXCL1 production.
Results
PARP-1 expression and activity state in melanoma cells
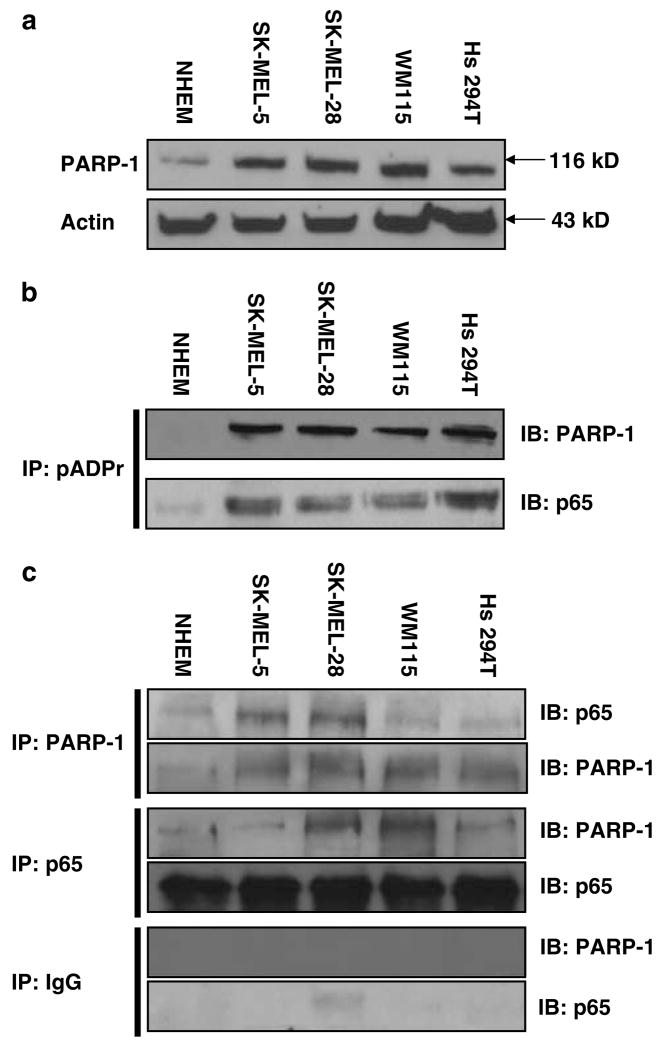
Cultured melanoma cells show significantly higher activation of NF-κB compared to normal melanocytes (Dhawan and Richmond, 2002). However, levels of expression of NF-κB target genes vary significantly, indicating that there are other factors involved in the transcriptional regulation of these genes. Having identified PARP-1 as a CXCL1 promoter-binding protein (Nirodi et al., 2001), we wanted to determine whether the level of PARP-1 expression was changed in melanoma cells compared to normal melanocytes. Western blot analysis of whole-cell extract from normal melanocyte cultures and a panel of melanoma cell lines showed an elevated level of PARP-1 expression in the melanoma cells (Figure 1a). To determine whether this elevated protein level resulted in elevated ADPribosylation of PARP-1 and/or NF-κB p65 and hence elevated PARP activity, immunoprecipitations with anti-poly(ADP-ribose) polymers were performed, and ADP-ribosylated proteins were resolved on 8% polyacrylamide gel electrophoresis (PAGE). Melanoma cells showed higher PARP activity than normal cells and both PARP-1 and p65 proteins could be detected in poly(ADP-ribose) polymer-immunoprecipitated samples, indicating that p65 is a potential substrate for PARP-1 in melanoma cells (Figure 1b). To determine whether the elevated ADP-ribosylation state in melanoma cells results in altered interaction between the two proteins, immunoprecipitation assays were performed (Figure 1c). Immunoprecipitation with either antibody showed no significant difference in the ratio of co-association of p65 and PARP-1 between the normal and the melanoma cell lines, indicating that alteration in ratio of the physical interaction between PARP-1 and p65 is not responsible for increased CXCL1 production and the consequent melanocyte transformation.
Figure 1.
Expression and activity of PARP-1 protein in melanoma cell lines is elevated. (a) Western blot analysis with PARP-1 and actin antibodies in NHEM and melanoma cells. (b) Whole cellular extracts (300 μg) immunoprecipitated with the pADPr antibody and subjected to immunoblot analysis with PARP-1 and p65 antibodies. IP: immunoprecipitation; IB: immunoblotting; IgG: immunoglobulin G. (c) Immunoprecipitation with the PARP-1 or p65 antibodies and immunoblot analysis with p65 and PARP-1 antibodies, respectively. Immunoprecipitation with IgG was performed as negative control. Control cells were NHEM and melanoma cell panels include: SK-MEL-5, SK-MEL-28, WM 115 and Hs 294T. This figure is a representative of three separate experiments.
