Abstract
Understanding the principles of calmodulin (CaM) activation of target enzymes will help delineate how this seemingly simple molecule can play such a complex role in transducing Ca2+-signals to a variety of downstream pathways. In the work reported here, we use biochemical and biophysical tools and a panel of CaM constructs to examine the lobe specific interactions between CaM and CaMKII necessary for the activation and autophosphorylation of the enzyme. Interestingly, the N-terminal lobe of CaM by itself was able to partially activate and allow autophosphorylation of CaMKII while the C-terminal lobe was inactive. When used together, CaMN and CaMC produced maximal CaMKII activation and autophosphorylation. Moreover, CaMNN and CaMCC (chimeras of the two N- or C-terminal lobes) both activated the kinase but with greater Kact than for wtCaM. Isothermal titration calorimetry experiments showed the same rank order of affinities of wtCaM > CaMNN > CaMCC as those determined in the activity assay and that the CaM to CaMKII subunit binding ratio was 1:1. Together, our results lead to a proposed sequential mechanism to describe the activation pathway of CaMKII led by binding of the N-lobe followed by the C-lobe. This mechanism contrasts the typical sequential binding mode of CaM with other CaM-dependent enzymes, where the C-lobe of CaM binds first. The consequence of such lobe specific binding mechanisms is discussed in relation to the differential rates of Ca2+-binding to each lobe of CaM during intracellular Ca2+ oscillations.
Ca2+/calmodulin-dependent protein kinase II (CaMKII) is found in every tissue in the body and is the most abundant protein kinase in neurons where it is widely recognized for its important role in regulating diverse neuronal functions (1). CaMKII is a dodecameric complex (twelve subunits) (2) and one of its most interesting properties is the ability to phosphorylate itself (autophosphorylation) following Ca2+/CaM activation, a process that confers Ca2+-independent activity upon the kinase (3). This autophosphorylation of CaMKII also increases its affinity for Ca2+/CaM by more than 1,000-fold, commonly referred to as “CaM-trapping” (4). Thr-286 in the α subunit and Thr-287 in the β, γ and δ subunits has been identified as the phosphorylation site that lead to these functions changes (1). Substitution of an Ala residue at Thr-286 in the α subunit abrogates both Ca2+-independent activity (5) and Ca2+/CaM-trapping (4).
The interactions between CaMKII and Ca2+/CaM are complex due in part to the association of subunits into the dodecameric structure. Hanson et al. (6) elegantly documented that autophosphorylation of Thr-286 occurs through an intersubunit interaction within a holoenzyme and requires that Ca2+/CaM be bound to neighboring subunits. One subunit serves as the phosphotransferase reaction and requires the binding of Ca2+/CaM to be active, while the second adjacent subunit requires binding of Ca2+/CaM to permit Thr-286 to be phosphorylated. In the case of autophosphorylation, it is clear that two Ca2+/CaMs must bind simultaneously, and we showed previously that Ca2+/CaM-binding and autophosphorylation of CaMKII shows cooperativity in all four of the mammalian isoforms (7). In addition, the initial interaction of Ca2+/CaM with the CaM-binding domain of each subunit imposes additional kinetic constraints. Using a series of fluorescent derivatives of CaM, Torok et al. (8) demonstrated that Ca2+/CaM interactions with CaMKII can be divided into at least three steps. CaM initially binds in an extended conformation via one lobe only. If nucleotide is present, the second lobe of CaM can interact with CaMKII which leads to a partial collapse of the two lobes relative to each other and this interaction is apparently sufficient for the bound subunit to be active. Expression of enzyme activity against an exogenous substrate would not be expected to exhibit cooperativity for Ca2+/CaM and this was shown to be the case (7). If two adjacent subunits of CaMKII are bound to Ca2+/CaM, autophosphorylation of Thr-286 ensues and this leads to a third step of very tight binding of Ca2+/CaM that is correlated with an additional collapse of the two domains of CaM (8). It is this third step that produces the CaM-trapped form of the kinase. Defining the details of Ca2+-CaM interaction with the kinase are critical to understand the rate-limiting step that leads to activation and autophosphorylation of CaMKII in cells.
CaM is a ubiquitous 16.8-kDa (148 amino acid) protein that binds to multiple protein targets, some in a Ca2+-dependent manner and others in a Ca2+-independent manner (9, 10). CaM is composed of an N- and C-terminal lobe tethered by a flexible linker region that, in total, resembles a dumbbell when in the non-target bound state. The N- and C-lobes each contain two EF-hand Ca2+-binding domains separated from each other by the alpha-helical tether (11) and Ca2+ binding is cooperative within each lobe (12). The C-terminal lobe binds Ca2+ with higher affinity but has slower association and dissociation rates while the N-terminal lobe has a lower affinity and faster on- and off-rates for Ca2+ (13, 14). From these kinetic constants, one can deduce that during the rising phase of intracellular Ca2+ concentration ([Ca2+]i), the faster association rates of the N-lobe would dictate that they bind Ca2+. If Ca2+/CaM-binding targets can interact with the N-terminal lobe of CaM, they would gain a preferential advantage during the rising phase of a Ca2+-transient over those that bind the C-terminal domain. Depending on the rate of this association event, the C-terminal lobe may or may not saturate with Ca2+. As the system relaxes to steady state the C-lobe is more likely to remain Ca2+ saturated because of its higher affinity. This important difference between the Ca2+ binding affinities in the two lobes imparts on CaM the potential for lobe-specific regulation of its interactions with target proteins during variations of [Ca2+]i.
It is well documented that target protein interactions significantly impact the Ca2+-binding properties of CaM. Protein binding can either decrease the binding affinity of CaM for Ca2+ (15, 16) or increase the Ca2+-binding affinity (17, 18) as anticipated from coupled reactions. The magnitude of these effects has been documented for several protein enzyme systems, including Ca2+/CaM interactions with CaMKII (19). We previously identified that the dissociation rates of Ca2+ from CaM are slowed when bound to CaMKII and that the differences seemed to correlate with alterations in the release of Ca2+ predominantly from the N-lobe (16). When bound to the phosphorylated form of CaMKII, Ca2+ dissociation of all four Ca2+ binding sites is slowed, consistent with the high affinity interaction of CaM bound to phospho-CaMKII (16).
While we presently lack detailed structural information for Ca2+/CaM bound to native CaMKII, a synthetic peptide mimicking the CaM-binding domain of the kinase (amino acids 290 to 314) has been crystallized with Ca2+/CaM (20). A similar peptide containing amino acids 293 to 312 of αCaMKII, was shown to kinetically mimic the association and dissociation kinetics of the phosphorylated form of CaMKII (21) and we presume the crystal structure best reflects the contacts made between Ca2+/CaM and autophosphorylated CaMKII. The structure identifies the typical antiparallel arrangement where the N-terminal domain of the peptide interacts predominantly, although not exclusively, with amino acids on the C-domain of CaM, and vice versa (20). This structural data suggests that the two domains of CaM exhibit a preferred orientation for binding to the CaM-binding domain of CaMKII but how domain specific effects control enzyme activation and autophosphorylation are not known.
In the present study, we build on earlier kinetic and structural work to investigate domain specific interactions between CaM and CaMKII. We first analyze the kinetics and steady-state binding of wtCaM (wild-type CaM) with CaMKII in the presence and absence of ADP to assess whether preferential binding of the N- or C-domain can be detected. Experiments measuring Ca2+-dissociation from CaM bound to CaMKII were consistent with the C-domain of CaM being free, while the N-domain was bound. In the presence of ADP, results obtained were consistent with the binding of both lobes of CaM to CaMKII. Isothermal titration calorimetry (ITC) revealed a remarkably weak binding affinity between Ca2+/CaM and CaMKII that increased significantly in the presence of ADP, consistent with the potential for CaM interactions via only one lobe in the absence of nucleotide. ITC also showed that Ca2+/CaM interact with CaMKII subunits at a stoichiometry of close to 1:1 in the presence or absence of ADP. We then asked whether CaMKII activity and autophosphorylation requires that the two halves of CaM be physically connected. Genetically engineered half-CaMs, which include CaM-N(1–75), CaMN(1–80), CaM-C(76–148) (22, 23) and CaM-C(81–148) were examined. CaMN(1–75) with CaMC(76–148) or CaMN(1–80) with CaMC(81–148) were both capable of supporting activation and autophosphorylation, but CaM-N(1–75) was the only fragment that, by itself, was sufficient to support the activation and autophosphorylation of CaMKII. ITC using a high affinity peptide mimetic of the CaM-binding domain of CaMKII revealed that CaMN(1–75) was the only fragment that bound in a simple manner which suggests that the inability of the other half-CaMs to activate may be due to inappropriate interactions with the CaM-binding domain of the kinase. To further assess the domain specificity for activation, constructs constituting chimeras of the two N- or C-terminal lobes, CaMNN and CaMCC (24, 25), were analyzed. One advantage of using such chimeras is that one maintains the impact of increasing the effective concentration of the second lobe upon binding of the first, more typical of wtCaM. Both of these chimeras were shown to activate and induce autophosphorylation of CaMKII, although CaMNN was ~2-fold more effective than CaMCC. Using ITC, we identified that wtCaM, CaMNN and CaMCC bound to CaMKII with a one-to-one stoichiometry and that their rank order of binding was similar to their rank order of CaMKII activation. Results using stopped-flow fluorimetry and quin-2 induced Ca2+-dissociation, insured that all of the constructs in this study were still capable of binding Ca2+, although some unexpected findings were revealed. Together, these results support a sequential model of CaM’s association to CaMKII with the N-lobe binding first causing an increase in the binding of nucleotide, leading to the C-lobe interactions and activation of each subunit.
Experimental Procedures
Protein Expression and Purification
wtCaM was expressed from a codon optimized cDNA in the pET-23d plasmid expressed in E. coli BL21(DE3)star as previously described (7). The CaMNN and CaMCC clones in pET9 were a gift from Dr. Guy Guillemette (26). CaMN(1–75), CaMN(1–80) and CaMC(76–148) in T7-7 were gifts from Dr. Madeline Shea (23, 27). CaMC81–148 was produced by PCR based mutagenesis starting with CaMC(76–148) as the template. All cDNAs were expressed in E. coli BL21(DE3)star (Invitrogen). CaM constructs were purified as described (7) with minor modifications. The half-CaMs were purified through a modified protocol. Briefly, the pellets were resuspended in 50 mM Tris, pH 7.5, and 1 mM EDTA, with protease inhibitor cocktail (Sigma) and sonicated before centrifugation at 18,000 × g for 1 hr at 4°C. The supernatant was collected and applied to a phenyl-Sepharose CL-4B column (GE Healthcare) previously equilibrated with 50 mM Tris, pH 7.5 and 2 mM CaCl2. After sample application, the column was first washed with 50 mM Tris, pH 7.5, 2 mM CaCl2 + 0.5 M NaCl and then with 50 mM Tris, pH 7.5 and 2 mM CaCl2. CaM was eluted with 50 mM Tris, pH 7.5, and 10 mM EGTA. Following evaluation with SDS-PAGE, the protein was extensively dialyzed into 50 mM MOPS, pH 7.0 and stored in this buffer at −20°C. The concentration of each protein was quantified by a BCA assay kit (Pierce).
