Nanoscale materials and structures hold great promise in various aspects of biomedical research, ranging from imaging, detection, and sensing to drug delivery and new therapies.[1,2] In addition to the intrinsic electronic, optical, or magnetic properties of the nanostructures, chemical methods used in fabrication and in controlling properties of the material surfaces and interfaces also play an important role in realizing desired nanoparticle functions.[3–6] Herein, we describe a surface-modification approach with proteins for the newly developed synthetic antiferromagnetic nanoparticles (SAFs),[7] and demonstrate for the first time that SAF nanoparticles enable magnetic detection of biomolecules at low analyte concentrations (10 pM) with better detection than conventional superparamagnetic materials. In addition, fluorescent labeling allows clear observations of tailorable single nanoparticle motion in response to a small external magnetic field gradient (10 Tm−1). By adjusting thickness of the magnetic layers (12 nm vs. 24 nm) within 100 nm diameter SAF, dramatically different magnetic motions were achieved. Such distinctive control represents a major advance that SAFs offer, as current magnetic manipulation and separation experiments often depend on superparamagnetic particles with micrometer sizes,[8] formation of particle aggregates,[6] or on the use of huge magnetic field gradients,[9,10] for which precise control over individual particles remains a big challenge. SAFs overcome these limitations through mono-dispersity and the use of very high magnetization ferromagnetic multilayers. Furthermore, interactions between magnetic layers within each single particle are exploited to eliminate remanence, and consequent magnetic aggregation at zero field, and to control magnetic responsiveness. Our surface functionalization approach utilizes the coordination chemistry between the intrinsic functional groups of proteins and inorganic elements of nanoparticles. The simplicity and success of this approach, evidenced by the biochemical specificity and the magnetic–fluorescence multifunctionality demonstrated herein, suggest that SAFs can serve as better labels for applications that include magnetic separation, manipulation, biomolecule detection, and magnetic lab-on-a-chip devices.[9–16]
High-moment, zero-remanence SAFs are trapezoidal disks with multilayer structures.[7] The essential magnetic feature of each SAF is two ferromagnetic layers separated by a nonmagnetic spacer layer. Herein, Co90Fe10 is used for the ferromagnetic layers and ruthenium metal is used for the spacer. At a spacer thickness of 2.5 nm used for these observations, the ferromagnetic layers are only weakly exchange-coupled and interact mostly through magnetostatic interactions. These interactions, with like poles repelling, result in a magnetostatic antiferromagnetic interlayer coupling.[7] The magnetic layers were deposited, along with a chemically etchable copper release layer and protective tantalum surface layers, using ion-beam deposition in a high vacuum. After deposition, template resists of polymethyl methacrylate (PMMA) and polymethyl glutarimide (PMGI) were removed by organic solvents and aqueous strippers. Nanoparticles were released by selective etching of the copper release layer with a copper ammine ([Cu(NH3)4])-based solution, and transferred to citrate buffer for particle collection through repeated cycles of centrifugation, aspiration, and re-suspension. These necessary processes leave the nanoparticle surfaces in a complex non-pristine state, and the utility of standard surface layers with well-known attachment chemistries (e.g. gold–thiol or silicon oxide–silane) is uncertain.