PARP-1 binds specifically to CXCL1 promoter
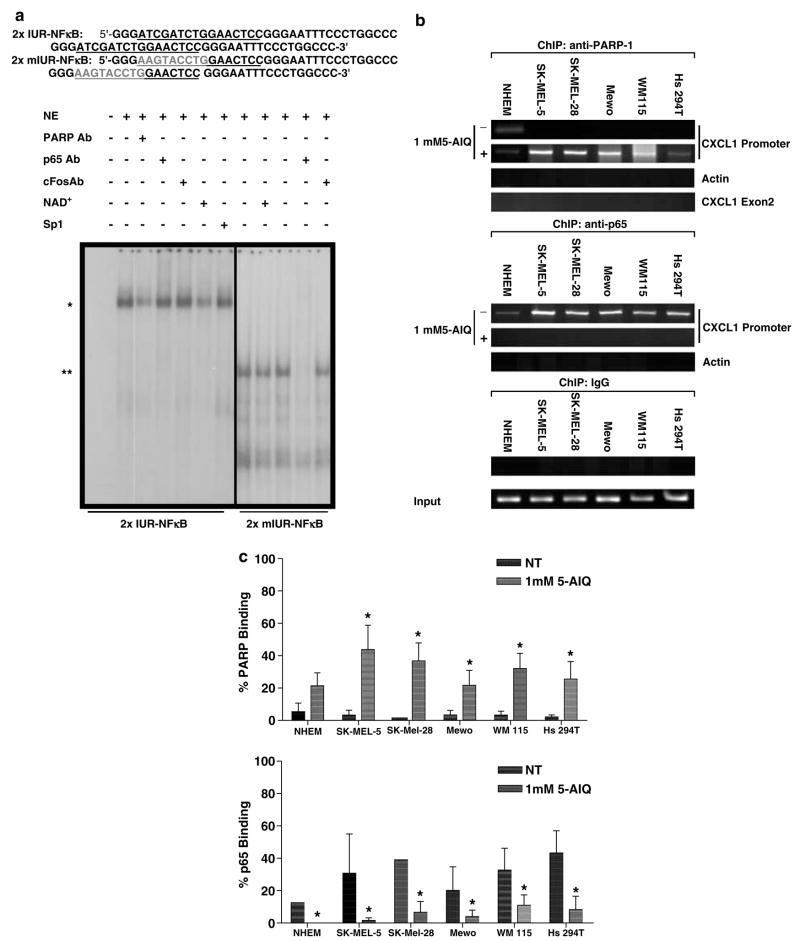
In order to confirm the interaction of PARP-1 with the CXCL1 promoter and determine whether this interaction was altered in melanoma cells, two approaches were undertaken. First, we analysed nuclear protein extracts from melanoma cells by electrophoretic mobility shift assay (EMSA) using DNA probes containing either wild-type (WT) 2 × IUR–NF-κB or mutated IUR sequences linked to NF-κB sequence. Incubation of Hs 294T nuclear cell extracts with WT 2 × IUR–NF-κB probe showed binding of a high molecular weight complex, and anti-PARP-1 antibody addition resulted in partial elimination of the shifted band, indicating that the complex binding to the IUR sequence contains PARP-1 (Figure 2a). Interestingly, enhancement of the automodification of PARP-1 by the addition of NAD+, its substrate, reduced the DNA binding of PARP-1 complex to the WT 2 × IUR–NF-κB probe. When the same extract was incubated with the mutated IUR sequence, the high molecular weight complex was no longer detected. Instead, a low molecular weight band appeared that was identified as NF-κB complex, as incubation with p65 antibody resulted in a band shift. Antibody against PARP-1 did not affect the intensity of this binding, demonstrating that mutation of the IUR eliminates PARP-1 DNA binding. The data indicate that PARP-1 binds to the CXCL1 promoter in a sequence-specific manner, and mutation in the IUR results in stronger p65 binding to the NF-κB element in the CXCL1 promoter. Addition of c-fos antibody and SP1 probe served as negative controls.
Figure 2.
PARP-1 binds specifically to the CXCL1 promoter. (a) Nuclear proteins (10 μg) were subjected to EMSA with the 2 × WTIUR–NFκB and 2 × mIUR–NFκB probes. (*) EMSA band that was partially eliminated by coincubation of PARP-1 antibody or NAD+; (**) EMSA band that eliminated by coincubation with the p65 antibody; Sp1: 50-fold excess unlabeled Sp1 consensus oligonucleotide; c-fos antibody served as negative control for super-shift assays. (b) The chromosomal DNA and nuclear proteins were immunoprecipitated with the PARP-1 or p65 antibodies. Immunoprecipitation with IgG was performed as negative control. Purified ChIP DNA was amplified with CXCL1 promoter-specific primers by PCR. Input and IgG ChIP DNA were amplified with CXCL1 primers. As control, the ChIP DNA was amplified by PCR with actin primers. (c) Densitometric quantitation of the gels using the Flourchem 8900 Imaging System (Alpha Innotech, San Leandro, CA, USA). Values from four independent experiments for the ChIP DNA were normalized against those for input DNA. The mean normalized values are shown ± s.e.m. The asterisks indicate P < 0.05.