For measuring steady-state binding and rates of dissociation, CaM was mutated to Cys at Lys75 and labeled with IAEDANS (Invitrogen) as described (28). CaM(C75) was incubated in 5 mM DTT for 20 min at room temperature and then desalted over a Biogel P2 column. The reduced CaM (0.5 mL of 200 µM) was incubated with 10-fold molar excess of IAEDANS in the dark at room temperature for 4 hr, and then reapplied to the Biogel P2 column. Peak fractions were pooled and dialyzed overnight against 50 mM MOPS, pH 7.0. The amount of label incorporated (in M) was calculated by dividing the absorbance at 336 nm (for IAEDANS) by the extinction coefficient of 5700 M−1cm−1. The concentration of the labeled CaM was determined using a BCA protein assay (Pierce) with bovine serum albumin as the standard. The dye to protein ratio in the CaM-C75-DANS preparations used in these experiments was 0.9.
CaMKII expression and purification
A rat αCaMKII cDNA was expressed in the baculovirus system using Sf21 cells as previously described (28). The kinase was purified from infected cells by phosphocellulose cation exchange column as described by Bradshaw et al. (29). Briefly, infected cell pellets were resuspended in lysis buffer (10 mM Tris pH 7.5, 5% betaine, 1 mM EGTA, 1 mM EDTA, 0.5 mM DTT, 0.1 mM PMSF, 5 mg/L leupeptin, and 20 mg/L soybean trypsin inhibitor) and disrupted by brief sonication. The material was clarified at 4°C at 24,000 rpm in a SW-28 rotor for 1 hour. The supernatant was applied by gravity flow onto a 2 g phosphocellulose column (P11 cation exchange resin, Whatman) at 4°C and washed with 50 ml of 50 mM PIPES pH 7.0, 100 mM NaCl. CaMKII was eluted with a linear gradient of 100 mM to 500 mM NaCl. The peak fractions were dialyzed into 10 mM HEPES, 200 mM KCl, and 20% glycerol at 4°C. CaMKII was quantified by measuring the absorbance at 280 nm and the concentration (in M) was calculated using an extinction coefficient of 66975 M−1cm−1. The extinction coefficient for CaMKII was determined using the ProtParam tool in ExPASy at http://www.expasy.org/tools/protparam.html (30) and was confirmed using values from a BCA protein assay.
Autophosphorylation assay
Autophosphorylation of CaMKII was assessed in 20 µl reactions containing 25 mM HEPES, 10 mM MgCl2, 50 mM KCl, 0.4 mM DTT, 100 µM ATP, 4 mM CaCl2 and 2 µCi of 32P-ATP in the presence of the indicated concentration of CaM. The reaction was carried out on ice for 10 min, initiated by adding 100 ng of CaMKII and the reactions were terminated by adding 5µl of 4× SDS-sample buffer. 40 ng of kinase were loaded on each lane of a 10% SDS-polyacrylamide gels. After staining with Coomassie blue and destaining overnight, the radioactivity incorporated into the enzyme was quantified by exposure of the gel to a phosphor screen for five hours and detected with a Typhoon scanner (Molecular Dynamics).
Western blot and Immunostaining
Autophosphorylation of CaMKII was also assessed by Western blotting as previously described (31). A rabbit polyclonal anti-phospho-Thr286-αCaMKII (Anti-ACTIVE CaM Kinase II pAB, rabbit, pT286, Promega) was used at a 1:5000 dilution. The bound antibody was visualized by measuring the fluorescence of a secondary antibody (anti-rabbit-IgG labeled with Alexa 568, dilution 1:5000; Molecular Probes) using the Typhoon scanner.
Activity assay
The activity of CaMKII was assessed in a 25 µl reaction containing 25 mM HEPES, 10 mM MgCl2, 50 mM KCl, 0.4 mM DTT, 50 µM syntide, 100 µM ATP, 4 mM CaCl2 and 2 µCi of 32P-ATP. To measure the CaM dependence of CaMKII activity the concentration of the wtCaM was varied from 1.25 nM to 5 µM, and the concentration of the CaMNN and CaMCC were varied from 10 nM to 100 µM. The ability of the isolated lobes of CaM to activate the CaMKII was assessed by varying the concentration of each domain from 25 µM to 500 µM (final concentrations), either by themselves or in combination of an N-domain and a C-domain. Reactions were pre-incubated for 1 min at 30°C, initiated by the addition of 10 ng of kinase and then incubated for 1 minute at 30°C. Reactions were terminated by spotting 20 µl onto P-81 filters and immersing in 75 mM H3PO4 acid. After air drying, the incorporated radioactivity was quantified by Cerenkov counting.
Isothermal titration calorimetry
ITC was performed in a Microcal VP-ITC. The buffer for experiments with full length CaMKII consisted of 25 mM HEPES, pH 7.4, 150 mM KCl, 1 mM MgCl2, 1 mM CaCl2 with or without 1 mM ADP. Purified CaMKII was added to the calorimetry cell at a concentration of 15–20 µM (subunit concentration). Aliquots of 200 µM wtCaM, CaMNN or CaMCC (10 µL) were automatically injected with constant mixing at 15°C. The data were fit with single or double site binding models using Microcal software. N reflects the binding stoichiometry, K is the association constant (K = 1/Kd), ΔH is the enthalpy and ΔS the entropy determined from the fits. While the fitting function is described as single or double sites, in our experiments, we use the word mode to encompass the fact that it is possible that ½ CaMs could bind to a single site in either an N- to C-terminal orientation or a C- to N-terminal orientation. This would represent two modes of binding to one target site.
For experiments with the CaMKII(291–312) peptide, experiments were performed in a buffer of 20 mM HEPES pH 7.15, 100 mM NaCl, and 2 mM CaCl2 as previously described (32). Experiments were performed with a concentration of CaMKII(291–312) of 400 to 600 µM in the reaction syringe and a concentration of each CaM lobe of 15 to 30 µM in the reaction cell. The biphasic nature of the titration curves indicated that binding was not always a simple 1:1 ratio. Fitting to the multiple binding site models was performed using the Origin software. The sequence of the CaMKII(291–312) peptide was: Ac-KKFNARRKLKGAILTTMLATRN-NH2.
Stopped-flow fluorimetry
All stopped-flow experiments were performed on an Applied Photophysics Ltd. (Leatherhead, UK) model SV.17 MV sequential stopped flow spectrofluorimeter with a dead time of 1.8 ms. For all experiments, data from five to six injections were averaged and then fit with either single, or double exponential functions. All solutions were made in a buffer containing 20 mM MOPS pH 7.5, 100 mM KCl and, where applicable, 1 mM MgCl2. The concentrations of reagents given below are those in the syringes, before mixing. For Ca2+ dissociation using quin-2, the excitation wavelength was set at 334.5 nm and the detection of fluorescence emission was controlled by a 435 nm cut-on filter (Oriel #51282). 4 µM CaM and 20 µM CaCl2 (along with CaM-target and/or nucleotide, as indicated in the figure legends) were mixed with 150 or 300 µM quin-2 (Molecular Probes, Eugene, OR). In experiments with peptide, 16 µM of Ac-FNARRKLKGAILTTMLATRN-NH2 (CaMKII(293–312)), a high affinity mimetic of the CaM-binding domain of CaMKII was included (21).
Measurement of CaM affinity
The affinity of CaMKII for CaM-C75-DANS was measured by titrating CaMKII into a solution of 100 nM (no nucleotide) or 25 nM (with ADP) CaM-C75-DANS in 25 mM MOPS pH.7.0, 150 mM KCl, 0.5 mM CaCl2, 0.1 mg/ml BSA with or without 5 mM MgCl2 and 0.25 mM ADP. After each addition of CaMKII, fluorescence was measured in a PTI fluorimeter, with excitation at 345 nm and emission monitored at 465 nm (5 nm bandwidths). For each point, fluorescence was averaged over a one-minute period. The fraction of CaM bound was calculated using the equation, , where I is the measured fluorescence intensity, Ifree is the fluorescence intensity in the absence of CaMKII, and Ibound is the fluorescence intensity of the fully bound CaM. The x-axis in each graph is the free concentration of CaMKII at each point, calculated by subtracting the concentration of bound CaM from the total concentration of CaMKII subunits. Each titration was performed at least three times, and the data was fit to the Hill equation (, where h is the Hill coefficient).
RESULTS
The influence of nucleotide on CaM binding to CaMKII
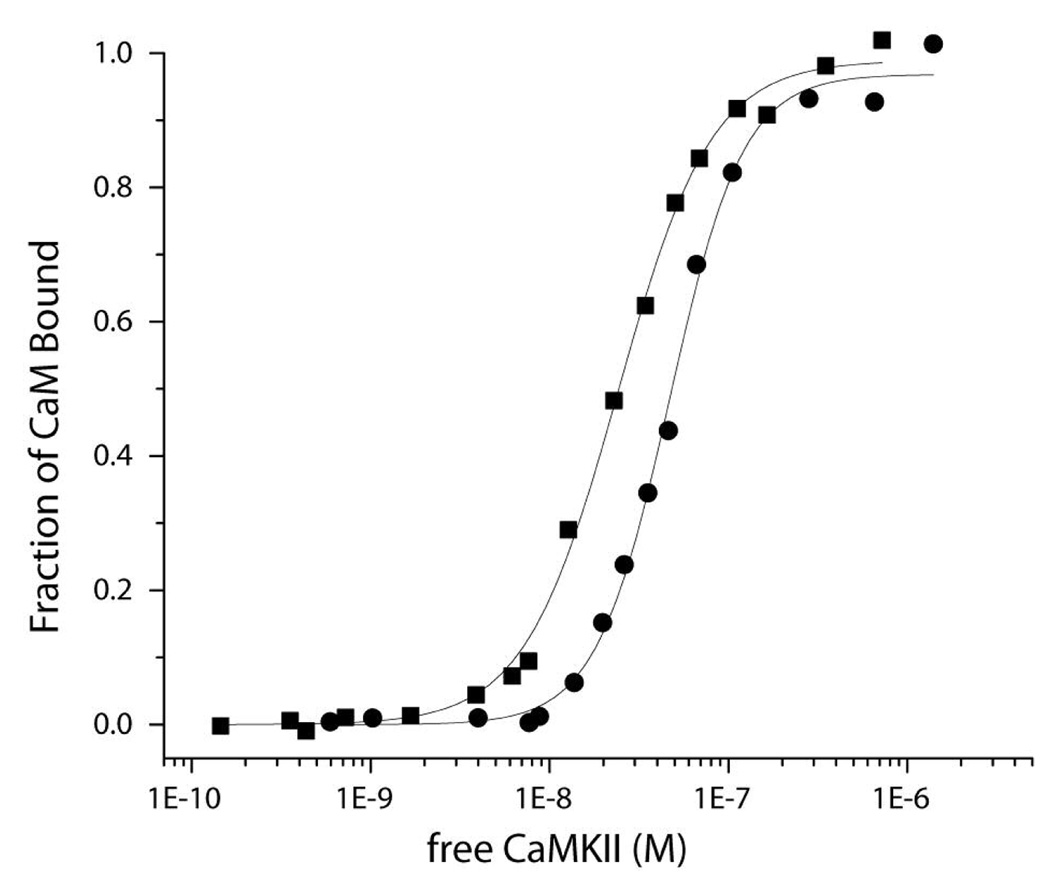
We previously established that the binding of fluorescently labeled CaM-C75-DANS to CaMKII in the absence of nucleotides exhibited a Hill coefficient of ~2 suggesting cooperativity in binding (7). Given the site of attachment of the fluorescent moiety, the changes reported by CaM-C75-DANS are likely dominated by associations of the N-terminal domain with target proteins. Torok et al. (8) established that under similar reaction conditions, CaM bound to CaMKII in an extended form, but when nucleotide was added CaM underwent a conformational rearrangement suggesting both lobes of CaM were now target associated. To further assess the impact of nucleotide binding on CaM interactions with CaMKII we completed a series of steady-state and stopped-flow fluorescence studies. Figure 1 shows a comparison of the steady state fluorescence of CaM-C75-DANS with CaMKII in the presence and absence of ADP. As reported previously, in the absence of nucleotides, the binding affinity of CaMKII using this fluorescent reporter for CaM is 62 ± 25 nM and the Hill coefficient was 1.9 ± 0.1. In the presence of a saturating concentration of ADP, the Kd for binding decreased to 19 ± 4 nM and the Hill coefficient dropped to 1.3 ± 0.3. These values are very similar to those reported by Torok et al. (8) who reported the Kd determined from kinetic measurements to be 74 nM and 17 nM in the absence and presence of nucleotide, respectively. Interestingly, there is also a decrease in cooperativity in CaM-binding to CaMKII in the presence of nucleotide.