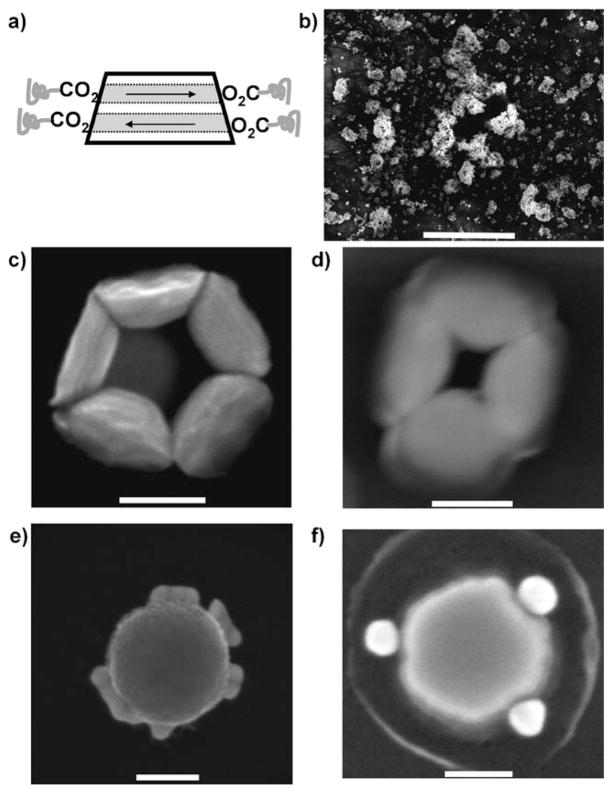
We thus used direct protein functionalization of the surface to render SAFs usable for biological applications. As all of the elements used to fabricate these SAFs can be bound to carboxylates by coordination chemistry,[17–19] proteins with many available carboxylates should be able to directly conjugate to SAF surfaces. To confirm this metal–ligand binding (Figure 1a), we first used carboxylate-modified polyethylene glycol (PEG) molecules (Quanta Biodesign, Ltd.) to functionalize SAF surfaces. As shown in the scanning electron micrograph (SEM; Figure 1b), PEG molecules with both ends modified by carboxylates (PEG-C2, MW 1338) could introduce strong clustering of SAFs, as expected for cross-linking of SAFs by PEG-C2. For SAF alone or SAF mixed with PEG molecules terminated at only one end by a carboxylate (PEG-C1, MW 459), particles remained separated from each other (see the Supporting Information, Figure S1). Interestingly, the particle cross-linking occurred mainly on the sidewalls of the SAFs, suggesting that the composition of nanoparticle surfaces may be different at top, bottom, and sidewall surfaces. This effect can be associated with variations in the tantalum layer thickness and possible interfacial intermixing during the aggressive deposition and etch processes employed. Preferential binding on the side walls is then consistent with previous work, in which carboxylate formed complexes with copper, iron, and ruthenium,[17,18,20,21] whereas tantalum bound strongly to phosphate.[22,23] It is likely that elements at SAF surfaces were naturally oxidized,[7] and the oxidation state of these metal elements should not affect the fundamentals of the metal–ligand binding.[24–26] A characteristic structure of sidewall cross-linking by PEG-C2 is shown in Figure 1c for a complex of 5 SAFs.
Figure 1.
Surface functionalization of SAFs examined using SEM. a) Surface functionalizaton of SAFs by coordination of metal elements with carboxylates. b) SAF clustered by PEG molecules, with both ends modified by carboxylates (PEG-C2). Scale bar: 5 μm. c) SAFs linked through sidewalls by PEG-C2. Scale bar: 100 nm. d) SAFs linked through sidewalls by streptavidin. Scale bar: 100 nm. e) SAFs, after streptavidin functionalization, specifically bound to BSA-biotin-coated gold particles on the sidewall. Scale bar: 100 nm. f) Unreleased SAFs (still on silicon substrate) after streptavidin functionalization also bound to BSA-biotin-coated gold particles on the sidewall. Scale bar: 50 nm.
We then used streptavidin as a model protein to directly functionalize SAFs (see the Supporting Information for details). Streptavidin–biotin binding is one of the strongest non-covalent interactions known that is widely applicable to biochemical research.[24] Dispersed single SAF–streptavidin conjugates were usually observed for samples that were exposed to constant vortexing. For samples that experienced settling, small clusters of SAF could be formed. Streptavidin also cross-linked some SAFs and formed the characteristic structure of SAFs with sidewall linkage (Figure 1d). Importantly, streptavidin retained biotin binding affinity after being conjugated to SAF surfaces. Both released (Figure 1e) and unreleased SAF (Figure 1 f), after streptavidin conjugation, formed recognizable superstructures in which gold nanoparticles, pre-functionalized by BSA-biotin, were consistently connected to SAF sidewalls.
