Abstract
Many aminoglycosidic antibiotics target the A-site of 16S RNA in the small ribosomal subunit and affect the fidelity of protein translation in bacteria. Upon binding aminoglycosides displace two adenines (A1492 and A1493 for E. coli numbering) that are involved in tRNA anticodon loop recognition. The major difference in the aminoglycosidic binding site between the prokaryota and eukaryota is an adenine into guanine substitution in the position 1408. This mutation likely affects the dynamics of near A1492 and A1493 and hinders the binding of aminoglycosides to eukaryotic ribosomes. With multiple 20 ns long all-atom molecular dynamics simulations we study the flexibility of a 22-nucleotide RNA fragment which mimics the aminoglycosidic binding site. Simulations are carried out for both, native and A1408G mutated RNA, as well as for their complexes with aminoglycosidic representative – paromomycin. We observe intra- and extrahelical configurations of A1492 and A1493, which differ between the prokaryotic and the mutated structure. We obtained configurations of the A-site that were also observed in the NMR and crystal structures. Our studies show the differences in the internal mobility of the A-site, as well as in ion and water density distributions inside the binding cleft, between the prokaryotic and mutated RNA. We also compare the performance of two force field parameters for RNA – Amber and Charmm.
Keywords: 16S ribosomal RNA, molecular dynamics, model A-site, aminoglycoside antibiotics, paromomycin, base flipping
Introduction
A ribosome is a large protein-synthesizing macromolecular complex whose proper functioning is crucial for the life of a cell. It translates the information encoded in mRNA into a polypeptide. The ribosome consists of two subunits: small (in prokaryota denoted 30S) and large (50S). Both subunits contain ribosomal RNA (rRNA) and proteins, e.g. the 30S subunit is composed of the so-called 16S rRNA and about 20 proteins. At the interface between subunits there are three tRNA binding sites (A, P and E) where subsequent amino acids are being attached during the polypeptide assembly. Several aminoglycosidic antibiotics bind to the A-site1,2 and affect the translation process by reducing its fidelity. These anti-bacterial drugs are particularly active against Gram-negative bacteria, however, the emerging bacterial resistance and toxicity for human ear and kidney cells limits their effectiveness in medical therapy. The understanding of their actions on a molecular level is needed to improve their specificity and selectivity.
Aminoglycosides are sugar derivatives3. Due to a number of amine groups they are positively charged in physiological pH3,4. For example, a model aminoglycosidic compound considered here — paromomycin (Figure 1a), carries a total charge of +5e. It binds to the bacterial 16S rRNA in the vicinity of the A-site, but is also known to bind to a corresponding location in the eukaryotic ribosome. The most important difference between these two sites is the nucleotide no. 1408 (numbering as in E. coli)5 — adenine in prokaryota and guanine in eukaryota (Figure 1b)6. It was proven experimentally that bacteria with a single A1408G mutation are immune to several amino-glycosides7–9.
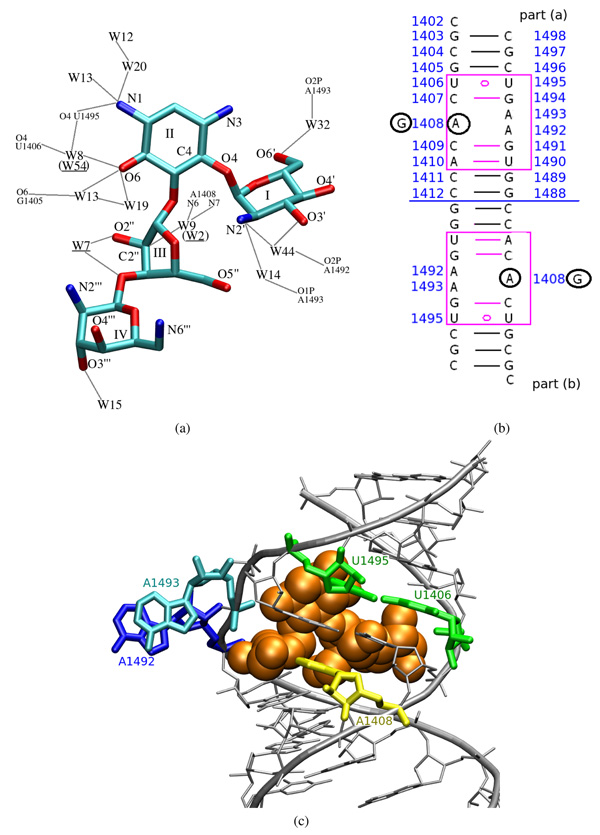
Figure 1.
(a) Structural view of paromomycin heavy atoms with ring numbering and marked crystal water molecules which mediate its binding to RNA. Colors correspond to atom types: cyan—carbon, blue—nitrogen, red—oxygen; “W” stands for the water oxygen. Gray lines symbolize hydrogen bonds. Water numbering is taken from the 1J7T PDB file with brackets denoting numbering in part (b) of the RNA duplex. (b) The sequence of the simulated 16S rRNA fragment with the added terminal cytidines (C1402(a) and C1402(b)). Adenines which were mutated to guanines are circled in black. Blue numbering indicates the sequence of the bases as in the original 16S rRNA of E. coli. (c) A model of one of the paromomycin binding sites complexed with the antibiotic shown as van der Waals spheres.
Two other crucial nucleotides in the paromomycin binding site are A1492 and A1493, situated across the helix from the nucleotide 1408 (see Figures 1b and 1c). Upon binding of paromomycin, these adenines protrude from the 16S rRNA helix and make space for the antibiotic3,10–12 — we will refer to this configuration as the flipped-out or extra-helical state. In contrast, when the antibiotic is absent, these two adenines tend to be hidden inside the RNA helix — in a flipped-in or intra-helical state13,14. Nevertheless, they still possess some degree of flexibility because their movability is critical for the incorporation of cognate tRNAs15–17. It was suggested that it is the degree of stabilization of A1492 in the flipped-out position, caused by aminoglycoside binding, that correlates with their antibacterial activity18.