Figure 1.
Steady-state fluorescence to assess the role of nucleotide binding of CaM to CaMKII. Shown are the binding curves produced by adding increasing concentrations of CaMKII into a cuvette containing CaM-C75-DANS (100 nM for no nucleotide and 25 nM with nucleotide) in 25 mM MOPS, pH 7.0, 150 mM KCl, 0.5 mM CaCl2 containing 0.1 mg/ml bovine serum albumin. The fraction of CaM bound is plotted against the concentration of free CaMKII. The black dots are titrations in the absence of ADP and the black squares are in the presence of 5 mM MgCl2 and 0.25 mM ADP. The fits are to the Hill equation as described in Methodology.
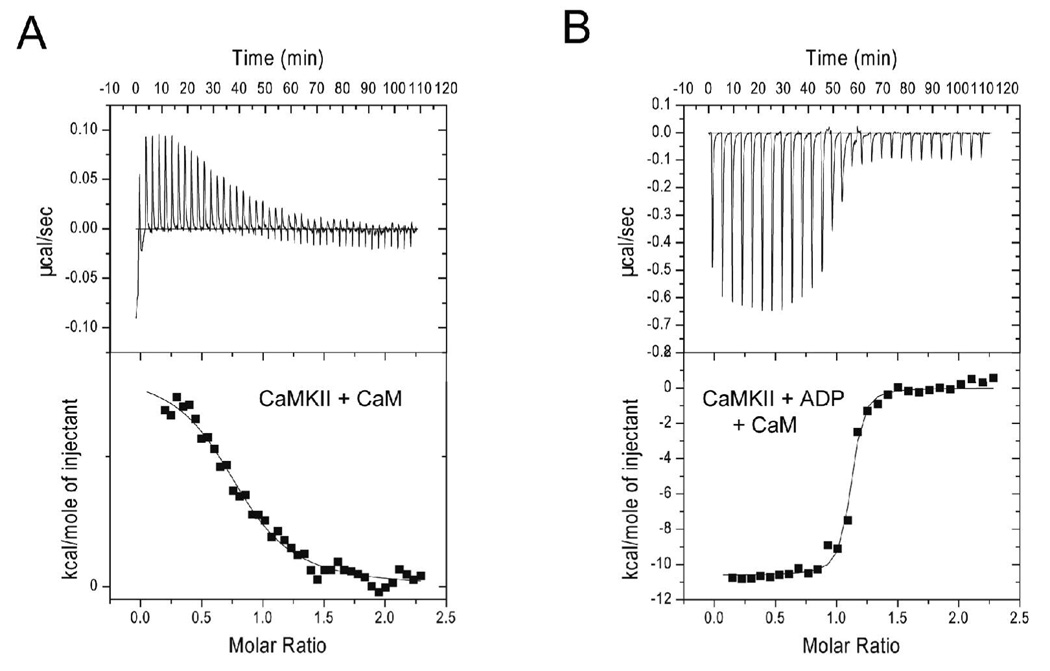
We confirmed that nucleotide binding increases the binding of CaM to CaMKII by analyzing their interaction with ITC. Figure 2 shows a representative example of such ITC results of CaM-binding to CaMKII without (panel A) and with (panel B) ADP and the summary data are presented in Table 1. The binding affinity measured in the presence of nucleotide was 35 ± 17 nM consistent with the titration curve generated with CaM-C75-DANS. The stoichiometry of binding (N) was somewhat variable but on average was very close to 1 (0.98 ± 0.2 mol of CaM/mol of CaMKII subunit). The Kd of Ca2+/CaM binding to CaMKII in the absence of nucleotide were surprisingly poor with a binding affinity of 1.3 ± 0.6 µM. The stoichiometry of binding was lower than in the absence of nucleotide, but close to 1 (0.83 ± 0.14 mol of CaM/mol of CaMKII subunit). It is not immediately apparent why the ITC results indicate a Kd for Ca2+/CaM binding nearly 20-fold worse than that reported using fluorescence from CaM-C75-DANS. However, given that IAEDANS is attached to the N-domain near the linker region of CaM this fluorescent technique may be reporting binding predominantly of the N-lobe. It is also plausible that the hydrophobic IAEDANS moiety increases the strength of interaction between CaM and CaMKII consistent with a modest increase in the capacity of CaM-C75-DANS to activate CaMKII (28). ITC does not depend on an altered form of CaM to report the binding interaction and is less prone to such potential artifacts. The weak binding reported for CaM in the absence of nucleotide would be consistent with interactions of CaM predominantly via one lobe additionally supported by the finding that CaM initially binds to CaMKII in an extended conformation (8).
Figure 2.
Calorimetric data for CaM-binding to CaMKII. The data shown represents the binding of wtCaM to CaMKII in the absence (A) or presence (B) of 1 mM ADP at 15°C. The top panel shows the raw power output (µcal/sec) per unit time. The bottom panel shows the integrated data. The solid line through the data represents the nonlinear least squares best fit of the data to a 1:1 binding model as described in Materials and Methods. The best fit parameters for these titration are the following: CaM with no ADP: N = 0.823, Kd = 1.9 µM, ΔH = 1.7 kcal/mol. CaM with ADP: N = 1.08, Kd = 38 nM, ΔH = −11.1 kcal/mol.
Table 1.
ITC data for binding of CaMKII to variants of CaM.
| Protein | N1 | Kd1 (µM) | ΔH1 (kcal/mol) | N2 | Kd2 (µM) | ΔH2 (kcal/mol) |
|---|---|---|---|---|---|---|
| CaMa | 0.83±0.14 | 1.32±0.57 | 1.9 ± 0.2 | - | - | - |
| CaM + ADPa | 0.98±0.20 | 0.035±0.17 | −12.8 ± 0.43 | - | - | - |
| CaMCC + ADPa | 0.84±0.20 | 0.37±0.07 | −6.5 ± 1.9 | . | - | - |
| CaMNN + ADPb | 0.26±0.02 | 0.064±0.004 | 0.70 ± 0.60 | 0.62 ± 0.08 | 0.34 ± 0.05 | −18.1 ± 0.2 |
ITC experiments with CaMKII were accomplished at 15°C in 25 mM HEPES, pH 7.4, 150 mM KCl, 1 mM CaCl2, 1 mM MgCl2 with or without nucleotide as indicated. Data is presented as averages ± SD with N of 3–7 for each condition.
Parameters obtained from a single binding mode model.
Parameters obtained from a multiple binding mode model.
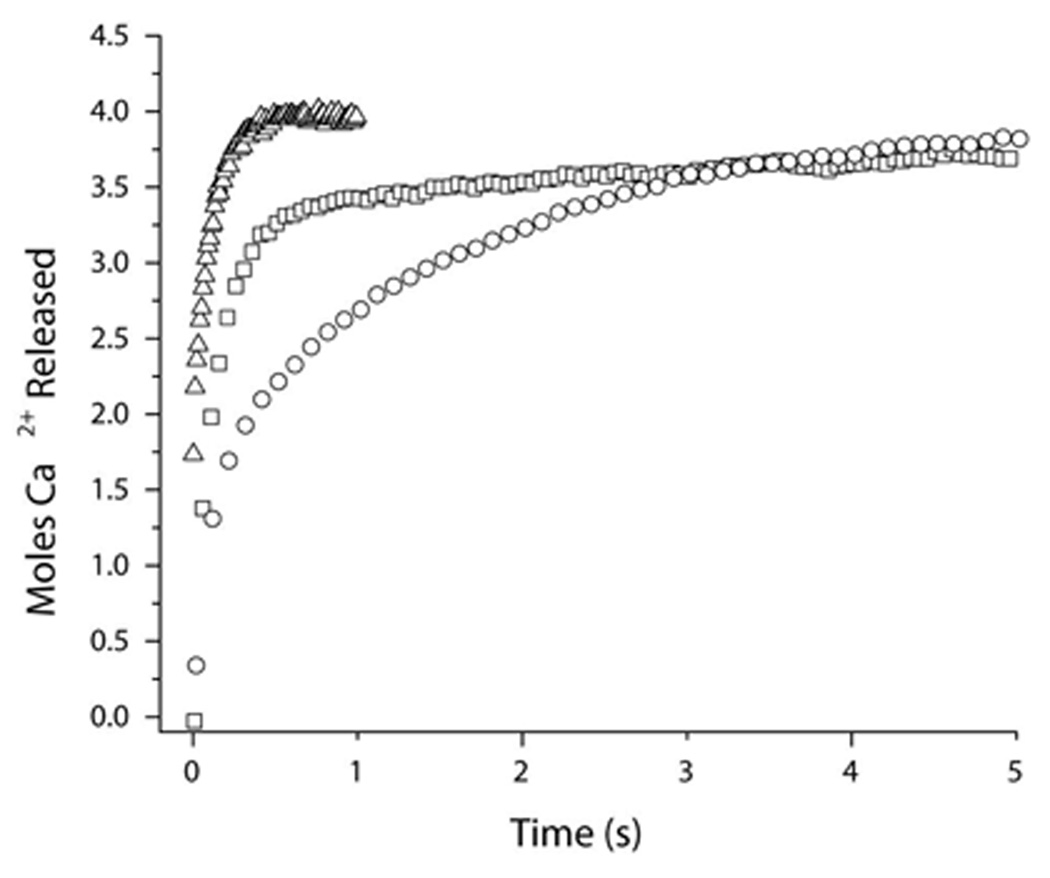
We demonstrated previously that when Ca2+/CaM binds to CaMKII in the absence of nucleotide or when phosphorylated at Thr-286 that there is a dramatic increase in CaM’s Ca2+-binding affinity (16). These effects can be assessed using the fluorescent Ca2+-chelator quin-2. As Ca2+ dissociates from CaM it binds to quin-2 producing an increase in fluorescence and the rates of dissociation can be quantified using stopped-flow fluorescence. Figure 3 demonstrates the relative impact of CaMKII in the absence and presence of ADP on Ca2+-dissociation from CaM. The Ca2+ dissociation from CaM in the absence of CaMKII shows only the slow release (9.1 s−1) of 2 mol of Ca2+ from the C-terminal lobe of CaM. The dissociation of Ca2+ from the N-terminal lobe is too fast to be measured in the absence of target. In the presence of CaMKII, ~3.3 moles of Ca2+ could be detected that are released at two different rates. Approximately 2.7 moles of Ca2+ are released at a rate of 9.9 s−1, and ~ 0.6 moles are released at 0.8 s−1. In the presence of ADP, there is a further slowing of Ca2+-dissociation from CaM. Approximately 1.5 moles of Ca2+ are released at 6.9 s−1, and 2.6 mol are released at 0.4 s−1 (see Table 2). This data supports the hypothesis that in the presence of ADP, CaM assumes tighter binding to CaMKII that additionally influences CaM’s affinity for Ca2+. It is interesting that 2.9 mol of Ca2+ are released from CaM when bound to CaMKII at a similar rate to the release of Ca2+ from the C-terminal lobe of CaM not in complex with CaMKII. This result would be consistent with a model where the Ca2+-bound N-terminal domain binds to CaMKII while the C-terminal lobe interacts little if at all with the enzyme as previously suggested by Torok et al. (8). These data imply an important asymmetry in the sequential manner in which the lobes of CaM initially interact with CaMKII, with the Ca2+-saturated N-lobe appearing to bind first to the kinase.