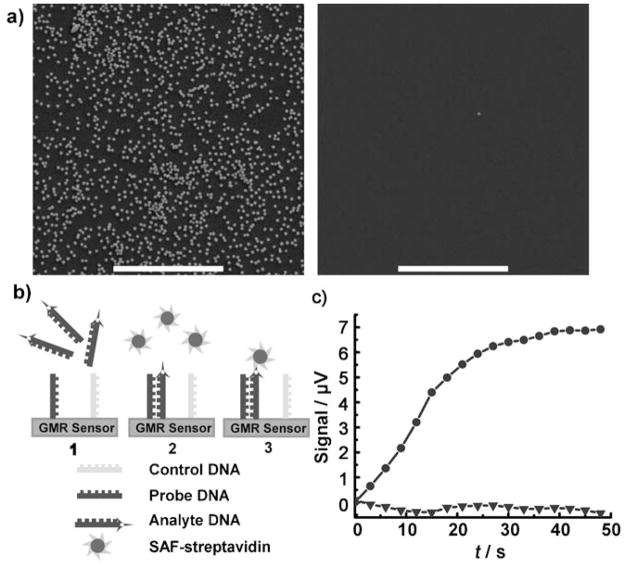
SAF–streptavidin conjugates were also able to recognize biotinylated substrates with high specificity (Figure 2a), which is an important prerequisite for nanoparticle biodetection experiments. DNA detection experiments using GMR spin-valve sensors[16,25–36] were conducted to examine the performance of SAF–streptavidin conjugates for biodetection (see the Supporting Information for a detailed description of the GMR sensor). Our detection scheme[37–40] utilizes a technique in which the sensor current and an in-plane, transverse magnetic tickling field are each modulated at different frequencies, and signal amplitudes at sum or difference frequencies are recorded.[41,42] In this way, we can greatly reduce 1/f noise and electromagnetic interference. Many magnetic nanoparticles, either commercially available or home developed, have been evaluated and failed to provide high sensitivity and specificity. We currently use superparamagnetic FeOx nanoparticle–streptavidin conjugates (MACSs, Miltenyi Biotech). Although MACSs have good chemical specificity, their saturation magnetization is small, so the GMR signal per particle is limited. SAFs with small sizes of 100 nm, high chemical specificity, and large and tunable magnetic moment should improve biodetection, and may serve as better magnetic labels for sensors developed by others by providing increased sensitivity, broader assay dynamic range, and allowing better quantification.[25–36,43]
Figure 2.
Streptavidin-functionalized SAFs used for biomolecule detection. a) SEM examination of the binding specificity of SAF–streptavidin to biotinylated substrate. Left image: many SAF particles on the biotinylated substrate (silicon substrate functionalized with 3-amino-propyltrimethoxysilane, then covalently modified with NHS-PEG-biotin). If the original silicon substrate is not biotinylated, negligible numbers of SAF were observed (right). Both experiments were performed in parallel under the same conditions. Scale bars: 5 μm. b) The DNA detection scheme: 1) Biotinlyated analyte DNA was applied and incubated to allow hybridization of analyte and probe DNA on the positive sensors. The negative sensor surface is coated with non-complementary poly-T DNA strands, and should not hybridize with analyte DNA. 2) After washing, the sensor was staged in the measurement system. SAF–streptavidin conjugate was then applied, and the corresponding time point was set as time 0. 3) SAF–streptavidin bound to the biotinylated analyte DNA on the positive sensor surface, and the resulting GMR signal was recorded in real time. No signal should be generated from the negative sensor. c) SAF–streptavidin was able to detect 10 pM analyte DNA. The analyte DNA is biotin-5′-TAT GGT AGA TGG GTA TTT GTG GAT TGG-3′, and the probe DNA sequence is 5′-TTT TTT TTT TCT ACC AAT CCA CAA ATA CCC ATC TAC CAT A-3′. The non-complementary control DNA sequence is 5′-TTT TTT TTT TTT-3′. ● GMR signal of the positive sensor; ▼ the negative sensor, with non-complementary control DNA strands on the sensor surface.