The mobility of A1492 and A1493 in small, model fragments of the A-site RNA was recently studied by means of replica-exchange19 and targeted molecular dynamics17. These studies confirmed that in the absence of the antibiotic the intra-helical state is the favored one. A molecular dynamics (MD) study was also performed for a larger fragment of 16S rRNA, known as the helix 4420. The authors observed the intra- and extra-helical configurations of A1492 on a nanosecond time scale. An MD simulation of a model RNA oligonucleotide in the complex with paromomycin was also performed21 but the behavior of the two adenines in the ligand-free state was not studied.
Our goal was to compare the dynamic properties of paromomycin binding site with and without the A1408G mutation in order to understand why such substitution prevents aminoglycosides from being active against the eukaryotic ribosomes. We performed six, 20-nanosecond long explicit solvent MD simulations of a model 16S rRNA fragment containing two A-sites22. Four simulations were carried out with the Charmm force field23,24 and included the prokaryotic 16S RNA fragment with and without paromomycin, and with and without the A1408G mutation. Two additional simulations with the Amber force field25, including the native and mutated 16S rRNA fragment without the drug, were performed to give insight into the dependence of the dynamics upon the force field parameters.
In this work we report the spontaneous flipping of A1492 and A1493 in and out of the RNA bulge, nucleotide dynamical correlation patterns, and the influence of the U1406○U1495 pseudo pair on the RNA internal dynamics. We also present the calculations of the water and sodium ion density in the paromomycin binding cleft and show the differences introduced by the mutation or by the choice of the force field. To the best of our knowledge, no prior MD simulations of the A1408G mutated RNA fragment were performed.
Methods
Starting structure preparation
For MD simulations we used the crystallographic structure of a 22-nucleotide 16S rRNA fragment which was solved at 2.5 Å resolution (PDB entry code 1J7T). This structure contains two mirrored, model prokaryotic ribosomal A-sites, each complexed with paromomycin. The choice of such RNA fragment can be justified in the light of experimental studies which demonstrate that the structural and thermodynamic aspects of aminoglycoside binding to the minimal A-site model are virtually the same as to the whole 30S subunit26. Moreover, root mean square deviation (RMSD) calculation shows that the positions of phosphorus atoms of the model RNA fragment (1J7T) differ from the positions of analogous phosphorus atoms in the crystal structure of the whole 30S subunit (1IBK) by only 1.2 Å. The two symmetric A-sites will be denoted here as part or site (a) and (b), respectively (see Figure 1b), hence the two copies of each nucleotide will be assigned either an (a) or (b) suffix (e.g. A1492(a) or A1492(b)).
The A1408G mutation, as well as the addition of the two terminal cytidines (C1402(a) and C1402(b)), which where missing in the original PDB file was performed with the SYBYL 7.2.5 program (Tripos). The complete sequence used in simulations is shown in Figure 1b. All hydrogen atoms were added using the hbuild function of Charmm27. All crystal water molecules were removed. To neutralize the total charge of the RNA, 44 Na+ ions were added with the use of a custom made Charmm script, which iteratively placed subsequent ions at points of the local minima of the potential energy.
The system was solvated with approximately 13,000 TIP3P28 water molecules which resulted in a simulation cell of 92×69×69 Å3, that provided at least 15 Å layer of solvent at each side of RNA. In order to mimic the intracellular salt concentration of 150 mmol per liter, 60 randomly chosen water molecules were replaced with 30 Na+ and 30 Cl− ions. Two different force fields were applied: Charmm2723,24 and Amber ff9925. The Charmm force field parameters for paro-momycin were assembled from, already available in Charmm, parameters for different sugar rings and amino alcohols. The partial atomic charges were determined on the basis of QM calculations, performed with the use of Gaussian29. The validity of the parameters was evaluated by comparing several inter-proton distances obtained during MD simulation of paromomycin in explicit solvent with the NMR data30 (for simulation details and parameters see Supporting Information).
Simulation protocol
All MD simulations were performed with the NAMD package31 in constant pressure (using the Langevin piston method32) and temperature (controlled by Langevin thermostat33), with periodic boundary conditions. Electrostatic interactions were calculated using the Particle Mesh Ewald summation method34. The SHAKE35 algorithm was used and a 2 fs integration time step.
In the thermalization stage, based on36, the system was heated from 30 K to 310 K with constraints applied to all heavy atoms of the RNA and, if applicable, the ligand. The constraint harmonic constant (k) was equal to 50 kcal/mol/Å2 in the first 85 ps of simulation and for another 35 ps it was lowered to 25 kcal/mol/Å2.
The equilibration stage was divided into two parts. First, the constraints were gradually diminished in 6 rounds of 50 ps and scaled as 0.1×0.5n, where n ∈ {0..5} is a round number. In the second, 600-ps long equilibration stage, the constraints were applied only to heavy atoms of terminal nucleotides C1402(a), C1402(b) (Figure 1b; k = 0,35 kcal/mol/ Å2), to P atoms of C1498(a) and C1498(b) (k = 0,3 kcal/mol/ Å2), and to P atoms of G1403(a) and G1403(b) (k = 0,25 kcal/mol/Å2). We adjusted k to obtain fluctuations of the termini corresponding to crystallographic temperature factors.
20 ns MD production phases were performed under the same conditions as the second stage of equilibration. Trajectories were collected every 5 ps. For a complete list of the performed MD simulations see Table 1.
Table 1.