Figure 3.
Stopped-flow analysis of Ca2+-dissociation from CaM in the presence of CaMKII with or without ADP. Ca2+ dissociation from CaM was measured by monitoring changes in Quin-2 fluorescence in an Applied Photophysics Ltd. model SV.17MV stopped-flow spectrofluorimeter at 22°C. 2 µM CaM, 20 µM CaCl2 in 20 mM MOPS pH 7.0, 100 mM KCl, 1 mM MgCl2, without (triangles) or with 2 µM CaMKII (squares) or CaMKII + 1 mM ADP (circles) was rapidly mixed with 150 µM Quin-2. Quin-2 was excited at 334.5 nm and emission was monitored through a 435 nm cut-on filter. The representative traces shown are averages of 5 runs each. The data were converted to mol Ca2+ released/mol CaM by assuming that the amplitude of Ca2+ release from CaM alone was equal to the release of 2 mol of Ca2+.
Table 2.
Ca2+-dissociation Rates from CaM analyzed by Quin-2 and Stopped-flow Fluorimetry
| Rate 1 | # Ca | Rate 2 | # Ca | Total # Ca | |
|---|---|---|---|---|---|
| CaMa | 9.09 ± 0.60 | 2 | TFTM | 2 | |
| CaM + CaMKIIb | 9.86 ± 0.81 | 2.70 ± 0.33 | 0.82 ± 0.38 | 0.57 ± 0.21 | 3.27 ± 0.26 |
| CaM + CaMKIIb + ADP | 6.87 ± 1.26 | 1.55 ± 0.16 | 0.43 ± 0.05 | 2.62 ± 0.24 | 4.17 ± 0.30 |
Rates and amplitudes were fit with a onea - or twob-rate binding model. Amplitudes reported are the mol of Ca2+ recovered assuming 2 mol of Ca2+ were released from CaM in the absence of target. TFTM (too fast to measure) means that fraction of the reaction fell below the temporal resolution of our instrument (1.8 ms) and could not be reliably detected.
Role of the isolated N- and C-terminal lobes in the activation and autophosphorylation of CaMKII
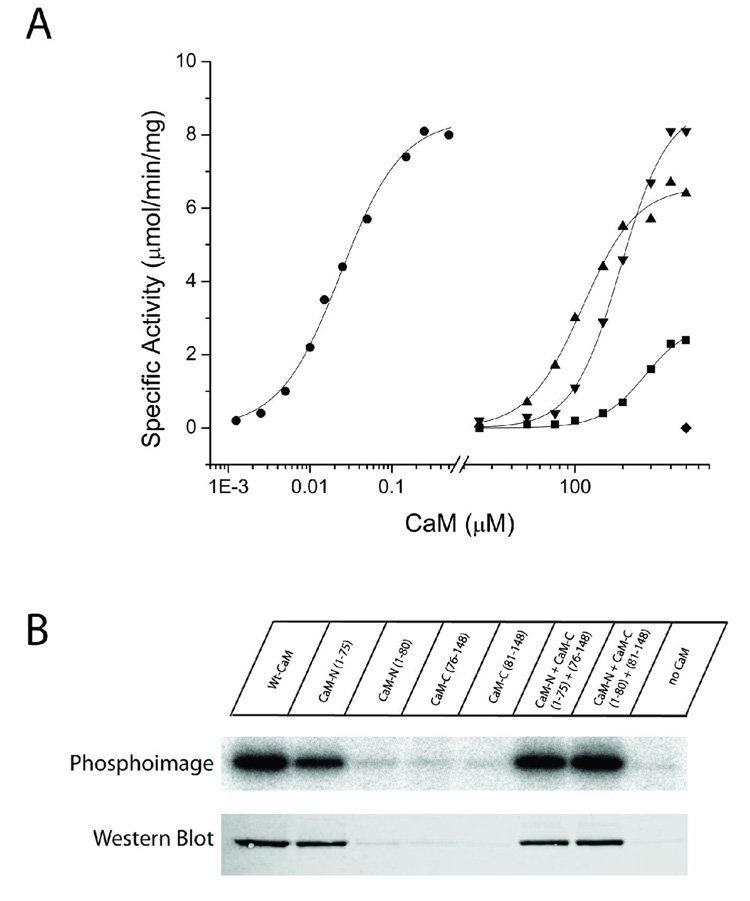
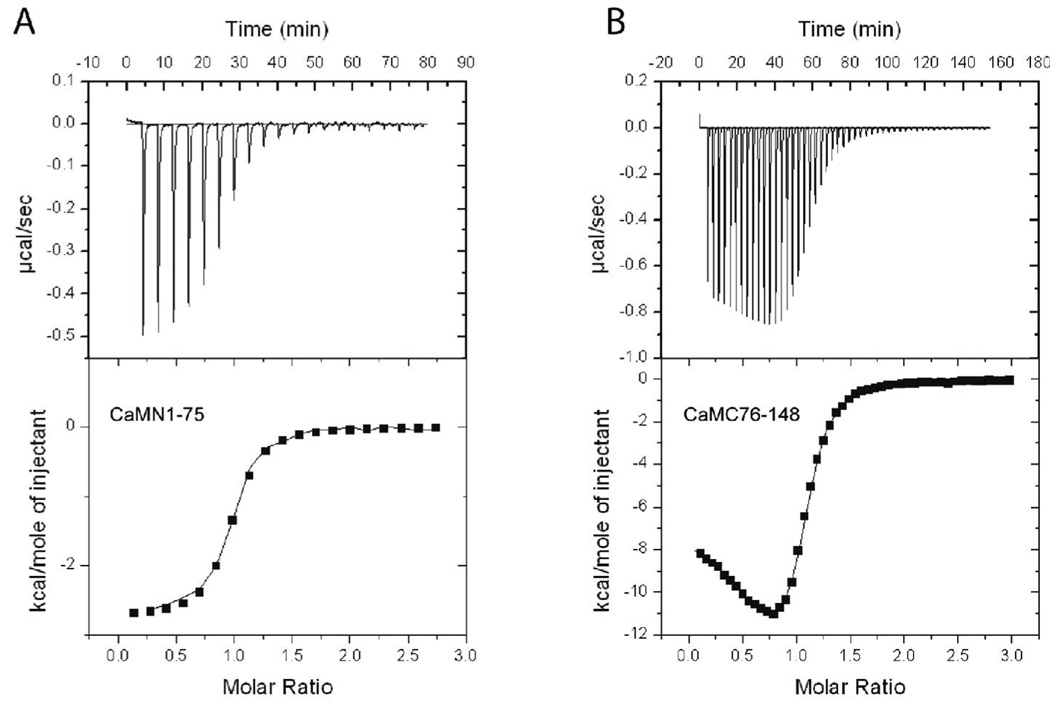
To assess the role of the N- and C-terminal lobes of CaM in their interaction with CaMKII genetically engineered domains of CaM were utilized. In experiments examining CaMKII activity towards a synthetic peptide substrate, only the CaMN1–75 fragment was shown to induce activity of CaMKII (Fig. 4A; Table 3). The response was dose-dependent with a half-maximal activation of approximately 290 µM; however, even at maximal concentrations (where activity appeared asymptotic) there was only 35% of maximal activity compared to wtCaM. Fits of the data to the Hill equation revealed a Hill coefficient of 2.8, indicating that the activation of CaMKII by the N-terminal domain exhibited significant cooperativity. Extending the domain to include 5 additional residues to create CaMN1–80 surprisingly eliminated the induction of CaMKII activity up to 500 µM. Amino acids 75–80 in CaMN1–80 were shown previously to interact with helix A of CaM that correlated with lowered Ca2+-binding affinity and increased thermal stability (33). The underlying reason of how these interactions would lead to a complete inability to activate CaMKII is presently not clear. All of our activation assays were accomplished at 4 mM Ca2+ to insure that all of the CaM fragments would be saturated with Ca2+. The C-terminal domains, CaMC76–148 and CaMC81–148 were also inactive up to 500 µM. Based on the differences in CaMKII activation between CaMN1–75 and CaMN1–80, it remains possible that further truncation might produce an active CaMC lobe by deleting additional residues beyond amino acid 81. We then assessed whether the isolated domains when mixed together could activate CaMKII. We found that both CaMN1–75 together with CaMC76–148 and CaMN1–80 with CaMC81–148 were capable of fully activating CaMKII. Both of these mixtures achieved maximal activation of the enzyme with the former pair exhibiting slightly lower half-maximal activation than the latter; 122 vs. 197 µM, respectively (Figure 4A and Table 3). Fits to the Hill equation showed that each pair activated the enzyme cooperatively with comparable Hill coefficients of approximately 2.6.
Figure 4.
Substrate phosphorylation and autophosphorylation induced by half-CaMs. A) Each CaM variant was assessed for its ability to activate CaMKII in reactions containing 25 mM HEPES, pH 7.4, 50 mM KCl, 0.4 mM DTT, 10 mM MgCl2, 4 mM CaCl2, 100 µM ATP, 50 µM syntide, and 2 µCi of 32P-ATP. Reactions were initiated by the addition of 10 ng of CaMKII, incubated for 1 min at 30°C, and terminated by spotting an aliquot on P81 paper and plunging the paper into phosphoric acid. Each reaction was performed in duplicate and separate experiments were repeated a minimum of 2 times. The CaM constructs shown are: wtCaM (circles), CaMN1–75 (squares), CaMN1–75 + CaM 76–148 (triangles), CaMN1–80 + CaMC81–148 (upside down triangles). CaMN1–80, CaMC76–148 and CaMC81–148 were inactive up to the indicated concentration (diamond). B) Each CaM variant was assessed for its ability to induce autophosphorylation of CaMKII. The reaction mixture contained 25 mM HEPES, pH 7.4, 50 mM KCl, 0.4 mM DTT, 10 mM MgCl2, 4 mM CaCl2, 100 µM ATP, and 2 µCi of 32P-ATP. Reactions were initiated by the addition of 100 ng of CaMKII and were allowed to react for 10 min on ice. The reactions were terminated by the addition of SDS-sample buffer and 40 ng of CaMKII from each reaction were loaded and run on a 10% SDS-PAGE gel. Following electrophoresis, staining and destaining, 32P incorporation was assessed by exposing the gel to a phosphor screen for 5 hr and then analyzing the screen on a Typhoon imaging system. Identical samples were also analyzed by Western blot with a rabbit polyclonal antibody specific for phospho-Thr286. The primary antibody was detected with an Alexa568-labeled anti-rabbit antibody and the signal detected using the fluorescence mode of the Typhoon imaging system.
Table 3.