Figure 2b depicts biotinylated target DNA selectively hybridized with specific sensors, and subsequently binding streptavidin-conjugated SAF nanoparticles to the sensor surface. The GMR signal of a positive sensor, with its surface modified by probe DNA strands that are complementary to the analyte DNA, is shown in Figure 2c as solid circles. The flat signal of the negative sensor indicates negligible non-specific binding of SAF–streptavidin conjugates to the sensor surface. A low concentration of analyte DNA of 10 pM was clearly detectable. This experiment demonstrates that synthetic antiferromagnetic nanoparticles can be used for biological detection with high sensitivity. Only about 400 pM SAF-streptavidin was used in the magnetic labeling step, whereas for the same DNA detection experiment using MACS, 10 nM magnetic nanoparticle solution was applied.[40] Furthermore, the signal achievable for 10 pM DNA detection with SAF–streptavidin conjugates is 7 μV, which is much larger than the typical value of 2–4 μV using MACS.[40] Our sensors can detect fM concentrations of several proteins,[44] and DNA detection at fM range[45] should also be possible with additional dependence on hybridized oligomer lengths. It should also be noted that the present measurements involved 1.5 μm-wide serpentine spin-valve sensors and magnetic fields of tens of Oe. Such small fields only weakly magnetize SAF or MACS nanoparticles, so the magnetic permeability and volume of the nanoparticles determines the signal strength. SAF-induced signals will further benefit from reduced saturation fields if ferromagnetic ruthenium coupling is used to reduce antiferromagnetic magnetostatic interactions. However, even at low fields, these data indicate that streptavidin-functionalized SAFs with both high magnetic moment and chemical specificity can be better magnetic labels for biological detection.
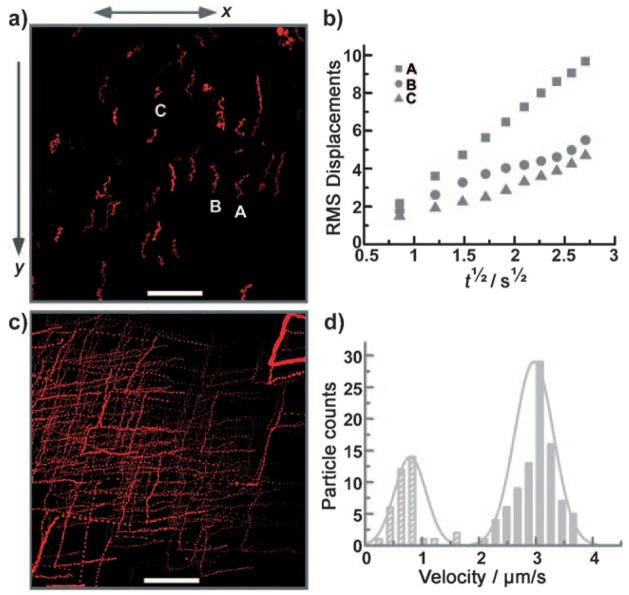
Protein functionalization also facilitated direct observation of the movement of SAFs under external magnetic field control. Fluorophore-modified streptavidins (Invitrogen Corporation) were also used to functionalize SAFs and provided excellent fluorescent imaging contrast. Continuous magnetic manipulation and fluorescent tracking of SAFs in a glass chamber slide revealed control of the velocity of individual SAF particles using a fairly small magnetic field gradient of 10 Tm−1 (Figure 3a,b, and corresponding movies in the Supporting Information).
Figure 3.
Magnetic manipulation of SAF–streptavidin using a small magnetic field gradient of 10 Tm−1. a) The path of A594s-SAF12 (corresponding to movie 1, Supporting Information), showing the response to the external field gradient in the y direction. Scale bar: 50 μm. b) The RMS displacements in the x direction (perpendicular to the direction of the magnetic field gradient) of the three labeled A594s-SAF12 particles A, B, and C. The slope of each curve was used to calculate the diffusion constant of the corresponding particle. c) The fluorescent path of the A594s-SAF24 sample (corresponding to movie 2, Supporting Information) demonstrating nanoparticle response to manipulation using a 2.54 cm permanent magnet. The sample turns when the magnet rotates, then moves straight in response to the field gradient. Scale bar: 50 μm. d) Distributions of the magnetically induced velocities of A594s-SAF12 (striped bars) and A594s-SAF24 (filled bars). For both A594s-SAF12 (a) and A594s-SAF24 (c), the end-to-end distances of particle motion along the vertical line versus time from (a) and (c) were used to calculate the velocity.