Types of MD simulations and their abbreviations used in the text. PRO denotes simulations performed for the prokaryotic RNA sequence with A1408 and EU—for the A1408G mutated structure corresponding to eukaryotic-like sequence.
| Amber | Charmm | ||
|---|---|---|---|
| prokaryotic (A1408) | with paromomycin | — | CHARMM_PRO_PAR |
| without paromomycin | AMBER_PRO | CHARMM_PRO | |
| eukaryotic (G1408) | with paromomycin | — | CHARMM_EU_PAR |
| without paromomycin | AMBER_EU | CHARMM_EU | |
Divalent ions
In the crystal structure of the entire small ribosomal subunit (PDB entry 1IBK) three Mg2+ ions are reported near the aminoglycoside binding site. In order to imitate the environment of the RNA fragment as in the 30S subunit, we initially included these ions in the simulated system.
This approach, however, was unsuccessful because in one part of the RNA destabilization of base pairs was observed after the first 3 ns of the production stage. We suppose that such instability was caused by divalent ions due to their proximity to the unstable base pairs. The reason may be that Mg2+ and other divalent ions are generally not correctly represented in the currently available nonpolarizable force fields due to the large positive charge of these ions and their capability to polarize the surrounding particles20,37. Furthermore, the initial positions of Mg2+ ions were taken from the structure of the whole 30S subunit, superimposed onto the model A-site. As a result, they might have been too close to the RNA helix and thus not properly solvated, leading to too strong interactions, that caused the instability of the proximate base pairs. In the other half of the molecule, the movements of A1492 and A1493 toward the inside of the helix were observed. We performed a similar simulation without Mg2+ ions to check whether the dynamics of the bases would be different. The entire helix appeared to be more stable and we observed very similar movements of A1492 and A1493. Based on our test simulations and literature20,37,38 we decided to perform simulations only with explicit monovalent ions.
Trajectory analysis
To check whether the MD preparation protocol led to a stable simulation, we calculated the RMSD43 of atomic positions from the crystal structure as a function of the simulation time. For this purpose we used the g_rms program of the GROMACS39–42 package.
| (1) |
where mi stands for standard mass of each atom, is the position of atom i at time t.
To analyze the average mobility of individual nucleotides, the root mean square fluctuations (RMSF)43 as a function of the nucleotide number were calculated with the g_rmsf program of the GROMACS package. First, it calculates the RMSF per atom (RMSF(i), eq. 2) and then averages those values for each nucleotide (RMSF(Ni))43.
| (2) |
where T denotes the total simulation time and — the mean position of atom i. To compare the MD derived nucleotide mobility with their temperature factors (beta factors, B(Ni)) from the original X-ray structure we applied the following relation: , where B(Ni) is the average beta factor for all atoms of the nucleotide no. Ni.
The analysis of the hydrogen bond network, glycosidic torsions, pseudo-rotation and opening angles44,45 in the RNA helix was performed with X3DNA program45.
A correlation matrix was calculated for all phosphorous atoms except for the unpaired termini C1402(a) and C1402(b) due to the applied constraints. Global translations and rotations of the entire RNA were eliminated under VMD46. The elements of the correlation matrix were derived using the following formula47, implemented in the Octave program48:
| (3) |
where denotes a deviation of atom i from its mean position at time t.
The distribution of water molecules and ions was calculated with the MolDyAna49 software. The algorithm divides the whole system into 1 Å3 cubes and in each simulation step increases the counts in the populated cubes. Eventually, the counter for each cube is multiplied by a local density (Gaussian distribution) and normalized over the whole trajectory. For this procedure the MD snapshots were aligned with the first frame independently for each of the binding sites.
Results and Discussion
Stability of the simulations
The RMSD from the crystal structure in the six 20 ns long MD simulations are leveled and fluctuate in the range 1.5–4 Å. Simulations of the bare oligonucleotide performed with the Charmm force field give higher average RMSD than those performed with the Amber force field (see Table 2). The paromomycin reduces the flexibility of the RNA fragment: average RMSD in the Charmm simulations with the antibiotic are about 50% smaller than the corresponding values obtained without paromomycin.
Table 2.
RMSD of atomic positions (with standard deviations) from the initial MD configuration averaged over 20 ns MD simulations. For labels denoting the simulation type see Table 1.