Activation of CaMKII by Mutant CaM Constructs.
| CaM mutants | Activity | ||
|---|---|---|---|
| Vmax (µMol/min/mg) | K0.5 (µM) | Hill coefficient | |
| Wt-CaM | 8.25 ± 0.42 | 0.025 ± 0.003 | 1.12 ± 0.07 |
| CaM-NN | 8.14 ± 1.79 | 0.71 ± 0.06 | 1.39 ± 0.08 |
| CaM-CC | 7.42 ± 1.38 | 1.40 ± 0.18 | 1.21 ± 0.02 |
| CaM-N (1–75) | 2.82 ± 0.39 | 289.87 ± 14.99 | 2.84 ± 0.28 |
| CaM-N (1–80) | no activity | ||
| CaM-C (76–148) | no activity | ||
| CaM-C (81–148) | no activity | ||
| CaM-N (1–75) + CaM-C (76–148) | 7.25 ± 0.66 | 122.10 ± 10.79 | 2.56 ± 0.02 |
| CaM-N (1–80) + CaM-C (81–148) | 8.13 ± 0.64 | 197.27 ± 3.90 | 2.69 ± 0.22 |
Reactions were performed in duplicate and repeated 2–3 times. Experimental data were fit with the Hill equation (where ‘’a’’ is the Vmax, ‘’b’’ is the K0.5 and ‘’h’’ is the Hill coefficient). The values reported are means ± standard deviations of the fits from at least two curves.
We next investigated if these domains of CaM could induce autophosphorylation of CaMKII. These experiments were carried out at the maximum amount of CaM that could be achieved in this assay which was 500µM. An identical pattern for CaMKII autophosphorylation was produced as observed for enzyme activation. The N-terminal lobe, CaMN1–75, was the only lobe that by itself was sufficient to cause the autophosphorylation of CaMKII (Fig. 4B). Experiments with wtCaM were performed at 4°C, which limits autophosphorylation of CaMKII to predominantly Thr-286 and avoids other sites within the enzyme (e.g. Thr-305/306 or Thr-253). Using antibodies specific to phosphor-Thr-286, we confirmed that this site was phosphorylated (Fig. 4B). None of the other domains of CaM alone were able to active the enzyme. However, as with the substrate activity we found that both CaMN1–75 when mixed with CaMC76–148 and CaMN1–80 when mixed with CaMC81–148 both induced autophosphorylation of CaMKII that was confirmed to include Thr-286 with antibody staining (Fig. 4B). We can confidently conclude from these results that CaMKII activity and its autophosphorylation do not require that the two domains of CaM are covalently linked. They also reveal that CaMN1–75 contains unique amino acids that are necessary and sufficient for enzyme activation and autophosphorylation. It is possible that the C-domain of CaM has a reduced affinity for binding to CaMKII and that we simply could not achieve adequate concentrations for activation in our assays. Regardless, the data suggest that either through its affinity or specific amino acid interactions, the N-domain of CaM plays a critical role in CaMKII activation.
Titration calorimetry utilizing the N- and C-lobes of CaM
In order to better understand the interaction of the individual lobes of CaM with CaMKII, we also studied their binding to CaMKII using ITC. Experiments in which the lobes of CaM were titrated with full length CaMKII provided an insufficient heat signal to deduce conclusions. Hence, we studied binding of the lobes of CaM to a CaMKII-based peptide. It has previously been shown that the interaction of CaM with CaMKII-based peptides is a useful model for the different ways CaM can interact with full length CaMKII (32). For instance, CaM was previously shown to bind peptides based on the full CaM-binding domain of CaMKII (such as CaMKII(291–312) used here) with sub-picomolar affinity. Here we observed the interaction of CaMKII(291–312) with CaMN1–75, CaMN1–80 and CaMC76–148 to be dramatically weaker than with CaM (see Table 4), corroborating the weaker activation K0.5 values for these constructs compared to CaM determined above. The interaction of CaMN1–75 with CaMKII(291–312) at all temperatures studied (5, 15, 25, and 37°C) was well described by a single binding mode model and provided Kd values in the range of 100 to 1000 nM (Table 4). In contrast, ITC titrations of both CaMN1–80 and CaMC76–148 were not well described by a single binding mode model but were much better described by a two binding mode model (Fig 5). Like the data for CaMN1–75, both these constructs demonstrated their more potent Kd values in the range of 100 to 1000 nM (Table 4). Together, these experiments indicate 1) CaMN1–75, CaMN1–80, and CaMC76–148 all interact much more weakly with a CaMKII(291–312) peptide than full length CaM, 2) CaMN1–75 interacts with CaMKII(291–312) via a single binding mode but CaMN1–80 and CaMC76–148 are capable of interacting in multiple ways, and 3) the second mode of binding is weaker for CaMC76–148 compared with CaMN1–80. These results may explain why CaMN1–75 activates full length CaMKII but CaMN1–80 and CaMC76–148 can not. It is feasible that CaMN1–75 binds full length CaMKII using a single binding mode that is compatible with CaMKII activation while CaMN1–80 and CaMC76–148 bind in multiple ways, some of which are nonproductive for CaMKII activation and hence compete with the binding mode that is required for CaMKII activation.
Table 4.
ITC Data for Binding of CaMKII Peptide to Variants of CaM
| CaM protein | Temperature (°C) | Kd1 (nM) | ΔH1 (kcal/mol) | Kd2 (nM) | ΔH2 (kcal/mol) |
|---|---|---|---|---|---|
| Wild type | 25 | 0.00007 ± 0.00004 | −6.1 ± 0.8 | - | - |
| CaMN1–75a | 25 | 360 ± 60 | −2.5 ± 1.0 | ||
| CaMN1–80b | 25 | 330 ± 170 | −3.1 ± 1.5 | 280 ± 170 | −3.8 ± 1.9 |
| CaMC76–148b | 25 | 140 ± 33 | −11.8 ± 0.1 | 18600 ± 9500 | 5.4 ± 1.1 |
| CaMN1–75a | 5 | 280 ± 50 | 1.9 ± 0.4 | - | - |
| CaMN1–75a | 15 | 500 ± 80 | −0.2 ± 0.1 | - | - |
| CaMN1–75a | 37 | 710 ± 100 | −5.1 ± 0.5 | - | - |
| CaMN1–80b | 5 | 110 ± 27 | 3.9 ± 0.3 | 1270 ± 32 | 5.7 ± 0.1 |
| CaMN1–80b | 15 | 280 ± 240 | −0.3 ± 0.9 | 1140 ± 200 | 2.4 ± 0.9 |
| CaMN1–80b | 37 | 990 ± 580 | −6.5 ± 2.1 | 200 ± 170 | −8.5 ± 2.2 |
| CaMC76–148b | 5 | 120 ± 9 | −3.6 ± 0.4 | 36000 ± 9900 | 6.8 ± 0.6 |
| CaMC76–148b | 15 | 160 ± 63 | −7.7 ± 0.7 | 28500 ± 20000 | 7.6 ± 3.4 |
| CaMC76–148b | 37 | 100 ± 44 | −15.5 ± 0.3 | 9430 ± 4800 | 1.7 ± 0.4 |
Uncertainties represent the standard deviation of independent experiments.
Parameters obtained from a single binding mode model.
Parameters obtained from a multiple binding mode model.
Figure 5.
Calorimetric data for binding of lobes of CaM to CaMKII peptide. The data shown represent binding of (A) CaMN1–75 to CaMKII(291–312) at 25°C and (B) CaMC76–148 to CaMKII(291–312) at 25°C. The top panel shows the raw power output (µcal/sec) per unit time. The bottom panel shows the integrated data. The solid line through the data represents the nonlinear least squares best fit of the data to a 2:1 binding model as described in Materials and Methods. The best fit parameters for these titration are the following: CaMN1–75: Kd = 360 nM, ΔH = −2.5 kcal/mol. CaMC76–148: Kd1 = 120 nM, Kd2 = 11900 nM, ΔH1 = −11.8 kcal/mol, ΔH2 = 4.7 kcal/mol.
Chimeras composed of two N- or two C-lobes of CaM in the activation and autophosphorylation of CaMKII
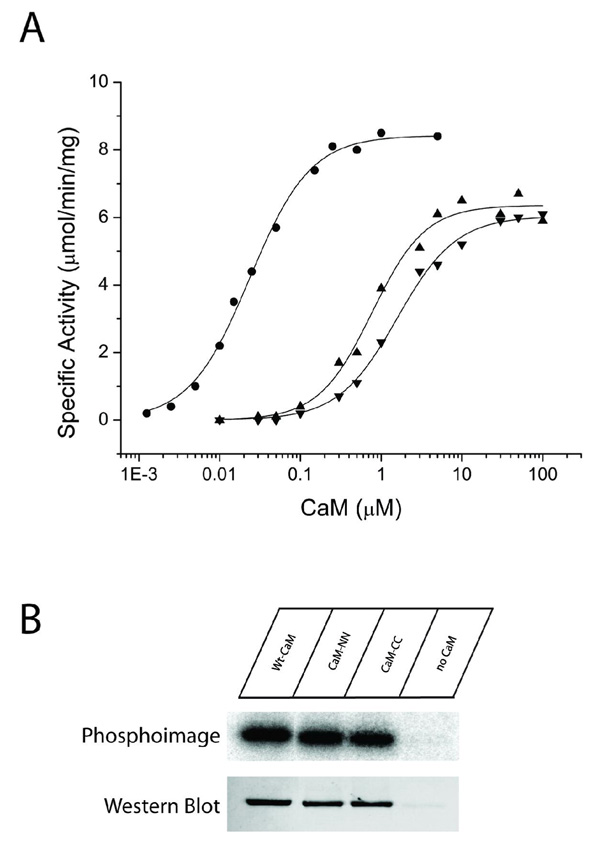
We found that the isolated C-lobe of CaM was incapable of supporting activation and the autophosphorylation of CaMKII. One possible reason for this observed inactivation would be that the isolated domain had a binding affinity too weak to achieve the necessary occupancy of the CaM-binding sites on CaMKII. We investigated this possibility further using chimeras composed of two copies of either the C-lobe (CaMCC) or the N-lobe (CaMNN) of CaM which capitalize on the increased effective concentration of the second lobe following binding of the first. We first assayed these chimeras for their ability to activate CaMKII. The dose-response relationships shown in Fig. 6 demonstrate both CaMNN and CaMCC activate CaMKII (summarized in Table 3). CaMNN had a half-maximal activation of 0.7 µM, CaMCC had a half-maximal activation of 1.4 µM, and wtCaM had a half-maximal activation of 0.025 µM. Fits of the data to the Hill equation revealed Hill coefficients of 1.2, 1.4, and 1.1 for CaMNN, CaMCC, and wtCaM, respectively.
Figure 6.
Substrate phosphorylation and autophosphorylation induced by full-length CaM variants. A) Each CaM variant was assessed for its ability to activate CaMKII exactly as previously described in figure legend 3. The CaM variants shown are: wtCaM (circles), CaMNN (triangles), and CaMCC (upside down triangles). B) Each CaM variant was assessed for its ability to induce autophosphorylation of CaMKII exactly as previously described in figure legend 3. The top panel shows an autoradiograph of 32P incorporation and the bottom panel is a blot immuno-stained with a phospho-Thr286 specific monoclonal antibody.
We also examined the capacity of CaMNN and CaMCC to induce autophosphorylation of CaMKII. Both mutant CaMs (at 100 µM final concentrations) induced autophosphorylation and at least one of the sites autophosphorylated was confirmed to be Thr-286 (Fig. 6B). In total, these data demonstrate that the C-terminal lobe, when physically combined with a second C-terminal lobe is fully capable of activating CaMKII. This implies that the C-domain of CaM is capable of binding to both of the target sites on a subunit of CaMKII to induce activation. This conclusion can also be drawn from the CaMNN results. A probable explanation for the lack of activation of CaMKII by the C-terminal lobe alone (CaMC76–148 or CaMC80–148) is poor binding affinity or that they bind in a mode that does not lead to activation.