EMSA demonstrated that mutation of the IUR element eliminated the PARP-1 association with CXCL1 promoter. However, since in EMSA assays where binding reactions are forced under extreme conditions, that is, high concentration of DNA probe, the question was then: ‘Is there a difference in the nature of PARP-1 interaction with the CXCL1 promoter in normal melanocytes vs melanoma cells at intracellular conditions?’ To investigate this matter, chromatin immunoprecipitation (ChIP) assay was used to compare NF-κB and PARP-1 binding to chromatin in normal melanocyte cultures and the panel of melanoma cell lines. Qualitative and quantitative analyses of these experiments are shown in Figure 2b and 2c. In these assays, the proteins binding to chromatin were crosslinked to the chromatin and the DNA was sheared to smaller fragments averaging 600 bp. Then, the chromatin was immunoprecipitated with either p65 or PARP-1 antibodies. The protein–DNA crosslinking was then reversed, and after DNA purification, the DNA was used to amplify the CXCL1 promoter sequence by polymerase chain reaction (PCR). In non-treated cells, PCR amplification revealed a band for CXCL1 promoter in p65-immunoprecipitated samples, but not in PARP-1-immunoprecipitated samples. However, when the cells were treated with the PARP inhibitor, 5-aminoisoquinolinone. HCl (5-AIQ), at 1mM concentration, the p65-immunoprecipitated samples resulted in much reduced amplification of the CXCL1 promoter and the PARP-1 immunoprecipitated samples showed enhanced amplification. As control, the ChIP DNA was amplified by PCR with CXCL1 exon 2 and actin primers. The negative control samples immunoprecipitated with rabbit immunoglobulin G (IgG) showed no amplification with the specific CXCL1 promoter primers. The input DNA represents the total DNA of the cells and is used as control for the PCR reactions. The results from the ChIP assays indicate that silencing of PARP-1 activity results in increased interaction of PARP-1 with the CXCL1 promoter at a cost of less p65 binding to the CXCL1 promoter (Figure 2c).
Ablation of PARP-1 expression leads to an increase in CXCL1 level
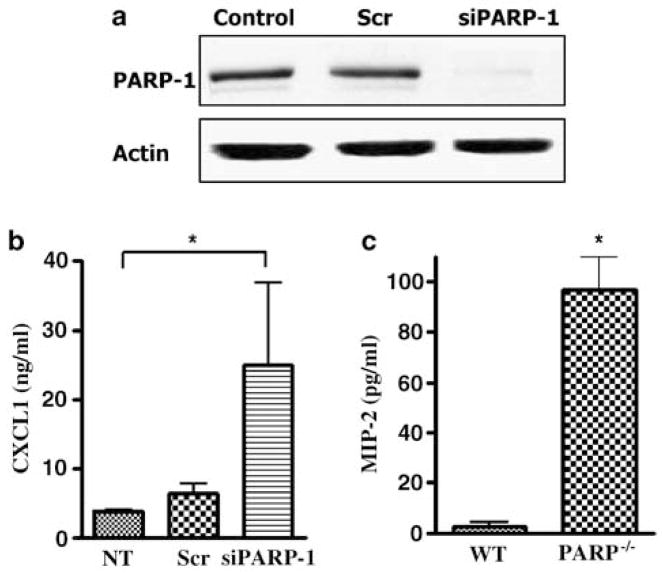
In order to establish whether PARP-1 physical presence was important for the regulation of CXCL1 transcription, PARP-1 expression was ‘knocked down’ using small interfering RNA (siRNA), targeting PARP-1 (Figure 3a). Interestingly, when PARP-1 was knocked down in the Hs 294T cells, the level of CXCL1 production increased significantly (Figure 3b). To test whether these observations were owing to artifacts resulting from siRNA, primary melanocytes were isolated from WT and PARP-1−/− pups and the CXCL1 mouse homolog, MIP-2, levels were measured from the conditioned medium. The data are in agreement with the siRNA results and also show an increase in CXCL1 production in PARP-1−/− melanocytes when compared to their WT counterparts (Figure 3c). Thus, loss of PARP-1 protein expression in melanocytes can have a stimulatory effect on CXCL1 transcription.
Figure 3.
Ablation of PARP-1 expression leads to an increase in CXCL1 protein level. (a) Cells were transfected with siRNA against PARP-1 and nonspecific scrambled RNA using the Oligofectamine transfection reagent. Whole-cell extracts were made after 48 h incubation with the siRNAs and immunoblot analysis for PARP-1 and actin was performed. This figure is a representative of three separate experiments. (b) The supernatant for the above transfected cells was collected after 48 h incubation with the siRNAs and was subjected to CXCL1 ELISA. (c) Primary melanocytes were isolated from WT and PARP −/− pups and 1.5 × 105 cells were seeded in 12-well plates in normal growth media. The level of MIP-2 protein was measured after 48 h using mMIP-2 ELISA. The data shown are the mean of three different experiments ± s.d.