| average RMSD [Å] | part (a) | part (b) |
|---|---|---|
| AMBER_PRO | (1.56 ± 0.39) | (2.45 ± 0.39) |
| AMBER_EU | (2.16 ± 0.34) | (2.39 ± 0.25) |
| CHARMM_PRO | (3.04 ± 0.37) | (3.10 ± 0.33) |
| CHARMM_EU | (3.33 ± 0.32) | (3.33 ± 0.28) |
| CHARMM_PRO_PAR | (1.66 ± 0.22) | (2.28 ± 0.12) |
| CHARMM_EU_PAR | (1.50 ± 0.18) | (1.56 ± 0.12) |
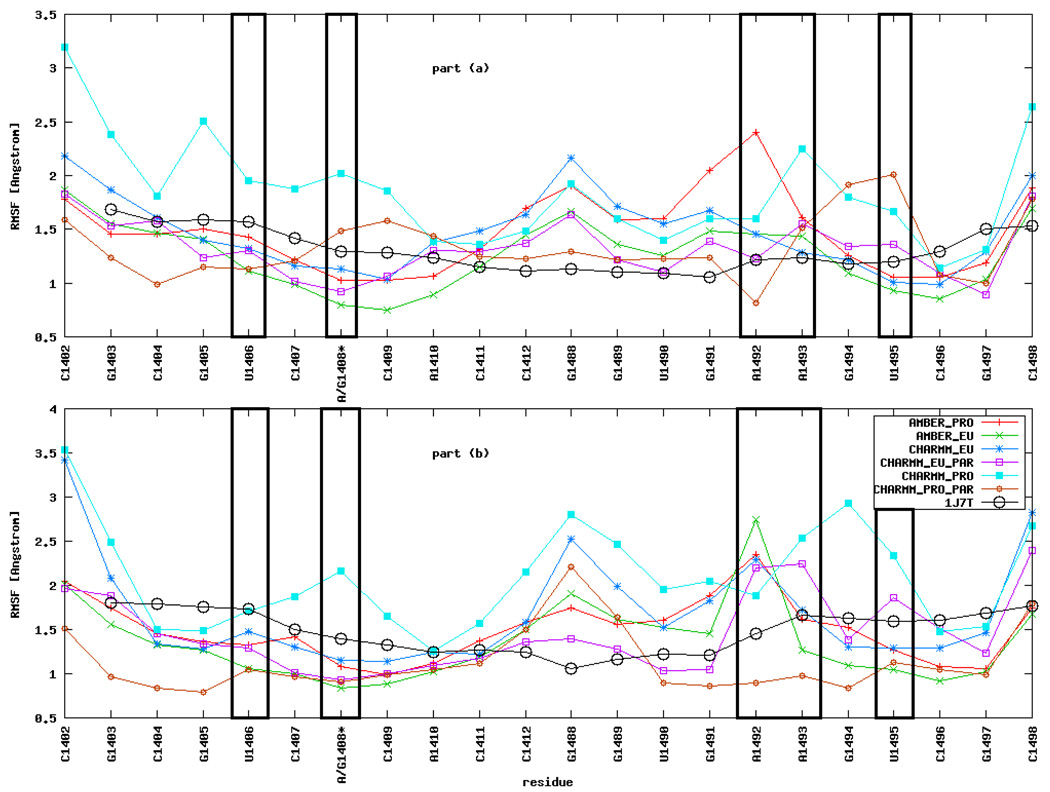
Figure 2 shows that RMSF are slightly higher for nucleotides in part (b) of the RNA fragment than for those in part (a), especially for A1492 and A1493. A possible reason for the observed RMSF discrepancies may arise from small differences in the initial positions of analogous atoms in parts (a) and (b) in the crystal structure. In the original PDB file, atomic positions in part (b) were determined with less accuracy than in part (a): they have higher temperature factors, and are surrounded by a smaller number of localized water molecules. It was shown that even a difference of 0.2 Å can lead to evident changes in the subsequent RNA dynamics21. Although, in general, two long enough MD simulations of the same system should converge to the same results, the limited sampling does not allow to cover all the accessible configurations.
Figure 2.
Root mean square fluctuations (RMSF) of nucleotide positions averaged over 20 ns MD simulations. See Figure 1b for the nucleotide numbering in parts (a) and (b). Asterisk (*) denotes that the base in this position is either guanine (G), as in eukaryota, or adenine (A), as in prokaryota. The comparison with the RMSF of the crystal structure of the complex is also shown.
Overall, the CHARMM_PRO simulation gives the highest RMSF for almost all nucleotides (see Table 1 for types of simulations) whereas the lowest values are observed in the AMBER_EU simulation. The high RMSF corroborate with the RMSD analysis and show that in our case the Charmm force field parameters allow for more mobility of nucleotides than the Amber parameters. In most of the MD simulations RMSF of A1492 and A1493 are higher than for the neighboring nucleotides due to their special role in the considered A-site RNA fragment (see Section Mobility of A1492 and A1493 for a detailed description of the observed moves).
As shown in a recent study21, an important factor for the stability of paromomycin binding site is the pair formed by U1406 and U1495. These nucleotides are in proximity to the adenine triplet and form an uridine pseudo pair (see Figure 1b). Among simulations of the bare RNA, their RMSF is the highest in the CHARMM_PRO and the lowest in AMBER_EU simulation. We do not observe any significant RMSF change of U1406 and U1495 upon antibiotic binding but we notice various configurations of this pair which are described in The U1406○U1495 pair Section.
The RMSF obtained from MD simulations are of the same order as the RMSF calculated from the crystallographic temperature factors presented in the original structure 1J7T (Figure 2, black circles). One must, however, note that the temperature factors were determined for the structure in the crystal environment i.e., with crystal packing forces and in the complex with a ligand. There-fore, in the simulations of the bare RNA in solution, the MD derived RMSF are expected to be higher than the crystallographic ones especially for residues forming the binding cleft and the termini. In our case this is particularly visible for A1492, A1493, G1488, G1489, U1490, G1491, and C1498 because the range of movement available for A1492 and A1493 increases after removing the paromomycin; therefore, these and neighboring nucleotides have to significantly adjust their positions to find the new potential energy minimum.