Alexa Fluor 594-streptavidin-conjugated SAF, with a SAF composition of Ta5 nm/Ru2 nm/CoFe6 nm/Ru2.5 nm/CoFe6 nm/Ru2 nm/Ta5 nm (denoted A594s-SAF12), were subjected to a magnetic field gradient in the y direction (Figure 3a). This sample is mainly composed of monodis-persed nanoparticles, as seen from SEM (see the Supporting Information, Figure S2). These A594s-SAF12 clearly responded by translating along the external magnetic field gradient direction. Disturbance by Brownian motion was apparent with the “jiggling” of the particle tracks. Analysis of the “jiggling” in the direction x perpendicular to the magnetic field gradient allowed determination of the diffusion constant D of individual A594s-SAF12 particles. Figure 3b shows the root-mean-square (RMS) displacements versus the square root of time for the corresponding particles marked in Figure 3a. Diffusion constants calculated from the relationship 〈Δx2〉1/2 =(2Dt)1/2 are 8, 2, and 2 μm2s−1 for particles A, B, and C, respectively. These values are in reasonable agreement with a calculation using the Stokes–Einstein equation for a single 100 nm spherical particle (D = 4 μm2s−1). Possible residues and protein surface functionalization can increase particle sizes and slow diffusion, and analogous effects can arise from surface interactions.
Observations of magnetic manipulation of single nanoparticles can be extremely useful for magnetic separation and manipulation in which biological targets or payloads are bound to SAF surfaces. Alexa Fluor 594-streptavidin-conjugated SAF (Figure 3c) with a SAF composition of Ta5 nm/Ru2 nm/CoFe12 nm/Ru2.5 nm/CoFe12 nm/Ru2 nm/Ta5 nm (A594s-SAF24) also exhibited magnetically induced motion using fields from a rotatable permanent magnet with magnetic field gradient of 10 Tm−1. A594s-SAF24 move in nearly linear paths along the external magnetic field gradient and turn as the magnet is rotated. Compared to the magnetically induced motion of A594s-SAF12, the movement of A594s-SAF24 appeared much less disturbed by Brownian motion, though the track of A594s-SAF24 still drifted from a straight line at certain time points. Brownian motion depends on the physical dimensions of the particles (D =kB T/6πηr for a spherical particle), which differ only slightly between A594s-SAF12 and A594s-SAF24. These SAFs do have similar Brownian motion, with an average squared displacement of 4–16 μm2s−1. In contrast, their magnetic motion also depends strongly on the particle magnetic layer thickness, which can be precisely adjusted. In our experiments, the saturation magnetic moment of each A594s-SAF24 particle with CoFe thickness of 24 nm is roughly twice as that of A594s-SAF12 with 12 nm thick CoFe layers (Supporting Information, Figure S3). The magnetically induced averaged velocities, determined from nanoparticle motion in corresponding video files, were 3 μms−1 for A594s-SAF24 and 1 μms−1 for A594s-SAF12, demonstrating dramatically different motions achievable using SAFs with different CoFe thicknesses. Figure 3d shows the histograms of the velocity distribution determined from these two experiments. The magnetically induced velocities for both samples are in agreement with the estimation using Stokes viscous drag forces 3πηdν =V▽−(MH) (see the Supporting Information for the calculation). The ability to tune, and clearly observe, distinctive magnetic nanoparticle responses is highly desired in many applications, such as multiplexed magnetic separation and manipulating molecular and cellular interactions.
In summary, we have developed a simple surface-functionalization method for conjugating lithographically fabricated antiferromagnetic nanoparticles to a model protein, streptavidin. The streptavidin-functionalized SAFs exhibited specific binding to biotin, allowed biomolecule detection with high sensitivity (10 pM), and tunable responses to a small external magnetic field gradient (10 Tm−1). These high moment and multifunctional (optical, magnetic, and specific targeting) magnetic nanoparticles are highly promising for a variety of biological applications, such as high-throughput biomolecule detection and multiplex magnetic manipulation. They could also potentially be used as in-vivo imaging and targeting agents, provided nontoxic biocompatible materials, such as iron oxide and titanium, can be exploited for SAF fabrication.