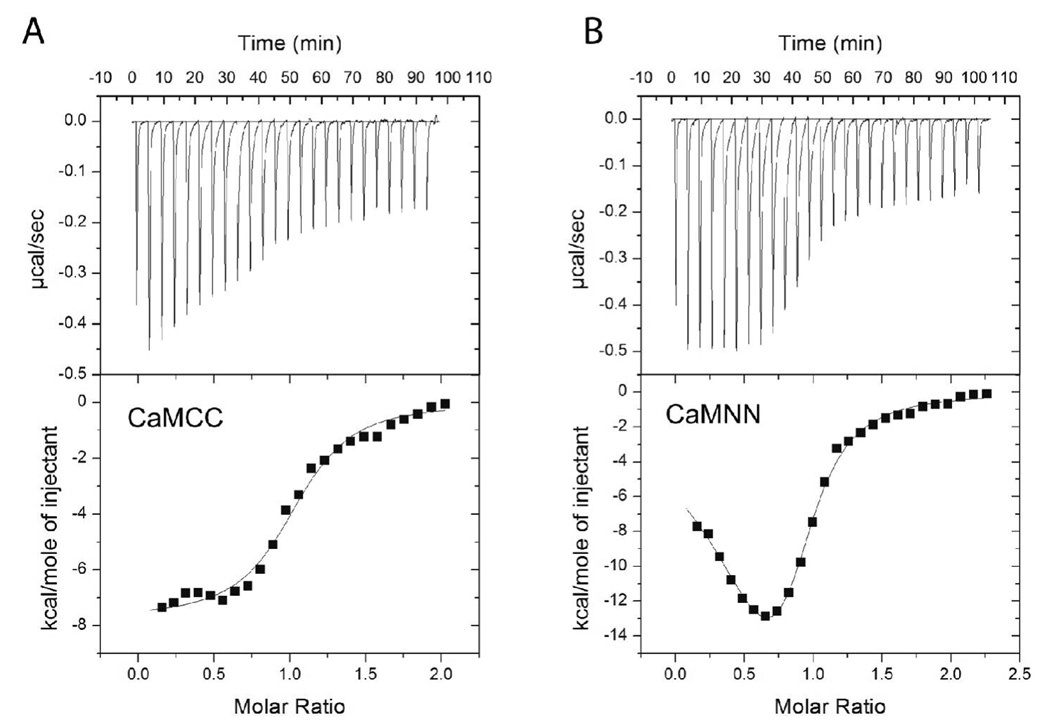
The activation curves showed that the rank order to achieve half-maximal activity was wtCaM, CaMNN, and CaMCC. These results could be explained either by decreased binding affinities, or subtleties in the specific interactions between the two domains required for most effective CaMKII activation. We therefore investigated the binding affinities of these forms of CaM using ITC. Because of the difficulties in maintaining soluble CaMKII at greater than 20 µM, we accomplished these experiments in the presence of 1 mM ADP to increase the overall binding affinity of CaM to CaMKII as described in Fig. 2. Example results are shown in Fig. 7 and the summary data can be found in Table 1. The data revealed that CaMNN and CaMCC, like wtCaM, bound with close to a 1:1 binding stoichiometry (subunit concentration). The data also revealed that CaMCC bound to CaMKII with a Kd of 370 ± 7 nM, while CaMNN bound with complicated properties requiring a two mode model. The tight binding mode had a Kd of 64 ± 4 nM while the weaker mode had a Kd of 340 ± 5 nM. This makes the rank order of binding affinities of these three forms of CaM, wtCaM>CaMNN>CaMCC, identical to the rank order of their ability to activate CaMKII.
Figure 7.
Calorimetric data for binding of CaMCC and CaMNN to CaMKII. The data shown represents the binding of (A) CaMCC or (B) CaMNN to CaMKII in the presence of 1 mM ADP at 15°C. The top panel shows the raw power output (µcal/sec) per unit time. The bottom panel shows the integrated data. The solid line through the data represents the nonlinear least squares best fit of the data to a one site binding model (for CaMCC) or a two site binding model (CaMNN) as described in Materials and Methods. The best fit parameters for these titration are the following: CaMCC: N = 0.979, Kd = 360 nM, ΔH = −7.3 kcal/mol. CaMNN: N1 = 0.36, N2 = 0.61, Kd1 = 18 nM, Kd2 = 360 nM, ΔH1 = −6.3 kcal/mol, ΔH2 = −14.7 kcal/mol.
Characterization of the Ca2+-binding properties of the CaM constructs
We assumed in interpreting the experiments above that all of the variants of CaM retained their ability to bind to Ca2+. However, it was possible that construction of truncations or chimeras of CaM may have disrupted Ca2+-binding. We therefore used stopped-flow fluorimetry to characterize the Ca2+-dissociation properties of the CaM constructs used in the present study and the results are summarized in Table 5. Because the Ca2+-binding affinity might have been poor in the CaM constructs, we monitored quin-2 induced dissociation in the absence and presence of the high affinity CaM-binding peptide which is known to increase the Ca2+-binding affinity of CaM (21). For each CaM construct, the amplitude associated with each rate was normalized by setting the total amplitudes to 100% in the presence of peptide and then calculating the percent contribution for each component. For example, wtCaM in the presence of peptide required a two component fit so the total amplitudes of both components were set to 100% (equivalent to 4 mol total Ca2+). Then the relative contribution of each component was adjusted to mol of Ca2+ recovered as summarized in Table 5. For wtCaM in the absence of peptide, only a one component fit was required which showed a rate of dissociation of 10 s−1, typical of that reported for Ca2+-dissociation from the C-terminal lobe of CaM (21). The amplitude of this single component was approximately equal to that of the C-terminal lobe in the presence of peptide. Dissociation of Ca2+ from the N-terminal domain of wtCaM is too fast to measure under the present experimental conditions unless peptide is present. CaMNN in the presence of peptide showed two components, one with a rate of 6.9 s−1 and the second with a rate of 0.35 s−1 with normalized amplitudes of 3.7 and 0.3, respectively. Surprisingly, in the absence of peptide, we could still recover significant amplitude of signal that also showed two rates; one of 226 s−1 and the other at 8.8 s−1 with normalized amplitudes of 0.7 and 0.5, respectively. This was unexpected, because Ca2+-dissociation from the N-terminal domain of wtCaM or from the N-domain alone (see below) is too fast to measure under the present conditions. This indicates that the Ca2+-binding properties of CaMNN are altered from that theoretically predicted from the fusion of two N-terminal domains. CaMCC also showed altered properties. In the presence of peptide, two measured rates of 1.3 and 0.32 s−1 with amplitudes of 1 and 3, respectively were determined. In the absence of peptide, two rates were measured - 16.15 and 3.6 s−1 with amplitudes of 3.2 and 0.7, respectively. The theoretically predicted result for CaMCC, if its binding properties were unperturbed, would have been a rate of 10 s−1 with an amplitude of 4 mol of Ca2+ released.
Table 5.
Quin-2 induced rates of Ca2+ dissociation from CaM constructs
| Construct | Rate 1 (s−1) | Amp. 1 (mol of Ca2+) | Rate 2 (s−1) | Amp. 2 (mol of Ca2+) |
|---|---|---|---|---|
| wtCaM | TFTM | 10 | 1.8 | |
| wtCaM + pep | 9.97 | 1.8 | 0.19 | 2.2 |
| CaMNN | 226 | 0.7 | 8.8 | 0.5 |
| CaMNN + pep | 6.9 | 3.7 | 0.35 | 0.3 |
| CaMCC | 16.15 | 3.2 | 3.6 | 0.7 |
| CaMCC + pep | 1.3 | 1.0 | 0.32 | 3.0 |
| CaM76–148 | 13.4 | 2.1 | ||
| CaM76–148 + pep | 0.56 | 2 | ||
| CaM81–148 | 43.3 | 1.9 | ||
| CaM81–148 + pep | 1.5 | 2 | ||
| CaM1–75 | TFTM | |||
| CaM1–75 + pep | 10.5 | 2 | ||
| CaM1–80 | TFTM | |||
| CaM1–80 + pep | 11.4 | 2 | ||
Rates and amplitudes were fit with a one- or two-rate binding model as necessary. Amplitudes reported are the mol of Ca2+ released per mol of protein assuming 100% of the amplitude was recovered in the presence of peptide. TFTM (too fast to measure) means that fraction of the reaction fell below the temporal resolution of our instrument (1.8 ms) and could not be reliably detected.
We also measured Ca2+-dissociation rates from the half-CaMs using an identical protocol (Table 5). The C-terminal domain CaMC76–148 showed a single dissociation rate of 13.4 s−1 in the absence of peptide, close to what would be predicted from the C-terminal domain of wtCaM (10 s−1). Deleting amino acids 76–80 (CaM81–148) produced a significant increase in the rate of Ca2+-dissociation to 43.3 s−1. Clearly, subtle deletions in the linker region of CaM can significantly impact the Ca2+-binding properties of the molecule. CaMN1–75 and CaMN1–80 were not measurably different from each other, but the release of Ca2+ in the absence of peptide was too fast to measure, so subtle differences as detected above for the isolated C-terminal domain might also exist, but could not be assessed in the present experimental set-up. From this data, we conclude that the lack of activation of the CaMN1–80, CaMC76–184, and CaMC81–148 cannot be ascribed to a lack of Ca2+-binding.
DISCUSSION
Autophosphorylation of CaMKII conveys unique properties on the kinase including Ca2+-independent activity and CaM-trapping. It is well established that autophosphorylation occurs through an intersubunit reaction mechanism within each holoenzyme (6, 29), and that Ca2+/CaM binding must occur between neighboring subunits within a holoenzyme (6). It is these properties that lead to the enzyme’s unique ability to decode the frequency of Ca2+ spikes in neurons (34). The rate of autophosphorylation is estimated to be ~20 s−1 at 37°C (29) which is faster than the estimated rate of Ca2+/CaM dissociation from unphosphorylated CaMKII (1–2 s−1)(4, 8, 28). Given that Ca2+/CaM may well be limiting in neurons, Ca2+/CaM-association and dissociation from CaMKII become the rate limiting steps in autophosphorylation, and understanding the mechanistic details of CaM interacting with CaMKII is critical.
We have shown that one important factor that determines the affinity of Ca2+-CaM-binding to CaMKII is nucleotide binding. Our results are very similar to those reported by Torok et al. (8) who identified that ADP binding to the kinase produces a significant increase in steady-state binding affinity that is reflected in a decreased rate of Ca2+/CaM dissociation. Previous work identified that Ca2+/CaM-binding to CaMKII increased the binding affinity of nucleotides (35). Based on free energy coupling, it is not surprising that the presence of ADP also increases Ca2+/CaM binding affinity. An important issue is how this increased binding affinity impacts Ca2+/CaM interactions with unphosphorylated CaMKII in the intracellular setting. Intracellular nucleotide concentrations are several mM, well beyond what is needed to occupy the nucleotide binding site of CaMKII. In this circumstance, the relevant binding affinity for Ca2+/CaM is the 12 nM value, not the 60–120 nM value in the absence of nucleotide. Tzortzopoulus and Torok (36) reported that the binding affinity of Ca2+/CaM for CaMKII incapable of phosphorylation of Thr-286 (an Ala-286 mutant) was 4.6 nM in the presence of ATP, supporting that the binding affinity of CaMKII for Ca2+/CaM inside cells is much lower than previously considered. This increased binding affinity is largely due to slowed dissociation of Ca2+/CaM and impacts the lifetime of the Ca2+/CaM-CaMKII complex (4, 8, 28).