Inhibition of enzymatic activity of PARP-1 leads to a decrease in CXCL1 protein level
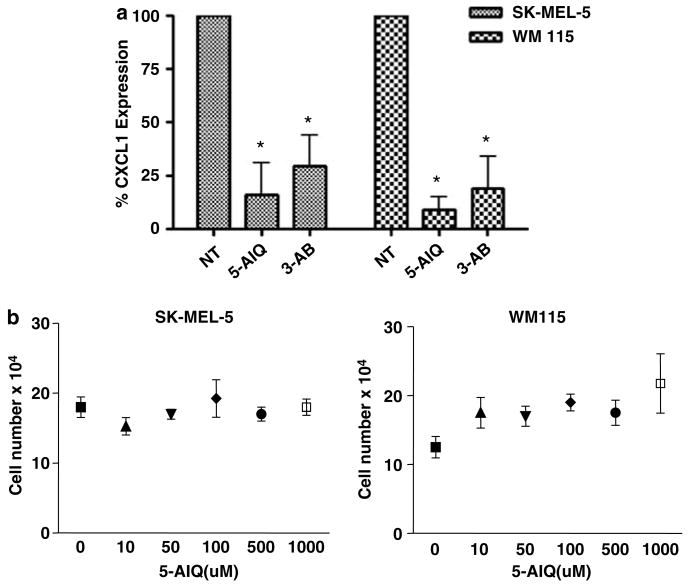
To determine the effect of inhibitors of PARP activity on the production of CXCL1 in melanoma cells, the cells were treated with two different PARP inhibitors, 5-AIQ at 1mM or 3-aminobenzamide (3-AB) at 10mM concentrations, for 48 h in order to monitor effects of inhibitors on CXCL1 protein expression and secretion (Figure 4a). Inhibition of PARP-1 enzymatic activity resulted in a decrease in the level of CXCL1 for all melanoma cells. In order to rule out nonspecific cellular toxicity effects owing to use of inhibitors, SK-MEL-5 and WM 115 cells were treated with increasing concentration of 5-AIQ (Figure 4b), and after 48 h of treatment, cell counts were performed after the addition of Trypan blue to the cells, using the hemocytometer. Treatment with the inhibitor had no effect on the viability of the cells even at high concentrations. These results indicate that the catalytic activity of PARP-1 provides an activating role in CXCL1 transcription. Thus, although the presence of PARP-1 protein may be inhibitory for CXCL1 transcription, PARP-1 in its enzymatically active state enhances CXCL1 transcription. This explains why, despite high levels of PARP-1 expression in melanoma cells, CXCL1 expression is elevated.
Figure 4.
Inhibition of enzymatic activity of PARP-1 leads to a decrease in CXCL1 protein level. Cells were incubated in SFM with the addition of indicated PARP-1 inhibitors, 5-AIQ at 1mM or 3-AB at 10mM, for 48 h. (a) The supernatant was collected and the level of secreted CXCL1 was measured using CXCL1 ELISA. (b) SK-MEL-5 and WM 115 cells were treated with increasing concentration of 5-AIQ and after 48 h of treatment cell counts were performed. The data shown are the mean of three different experiments ± s.d.
Discussion
The CXCL1 chemokine plays an important role in pathogenesis of inflammation and tumorigenesis. In normal cells, CXCL1 is an inducible chemokine that is expressed in response to exposure to various stimuli such as interleukin-1 and tumor necrosis factor-α (Shattuck et al., 1994). However, the expression of this chemokine becomes disregulated during melanomagenesis and malignant melanoma cells express constitutively high levels of CXCL1. The metastatic and angiogenic abilities of a number of tumors have been attributed to elevated levels of ELR+, angiogenic chemokines such as CXCL1 (Strieter et al., 1995; Keane et al., 1997). The role of these chemokines in tumor growth has also been implicated in many tumor types, including pancreas, head and neck, and non-small-cell lung tumors (Olbina et al., 1996; Richards et al., 1997; Takamori et al., 2000). Thus, it is imperative to discern the mechanisms by which these chemokines are regulated in order to develop targeted therapy in cancer and inflammation.
In this study, we demonstrated for the first time that PARP-1 can have a dual role as a transcriptional modulator for CXCL1 in normal melanocyte and melanoma cell lines. Our data support the previous report that inhibition of PARP-1 activity results in decreased CXCL1 production in melanoma cell lines. Moreover, we have gone on to show through ChIP assays that PARP-1 interacts with the IUR of the CXCL1 promoter in the non-(ADP-ribosylated) state and binding of PARP-1 to the IUR inhibits NF-κB binding to the CXCL1 promoter. Our data are in agreement with previous work by Soldatenkov et al. (2002), who reported a negative role for PARP-1 in transcription regulation. The data showed that direct interaction of PARP-1 protein with its own gene promoter resulted in suppression of transcription. However, in response to DNA damage, PARP-1 catalytic activity was stimulated and automodification of PARP-1 subsequently prevented its interaction with the promoter. This relieved the PARP-mediated block on the promoter and allowed for transcription of PARP-1 and other genes suppressed by PARP-1. Similarly, the nucleosome binding properties of PARP- 1 that aid in the formation of compact, transcriptionally repressed chromatin structures was described recently (Tulin and Spradling, 2003; Tulin et al., 2003; Kim et al., 2004). Interestingly, Kim et al. (2004) were able to show that PARP-1 occupies transcriptionally repressed chromatin domains and that PARP-1 incorporation into chromatin represses Pol II-dependent transcription, indicating its important role in the modulation of chromatin structure as well as transcription. This would explain greater affinity of the larger complex binding to the WT-IUR than free NF-κB as seen in the EMSA experiments. However, it is important to note that EMSA has limitations with regard to the ability to detect the PARP/NF-κB binding to the promoter for CXCL1. These interactions may be more apparent at the chromatin level where PARP activity affects the chromatin as well as p65, highlighting the complexities involved within the living cell that is not always apparent in in vitro experiments.