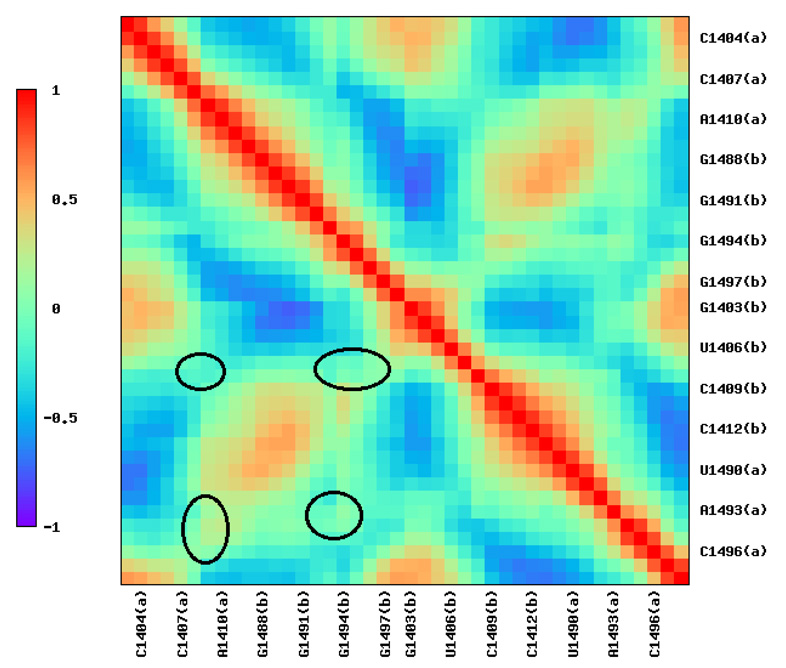
The overall dynamical correlation patterns, presented in Figure 3 for the selected AMBER_PRO simulation, are similar for all types of MD simulations. We observe a strong positive correlation between the subsequent nucleotides (red diagonal), a weaker positive correlation between nucleotides of the opposite strands forming base pairs (yellow counter-diagonal, i.e., the diagonal starting in the left bottom corner), and also a negative correlation between the nucleotides situated in the center and termini of the RNA helix (blue spots). Other positive and negative correlation spots indicate the central bending and twisting of the RNA helix. Spots of zero correlation denoted with black circles indicate the RNA bulge i.e., nucleotides A/G1408, A1492, A1493 and the neighboring ones whose mutual moves were not correlated with other nucleotides. Only some subtle differences in correlation patterns for the bare RNA, related to the use of different force fields and A1408G mutation, were noticed and were mainly connected to movements of A1492 and A1493. The correlations derived from the simulations of the complex with paromomycin (data not shown) suggest that the eukaryotic-like structure bends less dynamically than the prokaryotic one because the negative correlation areas are more intense and larger for the prokaryotic structure.
Figure 3.
Exemplary dynamical correlation matrix derived from the AMBER_PRO MD simulation.
Mobility of A1492 and A1493
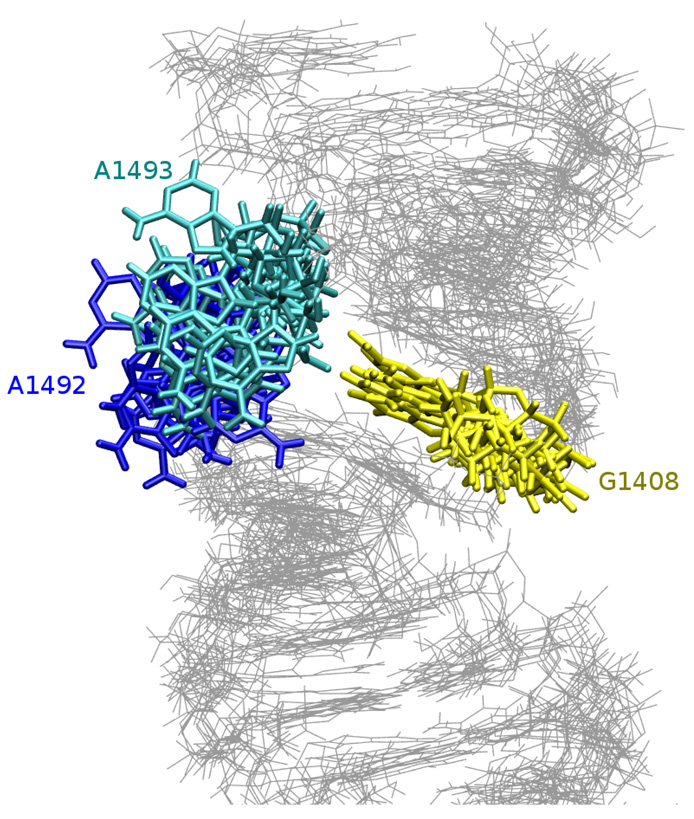