Torok et al. (8) also identified that Ca2+/CaM-binding to unphosphorylated CaMKII left CaM in an extended conformation, presumably because one of the two domains was unable to make appropriate contacts with the CaM-binding domain of CaMKII. We discovered that the Ca2+-dissociation rates of the C-terminal domain of CaM were largely unaffected when bound to CaMKII, suggesting that CaM is binding to unphosphorylated CaMKII via its N-lobe. In the presence of ADP, the two domains of CaM collapse indicating that both domains are interacting with their target binding sites on CaMKII (8). Consistent with this result is our finding that the Ca2+-dissociation rates from both lobes of CaM are slowed when bound to CaMKII in the presence of ADP. These results are consistent with a kinetic scheme where the N-terminal lobe of CaM binds to CaMKII first, followed by nucleotide binding and subsequent exposure of sites on CaMKII that can productively interact with the C-domain of CaM.
We evaluated the role of the individual domains of CaM in their interaction and activation of CaMKII with a number of engineered variants. Using this strategy it was possible to produce quantities of the isolated domains where they could be used at several hundred micromolar concentrations. These concentrations were required to reveal several interesting findings with these half-CaMs. We produced two versions of half-CaMs; one pair involved amino acids 1–75 and 76–148 and the other pair included amino acids 1–80 and 81–148. Both pairs of these half-CaMs were capable of fully activating CaMKII with half-maximal activation at concentrations of ~150–200 µM. Both pairs also induced autophosphorylation of CaMKII. We conclude that there is no requirement for the domains of CaM to be physically tethered to activate CaMKII or to induce autophosphorylation.
When tested individually, only the N-terminal domain fragment of 1–75 was found to activate CaMKII. The half-maximal concentration for activation was 290 µM, but even up to 500 µM CaMN1–75 could not maximally activate the kinase. This domain by itself could also lead to CaMKII autophosphorylation. Given this observation, it was somewhat surprising that the CaMN1–80 fragment showed no capacity for activating CaMKII even though when mixed with CaMC81–148 it was capable of supporting activity. This suggests that in the absence of the C-terminal lobe, the interaction of CaMN1–80 appears to sterically inhibit the binding of a second CaMN1–80 molecule. An alternative possibility is that CaMN1–80 adopts a conformation different from CaMN1–75 that either alters its ability to bind Ca2+ or alters its capacity to bind to the target, perhaps forming an autoinhibited conformation. In fact, Faga et al., reported that amino acids 76–80 in CaMN1–80 fold back and interact with the N-domain and alter its Ca2+ binding properties and its thermal stability (33). However, since CaN1–80 with CaM81–148 is fully capable of activating CaMKII, it is difficult to envision how these latter effects could explain the data.
Half-CaMs have been used previously to examine the activation of other CaM-dependent enzymes. The half-CaMs were most often produced via proteolytic digestion of wtCaM with subsequent purification of the resulting fragments (37, 38). A general conclusion drawn from these studies is that each CaM-binding enzyme exhibits unique interactions with the isolated lobes of CaM. For example, myosin light chain kinase (MLCK) from smooth or skeletal muscle and neuronal nitric oxide synthase (nNOS) also appear to bind through an ordered mechanism, although in this case the C-terminal domain of CaM binds first followed by the N-terminal domain (37). However, each of these enzymes also exhibited unique properties when interacting with the isolated N- and C-domains of CaM. In the case of skeletal muscle MLCK, the C-terminal domain of CaM alone was capable of producing 65% maximal activation but only 10% of activation was detected with smooth muscle MLCK. Either lobe independently was found to activate phosphorylase kinase with the C-terminal lobe being more effective (39). In contrast, neither lobe was sufficient to induce activity of nNOS (26, 37). An important point to note is that not all of these studies extended the concentration of the half-CaM to the extent we did in the present study. So, lack of activation could relate to insufficient concentrations of the half CaMs. Like with MLCK, maximal CaMKII activation appears to require both lobes of CaM. However, unlike MLCK which appears to bind CaM with an ordered mechanism of C-terminal lobe followed by the N-terminal lobe, the present results with CaMKII support the opposite sequence of binding.
A reasonable point of concern with the isolated lobes of CaM is that without the linker domain they could bind to CaMKII in many possible orientations, some potentially inhibitory, thus complicating interpretation of the results. We therefore examined chimeras created with two N- or two C-domains for their ability to activate CaMKII. As anticipated, CaMNN activated CaMKII and induced autophosphorylation; however, CaMCC also activated CaMKII with a half maximal activation only twofold higher than CaMNN. These data indicate that both the N- and C-domains have the necessary contact sites to interact with and activate CaMKII if provided in the appropriate orientations. The ITC results did reveal that CaMCC had a binding affinity for CaMKII (in the presence of ADP) poorer than CaMNN which indicates that the N-domain binding affinity is greater than the C-domain. We can thus interpret the inability of the isolated C-terminal domain of CaM to activate the kinase as either a poor binding affinity, or because it binds in an inappropriate orientation on CaMKII, producing an inhibitory effect. This inhibition can be relieved by the presence of the N-domain either because binding of the N-domain provides an increase in affinity for binding of the C-lobe or because it forces the C-domain out of an unproductive/inhibitory interaction with CaMKII. The results with the chimeric CaMs are distinct from those reported with other CaM-dependent enzymes. CaMNN was shown not to activate either skeletal or smooth muscle MLCK while CaMCC was active (24). In contrast, CaMNN was shown to activate neuronal NOS, while CaMCC was inactive against this target (24, 26). CaMKII appears to be the most promiscuous of this subset of CaM-dependent targets in being activated by both CaMCC and CaMNN.
Through elegant computational strategies, Shifman et al. (40) recently reported the design of mutants of CaM deficient in Ca2+-binding at the N-terminus (sites 1 and 2) or C-terminus (sites 3 and 4) that were additionally engineered to maintain a closed conformation. They showed that both of these constructs would bind to and activate CaMKII and interpreted the data to conclude that CaM with only two bound Ca2+ ions can activate the kinase. However, they did not consider in their interpretation that two CaM molecules might be interacting with the kinase to produce activation. This can clearly happen since we showed that the isolated N-terminal domain or isolated N- and C-domains together, were fully capable of activating CaMKII. Thus, it remains an open question of whether a partially Ca2+-saturated form of native CaM is capable of activating CaMKII or not.
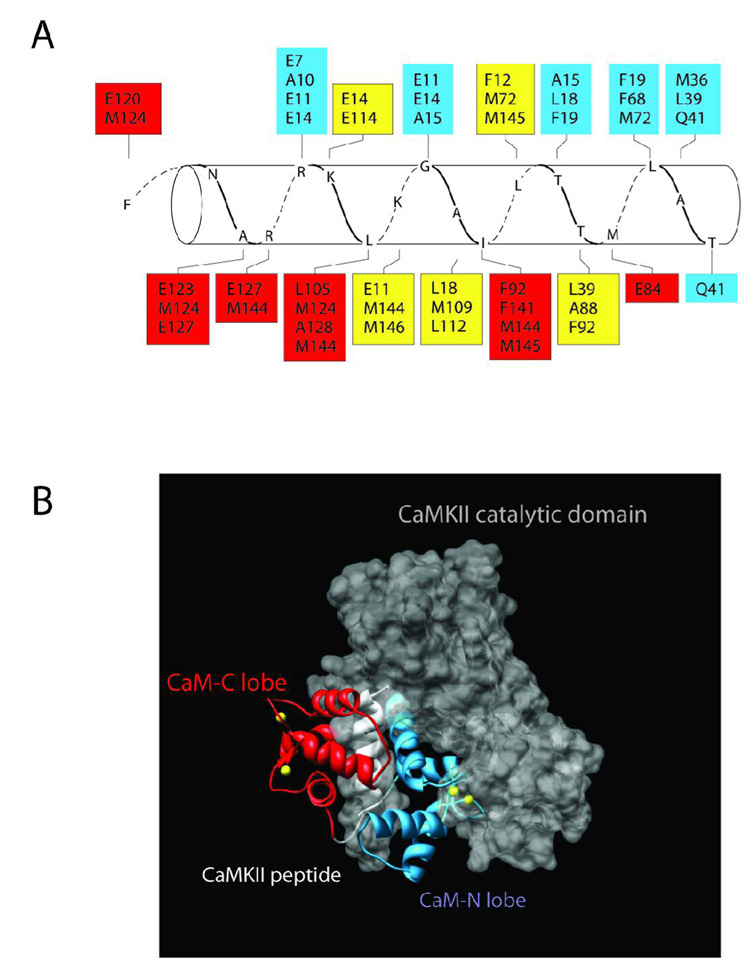
Our results can also be interpreted from a structural perspective. As noted in the Introduction, a crystal structure of Ca2+/CaM in association with a high affinity CaM-binding peptide mimicking the CaM-binding domain of CaMKII is available (20). A model of this structure (Fig. 8A) shows the typical anti-parallel orientation for Ca2+/CaM-target interactions with the amino acids of the N-domain of CaM (in blue boxes) interacting with the C-domain of the target peptide and vice versa. However, a clear sidedness is also evident with most of the N-domain residues in CaM binding on one side of the helical peptide while those in the C-domain (in red boxes) bind on the opposite side. Note also that there are amino acids in the target that react with residues in both the N- and C-lobe of CaM (in yellow boxes). When aligned into the crystal structure of the inactive catalytic domain of CaMKII (41) one can visualize that the N-terminal domain of CaM seems to lie in a crevice formed between the autoregulatory domain and the catalytic domain (Fig. 8B). The C-terminal domain of CaM appears to interact with the less obstructed face of the CaM-binding domain. This model suggests that the N-domain may play a role in orienting residues necessary for forming or making the nucleotide binding pocket accessible. Once nucleotide binds, the remainder of the CaM-binding domain would interact productively with the C-lobe of CaM leading to enzyme activation. This would be consistent with our results that the N-domain plays a dominant role in the initial interaction of CaM with CaMKII that ultimately leads to enzyme activation.
Figure 8.
Structure and Model of Ca2+/CaM bound to CaMKII. A) Amino acids in CaM are shown in boxes that interact with residues of the peptide (CKII 293–314) that mimics the CaM-binding domain of CaMKII. The residues in red are from the C-terminal domain of CaM, blue are from the N-terminal domain and yellow are from both N- and C-domains. The crystal structure that this figure is based upon is derived from PDB # 1CM1 (20). B) The complete crystal structure of PDB #1CM1 was aligned into the crystal structure of the autoinhibited catalytic domain of CaMKII (PDB# 2BDW) (41). The amino acids of the peptide were superimposed on those same residues in the CaM-binding domain of the catalytic domain as a means of constraining the alignment. The backbone of the C-lobe and N-lobes of CaM are shown in red and blue, respectively. The peptide (CKII 293–314) is in white and the catalytic domain is shown as a space filling model in grey. The yellow balls represent Ca2+ ions. Note that the N-lobe lies in a crevice between the autoregulatory domain and the catalytic domain. This figure was created using Chimera (http://www.cgl.ucsf.edu/chimera/).
In total, these results indicate that the activation of CaMKII by Ca2+/CaM likely proceeds through an ordered mechanism. The N-terminal domain binds with higher affinity and appears to bind first followed by binding of the C-terminal lobe. While the N-terminal domain of CaM has a poorer affinity for Ca2+ than the C-terminal domain, the N-lobe association rate kinetics is >50-fold faster (17). During the rising phase of Ca2+-influx, it is the N-terminal domain that would fill with Ca2+ first and become productive to interact with target proteins. Our data support that binding of the N-terminal domain is the first step of a sequential mechanism leading to the productive activation of CaMKII. This means that CaMKII would gain a competitive advantage over the other CaM-dependent enzymes noted above by utilizing preferential binding to the N-lobe instead of the C-lobe, since the C-lobe will require a longer period of time to become fully Ca2+ saturated. Additionally, it appears that nucleotide binding is an important step between binding of the N-terminal and C-terminal lobes of CaM. This conclusion is supported by the fact that the dissociation rates of Ca2+ from the C-lobe of CaM appear unaffected in the presence of CaMKII and that CaM binding to unphosphorylated CaMKII in the absence of nucleotide is in a fully extended conformation (8). Once both lobes of Ca2+/CaM are bound the enzyme is capable of supporting phosphorylation of its target substrates, or if a neighboring subunit is also in the Ca2+/CaM state, autophosphorylation ensues at Thr-286. Autophosphorylation leads to the exposure of amino acids on CaMKII that form additional contacts with CaM leading to a further collapse of the two domains of CaM (8) ultimately producing the high affinity or “CaM-trapped” state of CaMKII.