In the context of NF-κB target gene transcriptional regulation, Chang and Alvarez-Gonzalez (2001) reported that direct PARP-1 interaction with NF-κB inhibits the binding of NF-κB to its element and this inhibition is relieved by the auto-poly(ADP-ribosylation) of PARP-1. In primary cultured mouse glial cells, PARP-1 was shown to be in an automodified state and inhibition of PARP activity or antisense RNA for PARP-1 mRNA reduced the lipopolysaccharideinduced DNA binding of NF-κB. Taken together, the data demonstrate that PARP-1 may be a negative factor in the activation of NF-κB through its direct physical interaction with the transcription factor.
The cell/tissue- and pathway-specific roles of PARP-1 in transcription were demonstrated clearly in a study by Ha (2004), in which PARP-1 −/− glial cells were compared to PARP-1 −/− peritoneal macrophages. Whereas the glial cells showed diminished p38-mitogen- activated protein kinase activation as well as NF-κB DNA-binding and target gene expression, PARP-1 −/− macrophages only lacked in NF-κB activation. Furthermore, murine lymphocytic leukemia cells deficient in PARP-1 exhibited increased DNA-binding activity of NF-κB and transfection of these cells with a PARPexpressing plasmid decreased the high level of binding to normal levels (Kameoka et al., 2000). Inhibition of PARP-1 activity in airway epithelial cells, on the other hand, showed reduced NF-κB activation and reduced CXCL8 expression upon H2O2 induction and prevented lung inflammation in vivo (Boulares et al., 2003). Similarly, CXCL8 as well as intercellular adhesion molecule-1 expression were reduced by PARP inhibitors in HaCaT keratinocytes (Szabo et al., 2001). These data indicate that PARP-1 can act as both an inhibitor and activator of NF-κB-dependent transcription. These data are supportive of our results from the PARP-1 −/− melanocytes and from the PARP-1 siRNA ‘knockdown’ cells, where we observed markedly higher levels of MIP-2/CXCL1 than in cells expressing normal levels of PARP-1. In contrast, the expression of CXCL1 is decreased when the activity of PARP-1 is inhibited. These data are once more indicative of PARP’s convoluted role in transcriptional regulation, where a fine balance exists between the inactive and active state of PARP-1. Thus, any minute shift in this equilibrium could give rise to significant modifications with respect to transcription. Taken together, the data implicate a cell/tissue-specific role of PARP-1 in the regulation of transcription and one must be cautious in assigning a single role to this complex protein.
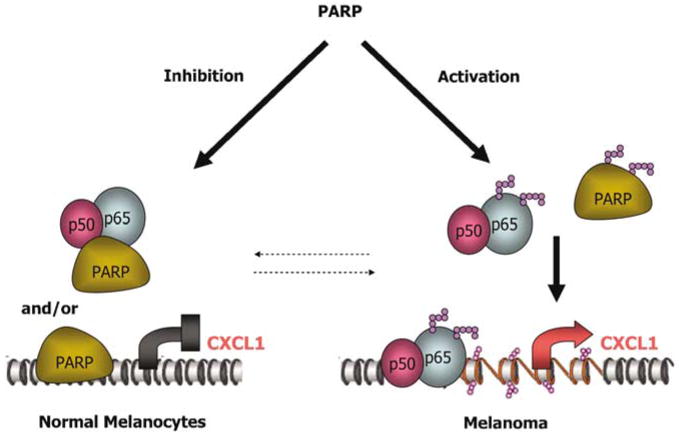
In summary, our study provides key insight into how aberrant activation of PARP-1 in melanoma cells can regulate the transcriptional activity of NF-κB. Based on the data we have presented here, we hypothesize that in normal melanocytes, PARP-1 activity is silent, leading to binding of PARP-1 to the promoter of CXCL1 and preventing NF-κB from binding to the promoter. However, in cancer cells exhibiting bioenergetic malfunction, this balance is shifted, resulting in more auto-poly(ADP-ribosylation) of PARP-1, whereby PARP-1 is dissociated from the promoter, allowing for an increased binding of NF-κB to the promoter and activated transcription (Figure 5). Thus, it appears that the physical interaction of PARP-1 with the CXCL1 promoter asserts a negative effect in transcription, whereas the activity of PARP-1 is important in promotion of CXCL1 transcription. This mechanism of transcriptional regulation creates a new venue by which we can target cancer cells and offers hope for more efficacious treatment combinations for melanoma.
Figure 5.
Schematic model for the role of PARP-1 in modulation of CXCL1 transcription. In normal melanocytes, enzymatically inactive PARP-1 interacts with IUR of CXCL1 promoter strongly and aid in the formation of transcriptionally inactive chromatin. PARP-1 may also bind NF-κB proteins, impeding their binding to the κB-site in CXCL1 promoter, resulting in the inhibition of CXCL1 transcription. In melanoma, PARP-1 is enzymatically active, leading to opening of the chromatin via PARP trans- and automodifications. The poly(ADP-ribose) modifications result in weaker PARP-1 interaction with the IUR element as well as with the NF-κB protein complex, leading to the activation of CXCL1 transcription.