Supplementary Material
Footnotes
This work was supported by grants from NIH (1U54 CA119367-01, S.S.G.), NSF (ECCS-0801385 and DBI-0551990, S.X.W.), a NIH Pathway to Independence Award (1K99 EB008558-01, A.F.), and a Stanford Graduate Fellowship (W.H.).
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200803994.
References
- 1.Alivisatos AP. Nat Biotechnol. 2004;22:47. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 2.Wagner V, Dullaart A, Bock AK, Zweck A. Nat Biotechnol. 2006;24:1211. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 3.Patolsky F, Zheng G, Lieber CM. Nanomedicine. 2006;1:51. doi: 10.2217/17435889.1.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Berry CC, Curtis ASG. J Phys D. 2003;36:R198. [Google Scholar]
- 5.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yavuz CT, Mayo JT, Yu WW, Prakash A, Falkner JC, Yean S, Cong L, Shipley HJ, Kan A, Tomson M, Natelson D, Colvin VL. Science. 2006;314:964. doi: 10.1126/science.1131475. [DOI] [PubMed] [Google Scholar]
- 7.Hu W, Wilson RJ, Koh A, Fu A, Faranesh AZ, Earhart CM, Osterfeld SJ, Han SJ, Xu L, Guccione S, Sinclair R, Wang SX. Adv Mater. 2008;20:1479. [Google Scholar]
- 8.Liu C, Lagae L, Borghs G. Appl Phys Lett. 2007;90:184109. [Google Scholar]
- 9.Dobson J. Nat Nanotechnol. 2008;3:139. doi: 10.1038/nnano.2008.39. [DOI] [PubMed] [Google Scholar]
- 10.Mannix RJ, Kumar S, Cassiola F, Zavala MM, Feinstein E, Prentiss M, Ingber DE. Nat Nanotechnol. 2008;3:36. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- 11.Pankhurst QA, Connolly J, Jones SK, Dobson J. J Phys D. 2003;36:R167. [Google Scholar]
- 12.Mirowski E, Moreland J, Zhang A, Russek SE. Appl Phys Lett. 2005;86:243901. [Google Scholar]
- 13.McCloskey KE, Chalmers JJ, Zborowski M. Anal Chem. 2003;75:6868. doi: 10.1021/ac034315j. [DOI] [PubMed] [Google Scholar]
- 14.Gelfand A. Economist.com. 2008 October 15; http://www.economist.com/science/tm/displaystory.cfm?story_id=12412184.
- 15.Brückl H, Panhorst M, Schotter J, Kamp PB, Becker A. IEE Proc Nanobiotechnol. 2005;152:41. doi: 10.1049/ip-nbt:20045015. [DOI] [PubMed] [Google Scholar]
- 16.Reiss G, Brueckl H, Huetten A, Schotter J, Brzeska M, Panhorst M, Sudfeld D. J Mater Res. 2005;20:3294. [Google Scholar]
- 17.Barth JV. Annu Rev Phys Chem. 2007;58:375. doi: 10.1146/annurev.physchem.56.092503.141259. [DOI] [PubMed] [Google Scholar]
- 18.Malik KZ, Robinson SD, Steed JW. Polyhedron. 2000;19:1589. [Google Scholar]
- 19.Fang JB, Sanghi R, Kohn J, Goldman AS. Inorg Chim Acta. 2004;357:2415. [Google Scholar]
- 20.Lim J, Eggeman A, Lanni F, Tilton RD, Majetich SA. Adv Mater. 2008;20:1721. [Google Scholar]
- 21.Turte KI, Shova SG, Spatar FA, Mazus MD, Malinovski TI. J Struct Chem. 1994;35:248. [Google Scholar]
- 22.Textor M, Ruiz L, Hofer R, Rossi A, Feldman K, Hahner G, Spencer ND. Langmuir. 2000;16:3257. [Google Scholar]
- 23.Brovelli D, Hahner G, Ruiz L, Hofer R, Kraus G, Waldner A, Schlosser J, Oroszlan P, Ehrat M, Spencer ND. Langmuir. 1999;15:4324. [Google Scholar]
- 24.Gonzalez M, Bagatolli LA, Echabe I, Arrondo JLR, Argarana CE, Cantor CR, Fidelio GD. J Biol Chem. 1997;272:11288. doi: 10.1074/jbc.272.17.11288. [DOI] [PubMed] [Google Scholar]