Adenines 1492 and 1493 are important for the binding of paromomycin because while flipping out to the solvent they make space for the antibiotic. Our simulations were based on the crystal structure of the complex, therefore, in the MD starting configuration, A1492 and A1493 occupied a flipped-out state. During the visualization of the MD trajectory we observed extensive motions of those adenines into and out of the RNA helix. While outside of the helix, the adenines moved generally as one pair but separate motions were observed towards the intra-helical states.
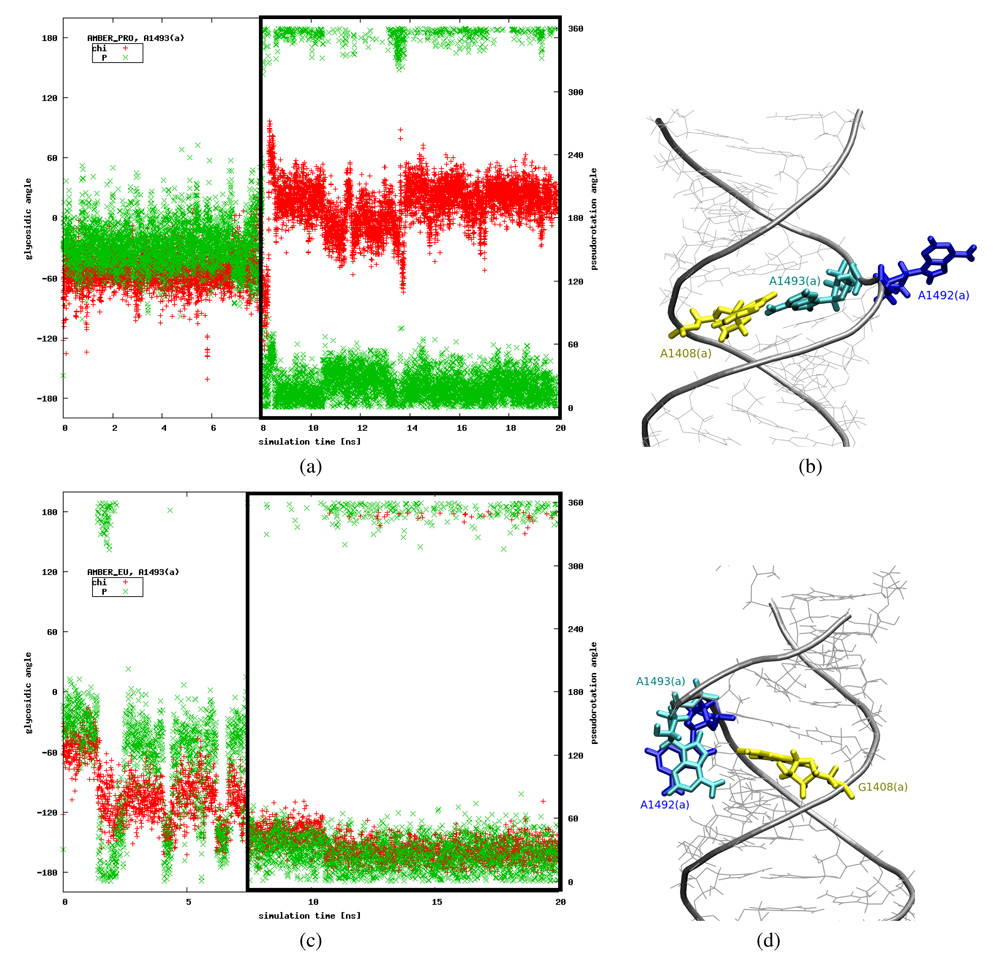
Glycosidic and pseudorotation angles
One possible way to quantify these movements is to analyze the nucleotide glycosidic (χ) and ribose sugar pseudorotation (P) angles20,44. For each trajectory we calculated the A1492 and A1493 χ and P angles. We found, however, that it may be an insufficient measure. Such special cases are presented in Figure 4. For example, a visualization of the AMBER_PRO trajectory reveals that A1493(a) moves toward the inside of the helix around 8th ns, however this fact is not reflected by an expected change in the glycosidic angle (see Figures 4a and 4b). Moreover, in the AMBER_EU simulation the χ and P angles indicate the flipped-in configuration of A1493(a), whereas the base, instead of being inside the helix, points toward the minor groove of RNA, in almost perpendicular plane to the G1408(a) base (see Figures 4c and 4d).
Figure 4.
Plots showing glycosidic (“chi”) and pseudorotation (“P”) angles changes in time [(a), (c)] in comparison with the configurations observed in MD trajectories [(b), (d)]. Black frames in plots (a) and (c) indicate the period in the simulation when the configurations presented in figures (b) and (d) were observed. Correct ranges of parameters for a base inside the helix are P ≈ 18° and χ ∈ {[–180; –90] ∪ [90; 180]}°.