Materials and methods
Cell culture
The human melanoma cell lines were obtained from ATCC. The cells were grown in 50% Dulbecco’s modified Eagle’s medium (DMEM), 50% F-12 supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids, 100 mg/ml penicillin and 100 mg/ml streptomycin. Normal human epidermal melanocytes (NHEM) were purchased from Cascade Biologies (Portland, OR, USA) and maintained in Medium- 154 with 1% human melanocyte growth supplement (HMGS). The primary mouse fibroblast and melanocytes were isolated from WT or PARP−/− pups (gifts from Dr ME Smulson at Georgetown University) and maintained in DMEM supplemented with 10% FBS and 1% non-essential amino acids, or with Medium-154 with 1% HMGS, respectively. Cell cultures were maintained at 37°C.
Chromosome immunoprecipitation
Cells were cultured in 15-cm plates to approximately 80–90% confluence. For PARP-1 inhibition, cells were treated with 1mM 5-AIQ (Axxora, LLC, San Diego, CA, USA). Cells were fixed with 1% formaldehyde in DMEM medium lacking FBS for 10 min and then collected. Nuclei were pelleted in hypotonic lysis buffer and lysed in 1 × sodium dodecyl sulfate (SDS) lysis buffer (50mM Tris, pH 8.1/10mM ethylenediaminetetraacetic acid (EDTA)/1% SDS with 10 μl/ml protease inhibitor cocktail and 10 μl/ml phosphatase inhibitors). Sonication conditions were tested to yield DNA fragments averaging 600 bp as assessed by agarose gel electrophoresis. Lysates were clarified, diluted 1:5 in ChIP dilution buffer (0.01% SDS/1.1% Triton X-100/1.2mM EDTA, 16.7mM Tris, pH 8.1/167mM NaCl with 10 μl/ml protease inhibitor cocktail and 10 μl/ml phosphatase inhibitors) and cleared with singlestranded DNA (ssDNA), bovine serum albumin (BSA), IgG serum and protein A/G sepharose beads for 2 h at 4°C. For each immunoprecipitation, 20 μg of the p65 or PARP-1 antibody was added to lysate prepared from the 15-cm plates. After incubation with antibody overnight at 4°C, 50 μl of a 50% protein A/G sepharose slurry containing ssDNA and BSA were added for an additional 1 h of incubation. Beads were then washed consecutively for 3–5 min on a rotating platform with 1ml of each solution: (a) low salt wash buffer (0.1% SDS/1% Triton X-100/2mM EDTA, 20mM Tris, pH 8.1/150mM NaCl), (b) high salt wash buffer (0.1% SDS/1% Triton X-100, 2mM EDTA, 20mM Tris, pH 8.1, 500mM NaCl), (c) LiCl wash buffer (0.25M LiCl/1% NP40/1% deoxycholate, 1mM EDTA/10mM Tris, pH 8.0) and (d) 1 × TE buffer twice. Protein–DNA complexes were eluted and purified and subjected to PCR for amplification of the CXCL1 promoter or β-actin for control. PCR primers for the CXCL1 promoter were 5′-GGCTGCATCAGCGGACCC (forward) and 5′-AGTGCCACTCGCAGGAGC (reverse). The primers for the PCR of β-actin were 5′-AGCCATG TACGTAGCCATCC (forward) and 5′-TTTGATG TCACG CACGATTT (reverse).
Immunoblot analysis and immunoprecipitation
Whole-cell extracts were obtained according to our standard protocol using radioimmunoprecipitation assay buffer. Briefly, cells were washed twice with ice-cold PBS and total cell lysates were isolated with a buffer containing 50mM Tris (pH 7.4), 150mM NaCl, 0.02% sodium azide, 0.1% SDS, 1% Nonidet P-40, 0.5% Na deoxycholate, 1mM EDTA with 10 μl/ml protease inhibitor cocktail (P-8340, Sigma, St Louis, MO, USA) and 10 μl/ml phosphatase inhibitors (P-2850/P5726, Sigma). The lysates were subjected to SDS–PAGE and probed with appropriate antibodies. Antibodies used were anti-p65 (A), anti-PARP-1 (H-250) and anti-actin (C-11) from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and anti-PAR (10 H) was purchased from Axxora, LLC (San Diego, CA). For secondary antibodies, horseradish peroxidase-conjugated anti-mouse, goat or rabbit IgG were obtained from Chemicon International (Temecula, CA, USA). The antibodies were visualized using enhanced chemiluminescence kit from Amersham Biosciences (Piscataway, NJ, USA). Immunoprecipitations were performed after preclearing cell lysates with protein A/G-agarose (Santa Cruz, CA, USA) for 2 h at 4°C as described previously.