- 25.Rife JC, Miller MM, Sheehan PE, Tamanaha CR, Tondra M, Whitman LJ. Sens Actuators A. 2003;107:209. [Google Scholar]
- 26.Sandhu A. Nat Nanotechnol. 2007;2:746. doi: 10.1038/nnano.2007.398. [DOI] [PubMed] [Google Scholar]
- 27.Gijs MAM. In: Magnetic Nanostructures in Modern Technology. Azzerboni B, et al., editors. Springer; Heidelberg: 2008. p. 153. [Google Scholar]
- 28.Edelstein RL, Tamanaha CR, Sheehan PE, Miller MM, Baselt DR, Whitman LJ, Colton RJ. Biosens Bioelectron. 2000;14:805. doi: 10.1016/s0956-5663(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 29.Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. Biosens Bioelectron. 1998;13:731. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 30.Schotter J, Kamp PB, Becker A, Puhler A, Reiss G, Bruckl H. Biosens Bioelectron. 2004;19:1149. doi: 10.1016/j.bios.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 31.de Boer BM, Kahlman JAHM, Jansen TPGH, Duric H, Veen J. Biosens Bioelectron. 2007;22:2366. doi: 10.1016/j.bios.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Janssen XJA, van LJ, Jzendoorn I, Prins MWJ. Biosens Bioelectron. 2008;23:833. doi: 10.1016/j.bios.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Graham DL, Ferreira H, Bernardo J, Freitas PP, Cabral JMS. J Appl Phys. 2002;91:7786. [Google Scholar]
- 34.Millen RL, Kawaguchi T, Granger MC, Porter MD. Anal Chem. 2005;77:6581. doi: 10.1021/ac0509049. [DOI] [PubMed] [Google Scholar]
- 35.Tamanaha CR, Mulvaney SP, Rife JC, Whitman LJ. Biosens Bioelectron. 2008;24:1. doi: 10.1016/j.bios.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Speetjens RW, Reekmans G, Palma RD, Liu C, Laureyn W, Borghs G. Sens Actuators B. 2007;128:1. [Google Scholar]
- 37.Li G, Sun S, Wilson RJ, White RL, Pourmand N, Wang SX. Sens Actuators A. 2006;126:98. doi: 10.1016/j.sna.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang SX, Li G. Adv Magn IEEE Trans Magn. 2008;44:1687. [Google Scholar]
- 39.Osterfeld SJ, Wang SX. In: Microarrays: New Development Towards Recognition of Nucleic Acid and Protein Signatures. Dill K, Liu R, Grodzinski P, editors. Springer; New York: 2009. [Google Scholar]
- 40.Xu L, Yu H, Akhrasb MS, Han SJ, Osterfeld SJ, White RL, Pourmand N, Wang SX. Biosens Bioelectron. 2008;24:99. doi: 10.1016/j.bios.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han SJ, Xu L, Wilson RJ, Wang SX. IEEE Trans Magn. 2006;42:3560. [Google Scholar]
- 42.de Boer BM, Kahlman JAHM, Jansen TPGH, Duric H, Veen J. Biosens Bioelectron. 2007;22:2366. doi: 10.1016/j.bios.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Shen W, Schrag BD, Carter MJ, Xie J, Xu C, Sun S, Xiao G. J Appl Phys. 2008;103:07A306. [Google Scholar]
- 44.Osterfeld SJ, Yu H, Gaster RS, Caramuta S, Xu L, Han SJ, Hall DA, Wilson RJ, Sun S, White RL, Davis RW, Pourmand N, Wang SX. Proc Natl Acad Sci. 2008;105:20637. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez JM, Josephson L, O3Loughlin T, Hogemann D, Weissleder R. Nat Biotechnol. 2002;20:816. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.