Hydrogen bonding
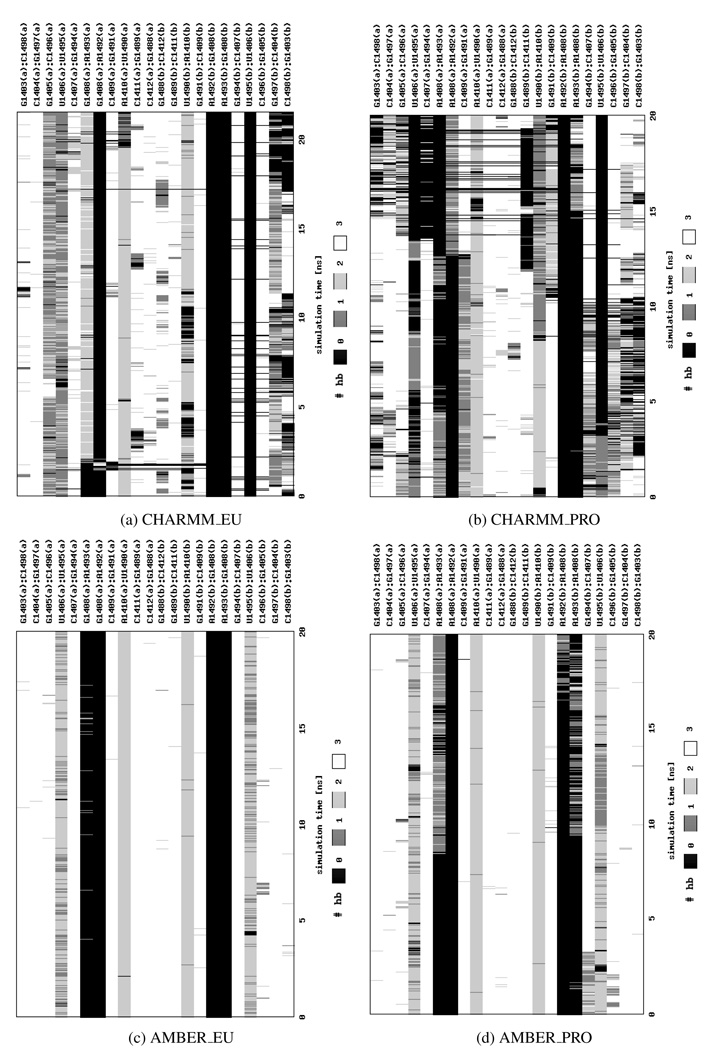
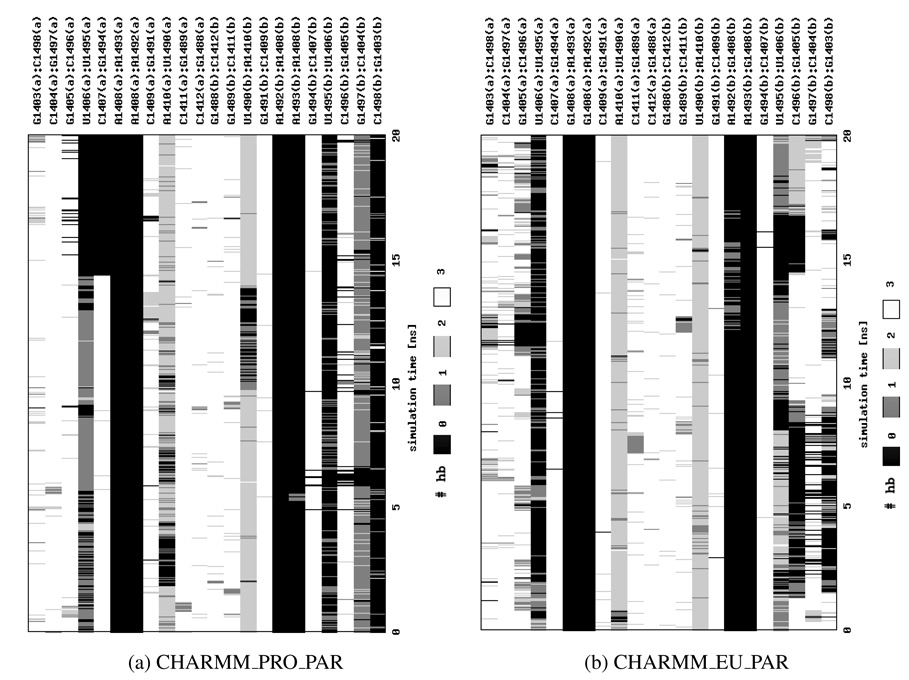
Therefore, to better classify adenine movements, we both carefully analyzed and visualized trajectories and applied an additional measure based on the number of hydrogen bonds that A1492 and/or A1493 formed with the opposite base 1408. This approach enabled us to differentiate the flipped-in and flipped-out states of the adenines. The changes in hydrogen bonding are presented in Figure 5 and show that, in general, adenines in parts (a) and (b) can behave differently. We also observed differences in the hydrogen bond network between the simulations of the native and A1408G mutated structure, which are more pronounced in the Charmm force field. In CHARMM_EU A1493(a) forms two or three hydrogen bonds with the opposite G1408(a) and this pair lasts throughout almost entire simulation (see Figure 5a). Whereas in CHARMM_PRO A1493(a) is connected to A1408(a) by one or two hydrogen bonds that break after approximately 5 ns (Figure 5b). By the end of the CHARMM_PRO simulation the A1493(b):A1408(b) pair is created but it is still far less stable than the one of the CHARMM_EU simulation. This observation suggests that the G:A interactions are more stable than the A:A ones which may be one of the causes of paromomycin selectivity toward bacterial RNA, where the pair A1408:A1493 can be more easily broken facilitating the antibiotic binding.
Figure 5.
The number of hydrogen bonds (hb) formed between base pairs in the RNA helix presented as a function of the simulation time. Calculations were performed using X3DNA45 program.