Enzyme-linked immunosorbent assay
For quantitation of hCXCL1 and murine macrophage inflammatory protein-2 (mMIP-2), cleared supernatants of cell culture medium were collected. Briefly, 1.5 × 105 cells/well in six-well plates were seeded in serum-free media (SFM) and incubated at 37°C for 12 h. After wash in SFM, the monolayers were incubated with 10mM 3-AB or 1mM 5-AIQ in SFM or left untreated for up to 48 h at 37°C. The supernatant was collected and hCXCL1 and mMIP-2 levels were determined by Quantikine enzyme-linked immunosorbent assay (ELISA) from R&D Systems Inc. (Minneapolis, MN, USA) according to the manufacturer’s protocol.
Cell growth response
Melanoma cell lines SK MEL5, and WM 115 (1.5 × 105 cells/well in six-well plates) were seeded in SFM and incubated at 37°C for 12 h. After wash in SFM, the monolayers were incubated with increasing concentration of 5-AIQ in SFM or left untreated for up to 48 h at 37°C. Cell counts were performed after addition of Trypan blue to the cells, using the hemocytometer on after 48 h of treatment.
EMSA
Cells were washed twice with ice-cold PBS and collected in a cell suspension buffer containing 10mM Hepes (pH 7.9), 10mM NaCl, 1.5mM MgCl2, 0.5mM dithiothreitol (DTT) and 5mM mercaptoethanol with 10 μl/ml protease inhibitor cocktail and 10 μl/ml phosphatase inhibitors. To separate cytoplasm/nuclear proteins, cells were lysed in 1% NP-40 in the cell suspension buffer described above. The destruction of cell membranes and the presence of intact nuclei were observed by staining with 0.04% Trypan blue. Cells were then centrifuged at 6000 g and the supernatant was collected as the cytoplasm protein fraction. The pellet was washed with a nuclei suspension buffer containing 20mM Hepes (pH 7.9), 10mM NaCl, 1.5mM MgCl2, 0.5mM DTT, 5mM mercaptoethanol, 0.2M EDTA and 1% NP-40. The nuclei were then lysed in 450mM NaCl hypertonic buffer in the nuclei suspension buffer described above. The following oligonucleotide probes were 32P-labeled using Prime-It II Random Primer Labeling Kit from Stratagene (La Jolla, CA, USA): the WT 2 × IUR–NFκB, had the upper strand sequence 5′-gggatcgatctggaactccgggaatttccctggcccgggatcgatctggaactccgggaatttccctggccc-3′ and the mutant IUR containing oligonucleotide, 2 × mIUR–NFκB, with mutations in the TCGAT motif of the IUR element had the upper strand sequence 5′-gggaAGTACctggaactccgggaatttccctggcccgggaAGTACctggaactccgggaatttccctggccc-3′. Uppercase characters indicate nucleotide replacements in the TCGAT motif, and the underscored sequences define the two copies of IUR element. Proteins (10 μg) were incubated with the oligos and protein/oligo complexes were electrophoresed in a 4% native polyacrylamide gel, transferred to 3 MM chromatography paper (Whatmann, Clinton, NJ, USA) and autoradiographed. For gel shift reactions, proteins were incubated with the specific antibody for 1 h at 4°C before oligo incubation. The antibodies used for observing the supershifted bands were Rel A (sc-109x, Santa Cruz, CA, USA) and PARP-1 (R&D Systems, MN, USA).
Establishment of siRNA against PARP-1
siRNA candidates directed against PARP-1 were designed according to the information found at the Dharmacon Inc. (Lafayette, CO, USA) web site. The nucleotide sequence of the siRNA target site in the PARP-1 gene was 5′-aaagucccaca cugguaccac-3′. This sequence was blast searched on the NCBI web site to verify the specificity for PARP-1 before manufacturing by Dharmacon Inc. As control, siCONTROL nontargeting siRNA no. 1, a scrambled non-targeting oligo designed by Dharmacon Inc. was used. The oligos (150 nM) were transfected into cells using the transfection reagent Oligofectamine, purchased from Invitrogen (Carlsbad, CA, USA), according to the manufacturer’s instructions. Supernatant was collected 48 h post-transfection for ELISA assays and cells were prepared for immunoblotting (see Immunoblot analysis).
Statistical analysis
Results are expressed as means ± s.d. from three independent experiments or representative replicate experiments. Statistical analysis was performed using the unpaired Student’s t-test. The value of P < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by grants from the National Institute of Health (NIH-CA-56704, AR), the VA Merit Award (AR), in part by grants from the National Cancer Institute (PO1CA- 74175, MES) and the US Air Force Office of Scientific Research (AF-FA9550-04-1-0395, MES).