ACKNOWLEDGEMENTS
The authors acknowledge the generous gifts of CaM constructs from Drs. Andy Hudmon (Indiana University), Madeline Shea (University of Iowa Sch. of Med.) and Guy Guillemette (University of Waterloo). The help of Dr. Vishwa Triveda (University of Texas at Houston) with ITC is acknowledged as are the many thoughtful discussions of this work with Drs. Yoshihisa Kubota and Dr. John Putkey (University of Texas at Houston).
Abbreviations
- CaM
calmodulin
- wtCaM
wild-type CaM
- CaMN
mutant of CaM composed of only the N lobe
- CaMC
mutant of CaM composed on only the C lobe
- CaMN1–75
CaMN composed of amino acids 1–75
- CaMN1–80
CaMN composed of amino acids 1–80
- CaMC76–148
CaMC composed of amino acids 76–148
- CaMC81–148
CaMC composed of amino acids 81–148
- CaMNN
a chimeric molecule composed of two N-lobes of CaM
- CaMCC
a chimeric molecule composed of two C-lobes of CaM
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- MLCK
myosin light chain kinase
- nNOS
neuronal nitric oxide synthase
- CaM-C75-DANS
IAEDANS-labeled CaM(C75)
- IAEDANS
5-((((2-iodoacetyl)amino)ethyl)amino)napthalene-1-sulfonic acid
- ITC
isothermal titration calorimetry
- [Ca2+]i
intracellular Ca2+ concentration).
Footnotes
This work was supported by NIH grant NS26086 (MNW). MSW gratefully acknowledges support from NIH training grant T32 NS07467.
Textual Footnotes: None
REFERENCES
- 1.Hudmon A, Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 2.Kolodziej SJ, Hudmon A, Waxham MN, Stoops JK. Three-dimensional reconstructions of calcium/calmodulin-dependent (CaM) kinase IIalpha and truncated CaM kinase IIalpha reveal a unique organization for its structural core and functional domains. J Biol Chem. 2000;275:14354–14359. doi: 10.1074/jbc.275.19.14354. [DOI] [PubMed] [Google Scholar]
- 3.Miller SG, Kennedy MB. Regulation of brain type II Ca2+/calmodulin-dependent protein kinase by autophosphorylation: a Ca2+-triggered molecular switch. Cell. 1986;44:861–870. doi: 10.1016/0092-8674(86)90008-5. [DOI] [PubMed] [Google Scholar]
- 4.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256:1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 5.Waxham MN, Aronowski J, Westgate SA, Kelly PT. Mutagenesis of Thr-286 in monomeric Ca2+/calmodulin-dependent protein kinase II eliminates Ca2+/calmodulin-independent activity. Proc Natl Acad Sci U S A. 1990;87:1273–1277. doi: 10.1073/pnas.87.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson PI, Meyer T, Stryer L, Schulman H. Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron. 1994;12:943–956. doi: 10.1016/0896-6273(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 7.Gaertner TR, Kolodziej SJ, Wang D, Kobayashi R, Koomen JM, Stoops JK, Waxham MN. Comparative analyses of the three-dimensional structures and enzymatic properties of alpha, beta, gamma and delta isoforms of Ca2+-calmodulin-dependent protein kinase II. J Biol Chem. 2004;279:12484–12494. doi: 10.1074/jbc.M313597200. [DOI] [PubMed] [Google Scholar]
- 8.Torok K, Tzortzopoulos A, Grabarek Z, Best SL, Thorogate R. Dual effect of ATP in the activation mechanism of brain Ca(2+)/calmodulin-dependent protein kinase II by Ca(2+)/calmodulin. Biochemistry. 2001;40:14878–14890. doi: 10.1021/bi010920+. [DOI] [PubMed] [Google Scholar]
- 9.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 10.Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 11.Babu YS, Sack JS, Greenhough TJ, Bugg CE, Means AR, Cook WJ. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 12.Linse S, Helmersson A, Forsen S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 13.Bayley P, Ahlstrom P, Martin SR, Forsen S. The kinetics of calcium binding to calmodulin: Quin 2 and ANS stopped-flow fluorescence studies. Biochem Biophys Res Commun. 1984;120:185–191. doi: 10.1016/0006-291x(84)91431-1. [DOI] [PubMed] [Google Scholar]
- 14.Teleman A, Drakenberg T, Forsen S. Kinetics of Ca2+ binding to calmodulin and its tryptic fragments studied by 43Ca-NMR. Biochim Biophys Acta. 1986;873:204–213. doi: 10.1016/0167-4838(86)90047-6. [DOI] [PubMed] [Google Scholar]
- 15.Putkey JA, Kleerekoper Q, Gaertner TR, Waxham MN. A new role for IQ motif proteins in regulating calmodulin function. J Biol Chem. 2003;278:49667–49670. doi: 10.1074/jbc.C300372200. [DOI] [PubMed] [Google Scholar]
- 16.Gaertner TR, Putkey JA, Waxham MN. RC3/Neurogranin and Ca2+/calmodulin-dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J Biol Chem. 2004;279:39374–39382. doi: 10.1074/jbc.M405352200. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JD, Snyder C, Walsh M, Flynn M. Effects of myosin light chain kinase and peptides on Ca2+ exchange with the N- and C-terminal Ca2+ binding sites of calmodulin. J Biol Chem. 1996;271:761–767. doi: 10.1074/jbc.271.2.761. [DOI] [PubMed] [Google Scholar]
- 18.Olwin BB, Storm DR. Calcium binding to complexes of calmodulin and calmodulin binding proteins. Biochemistry. 1985;24:8081–8086. doi: 10.1021/bi00348a037. [DOI] [PubMed] [Google Scholar]
- 19.Tzortzopoulos A, Best SL, Kalamida D, Torok K. Ca2+/calmodulin-dependent activation and inactivation mechanisms of alphaCaMKII and phospho-Thr286-alphaCaMKII. Biochemistry. 2004;43:6270–6280. doi: 10.1021/bi035449u. [DOI] [PubMed] [Google Scholar]
- 20.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 21.Waxham MN, Tsai AL, Putkey JA. A mechanism for calmodulin (CaM) trapping by CaM-kinase II defined by a family of CaM-binding peptides. J Biol Chem. 1998;273:17579–17584. doi: 10.1074/jbc.273.28.17579. [DOI] [PubMed] [Google Scholar]
- 22.Jaren OR, Kranz JK, Sorensen BR, Wand AJ, Shea MA. Calcium-induced conformational switching of Paramecium calmodulin provides evidence for domain coupling. Biochemistry. 2002;41:14158–14166. doi: 10.1021/bi026340+. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen BR, Faga LA, Hultman R, Shea MA. An interdomain linker increases the thermostability and decreases the calcium affinity of the calmodulin N-domain. Biochemistry. 2002;41:15–20. doi: 10.1021/bi011718+. [DOI] [PubMed] [Google Scholar]
- 24.Persechini A, Gansz KJ, Paresi RJ. Activation of myosin light chain kinase and nitric oxide synthase activities by engineered calmodulins with duplicated or exchanged EF hand pairs. Biochemistry. 1996;35:224–228. doi: 10.1021/bi952383x. [DOI] [PubMed] [Google Scholar]
- 25.Persechini A, Gansz KJ, Paresi RJ. A role in enzyme activation for the N-terminal leader sequence in calmodulin. J Biol Chem. 1996;271:19279–19282. doi: 10.1074/jbc.271.32.19279. [DOI] [PubMed] [Google Scholar]
- 26.Spratt DE, Newman E, Mosher J, Ghosh DK, Salerno JC, Guillemette JG. Binding and activation of nitric oxide synthase isozymes by calmodulin EF hand pairs. FEBS J. 2006;273:1759–1771. doi: 10.1111/j.1742-4658.2006.05193.x. [DOI] [PubMed] [Google Scholar]
- 27.Sorensen BR, Shea MA. Interactions between domains of apo calmodulin alter calcium binding and stability. Biochemistry. 1998;37:4244–4253. doi: 10.1021/bi9718200. [DOI] [PubMed] [Google Scholar]
- 28.Putkey JA, Waxham MN. A peptide model for calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J Biol Chem. 1996;271:29619–29623. doi: 10.1074/jbc.271.47.29619. [DOI] [PubMed] [Google Scholar]
- 29.Bradshaw JM, Hudmon A, Schulman H. Chemical quenched flow kinetic studies indicate an intraholoenzyme autophosphorylation mechanism for Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 2002;277:20991–20998. doi: 10.1074/jbc.M202154200. [DOI] [PubMed] [Google Scholar]
- 30.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudmon A, Aronowski J, Kolb SJ, Waxham MN. Inactivation and self-association of Ca2+/calmodulin-dependent protein kinase II during autophosphorylation. J Biol Chem. 1996;271:8800–8808. doi: 10.1074/jbc.271.15.8800. [DOI] [PubMed] [Google Scholar]
- 32.Tse JK, Giannetti AM, Bradshaw JM. Thermodynamics of calmodulin trapping by Ca2+/calmodulin-dependent protein kinase II: subpicomolar Kd determined using competition titration calorimetry. Biochemistry. 2007;46:4017–4027. doi: 10.1021/bi700013y. [DOI] [PubMed] [Google Scholar]
- 33.Faga LA, Sorensen BR, VanScyoc WS, Shea MA. Basic interdomain boundary residues in calmodulin decrease calcium affinity of sites I and II by stabilizing helix-helix interactions. Proteins. 2003;50:381–391. doi: 10.1002/prot.10281. [DOI] [PubMed] [Google Scholar]
- 34.De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 35.King MM, Shell DJ, Kwiatkowski AP. Affinity labeling of the ATP-binding site of type II calmodulin-dependent protein kinase by 5'-p-fluorosulfonylbenzoyl adenosine. Arch Biochem Biophys. 1988;267:467–473. doi: 10.1016/0003-9861(88)90052-5. [DOI] [PubMed] [Google Scholar]
- 36.Tzortzopoulos A, Torok K. Mechanism of the T286A-mutant alphaCaMKII interactions with Ca2+/calmodulin and ATP. Biochemistry. 2004;43:6404–6414. doi: 10.1021/bi036224m. [DOI] [PubMed] [Google Scholar]
- 37.Persechini A, McMillan K, Leakey P. Activation of myosin light chain kinase and nitric oxide synthase activities by calmodulin fragments. J Biol Chem. 1994;269:16148–16154. [PubMed] [Google Scholar]
- 38.Newton DL, Oldewurtel MD, Krinks MH, Shiloach J, Klee CB. Agonist and antagonist properties of calmodulin fragments. J Biol Chem. 1984;259:4419–4426. [PubMed] [Google Scholar]
- 39.Kuznicki J, Grabarek Z, Brzeska H, Drabikowski W, Cohen P. Stimulation of enzyme activities by fragments of calmodulin. FEBS Lett. 1981;130:141–145. doi: 10.1016/0014-5793(81)80683-7. [DOI] [PubMed] [Google Scholar]
- 40.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci U S A. 2006;103:13968–13973. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]