Abbreviations
- 3-AB
3-aminobenzamide
- ADP
adenosine diphosphate
- 5- AIQ
5-aminoisoquinolinone.HCl
- BSA
bovine serum albumin
- ChIP
chromatin immunoprecipitation
- CXCL1
CXC ligand 1
- DMEM
Dulbecco’s modified Eagle’s medium
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EMSA
electrophoretic mobility shift assay
- FBS
fetal bovine serum
- HMGI/Y
high-mobility group-I/Y
- HMGS
human melanocyte growth supplement
- IgG
immunoglobulin G
- IUR
immediate upstream region
- NAD+
β-nicotinamide adenine dinucleotide
- NF-κB
nuclear factor-kappa B
- NHEM
normal human epidermal melanocyte
- PAGE
polyacrylamide gel electrophoresis
- PARP-1
poly(ADP-ribose) polymerase-1
- PCR
polymerase chain reaction
- SDS
sodium dodecyl sulfate
- SFM
serum-free media
- SP1
stimulating protein-1
- ssDNA
single-stranded DNA
- WT
wild type
References
- Alvarez-Gonzalez R, Spring H, Muller M, Burkle A. J Biol Chem. 1999;274:32122–32126. doi: 10.1074/jbc.274.45.32122. [DOI] [PubMed] [Google Scholar]
- Balentien E, Mufson BE, Shattuck RL, Derynck R, Richmond A. Oncogene. 1991;6:1115–1124. [PubMed] [Google Scholar]
- Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Am J Respir Cell Mol Biol. 2003;28:322–329. doi: 10.1165/rcmb.2001-0015OC. [DOI] [PubMed] [Google Scholar]
- Burkle A. Bioessays. 2001;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- Cayuela ML, Carrillo A, Ramirez P, Parrilla P, Yelamos J. Biochem Biophys Res Commun. 2001;285:289–294. doi: 10.1006/bbrc.2001.5178. [DOI] [PubMed] [Google Scholar]
- Chang WJ, Alvarez-Gonzalez R. J Biol Chem. 2001;276:47664–47670. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Richmond A. J Biol Chem. 2002;277:7920–7928. doi: 10.1074/jbc.M112210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC. Proc Natl Acad Sci USA. 2004;101:5087–5092. doi: 10.1073/pnas.0306895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghnegahdar H, Du J, Wang D, Strieter RM, Burdick MD, Nanney LB, et al. J Leukocyte Biol. 2000;67:53–62. doi: 10.1002/jlb.67.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, et al. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- Huang K, Tidyman WE, Le KU, Kirsten E, Kun E, Ordahl CP. Biochemistry. 2004;43:217–223. doi: 10.1021/bi0301800. [DOI] [PubMed] [Google Scholar]
- Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, et al. Biochem J. 2000;346(Part 3):641–649. [PMC free article] [PubMed] [Google Scholar]
- Keane MP, Arenberg DA, Lynch JP, III, Whyte RI, Iannettoni MD, Burdick MD, et al. J Immunol. 1997;159:1437–1443. [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Li A, Varney ML, Singh RK. Clin Cancer Res. 2001;7:3298–3304. [PubMed] [Google Scholar]
- Meisterernst M, Stelzer G, Roeder RG. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirodi C, Nagdas S, Gygi SP, Olson G, Aebersold R, Richmond A. J Biol Chem. 2001;276:9366–9374. doi: 10.1074/jbc.M009897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbina G, Cieslak D, Ruzdijic S, Esler C, An Z, Wang X, et al. Anticancer Res. 1996;16:3525–3530. [PubMed] [Google Scholar]
- Owen JD, Strieter R, Burdick M, Haghnegahdar H, Nanney L, Shattuck-Brandt R, et al. Int J Cancer. 1997;73:94–103. doi: 10.1002/(sici)1097-0215(19970926)73:1<94::aid-ijc15>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Richards BL, Eisma RJ, Spiro JD, Lindquist RL, Kreutzer DL. Am J Surg. 1997;174:507–512. doi: 10.1016/s0002-9610(97)00165-7. [DOI] [PubMed] [Google Scholar]
- Schadendorf M, Algermissen W, Sticherling M, Czarnetzki BM. J Immunol. 1994;153:3360. [PubMed] [Google Scholar]
- Schadendorf D, Fichtner I, Makki A, Alijagic S, Kupper M, Mrowietz U, et al. Br J Cancer. 1996;74:194–199. doi: 10.1038/bjc.1996.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck RL, Wood LD, Jaffe GJ, Richmond A. Mol Cell Biol. 1994;14:791–802. doi: 10.1128/mcb.14.1.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Haddad BR, Rosenthal DS, Weaver Z, Coleman A, Luo R, et al. Proc Natl Acad Sci USA. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Samara R, Espinoza LA, Hassa PO, et al. Oncogene. 2003;22:8460–8471. doi: 10.1038/sj.onc.1206897. [DOI] [PubMed] [Google Scholar]
- Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, Iyer S, et al. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A. J Biol Chem. 2002;277:665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, et al. J Biol Chem. 1995;270:27348–27357. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- Szabo E, Virag L, Bakondi E, Gyure L, Hasko G, Bai P, et al. J Invest Dermatol. 2001;117:74–80. doi: 10.1046/j.0022-202x.2001.01388.x. [DOI] [PubMed] [Google Scholar]
- Takamori H, Oades ZG, Hoch OC, Burger M, Schraufstatter IU. Pancreas. 2000;21:52–56. doi: 10.1097/00006676-200007000-00051. [DOI] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Wood LD, Farmer AA, Richmond A. Nucleic Acids Res. 1995;23:4210–4219. doi: 10.1093/nar/23.20.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Richmond A. Cancer Res. 2001;61:4901–4909. [PubMed] [Google Scholar]
- Zhang Z, Hildebrandt EF, Simbulan-Rosenthal CM, Anderson MG. Virology. 2002;296:107–116. doi: 10.1006/viro.2002.1385. [DOI] [PubMed] [Google Scholar]