On the other hand, in the simulations with the Amber force field of the eukaryotic-like A-site, i.e., AMBER_EU, we did not observe the hydrogen bonding between A1492 or A1493 and G1408 in neither part of the molecule (see Figure 5c). In part (b) the adenines were in a flipped-out configuration throughout the entire simulation, whereas in part (a) A1493(a) was outside of the helix, stacking with A1492 and interacting with the minor groove of the RNA (as in Figure 4d). In contrast, in the AMBER_PRO simulation the hydrogen bonds between A1493 and A1408 were formed in both RNA parts (Figure 5d). In part (a) only one base, A1493(a), was in the flipped-in state, and A1492(a) was bulged out to the solvent, whereas in part (b) A1493(b) moved toward the inside of the helix at about 10th ns, A1492(b) followed it after another ~7 ns, and then both adenines stayed in the flipped-in configuration till the end of the simulation. In this conformation only one base at a time could form the hydrogen bonds with the opposite A1408(b), thus in Figure 5d the exchange of bonds is visible.
Intra-helical configurations
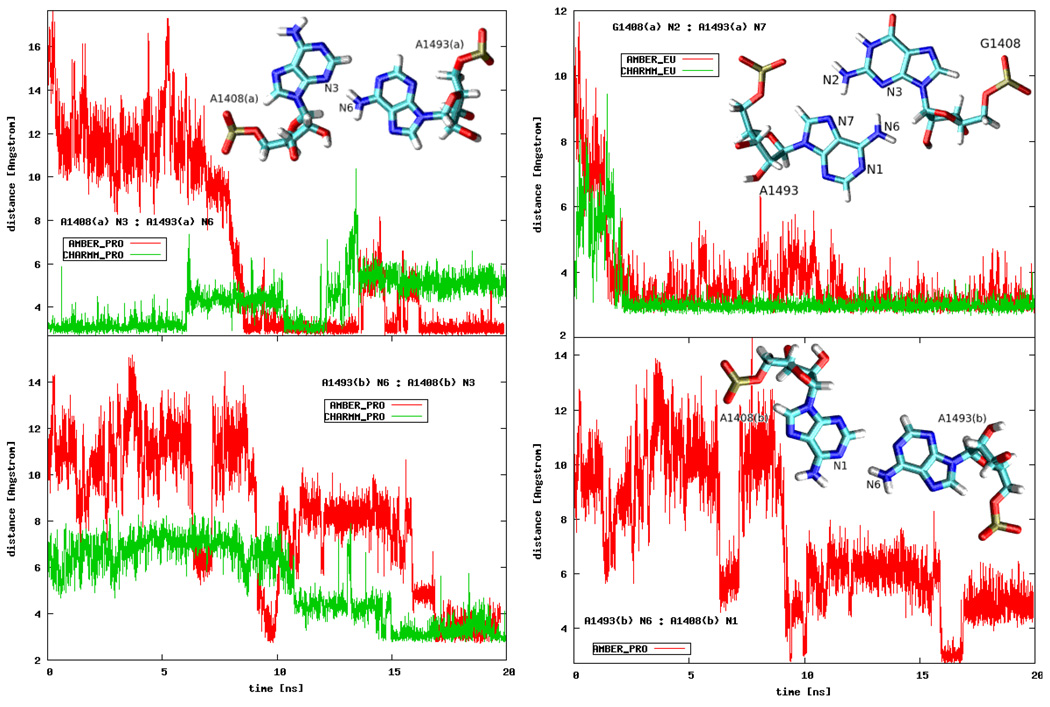
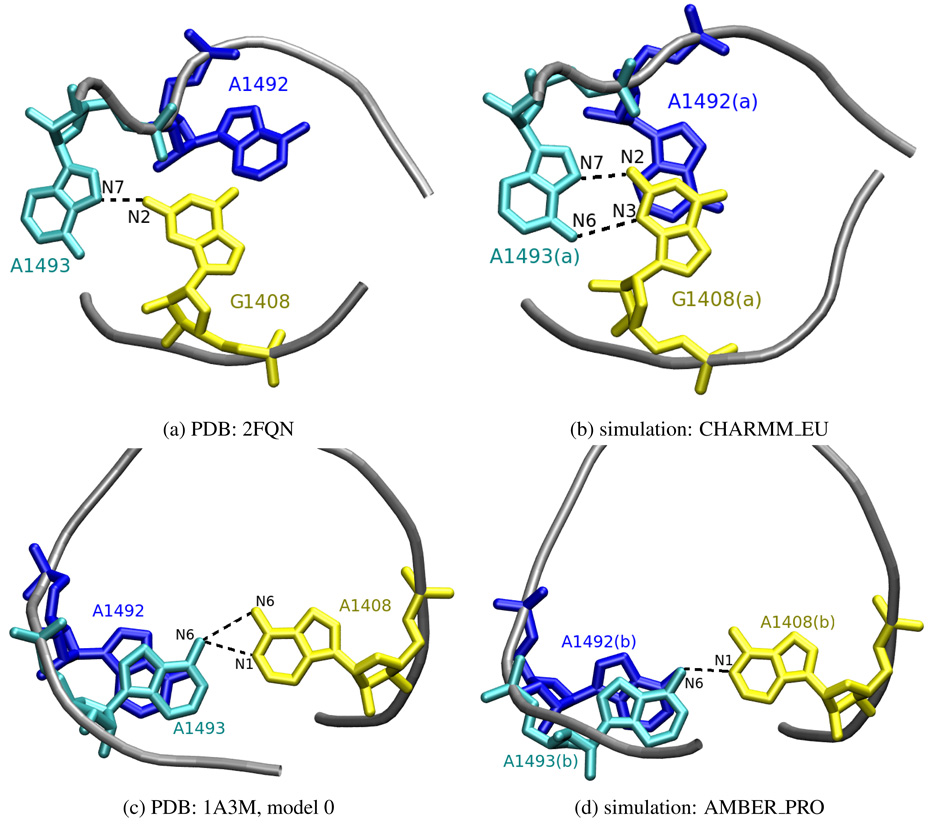
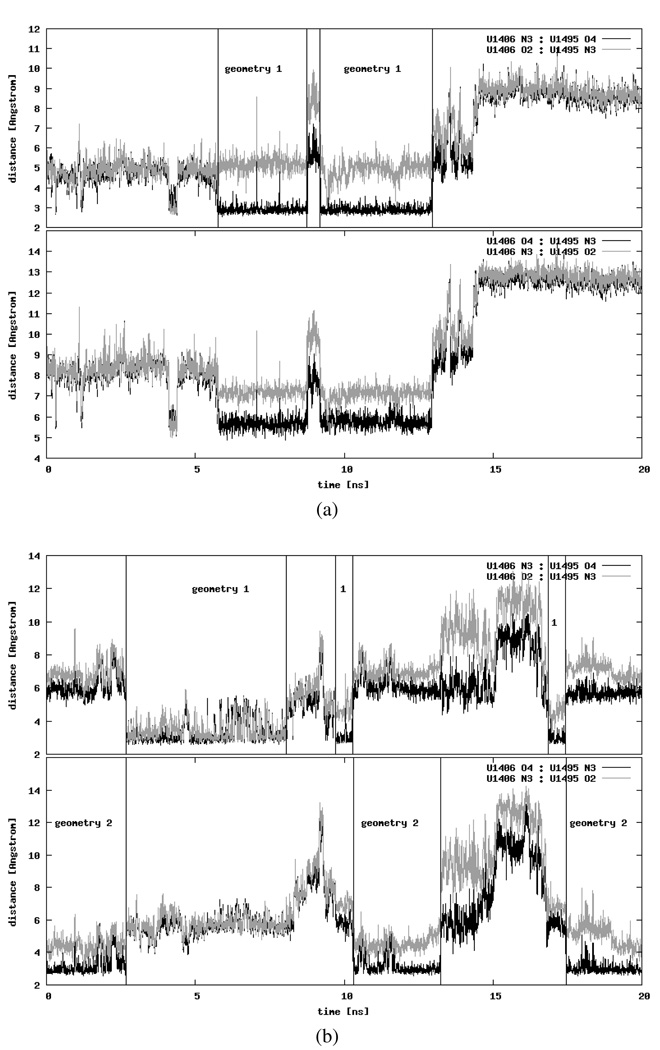
We detected several different intra- and extra-helical adenine configurations. In order to differentiate between the intra-helical states, apart from monitoring the number of hydrogen bonds formed with A1408 or G1408, we also analyzed the conformations of the three bases in relation to one another. Exemplary pairings are presented in Figure 6. In CHARMM_EU simulation the G1408(a):A1493(a) pair was well-defined in a conformation called the sheared GA pair44, where the A1493(a) N7:G1408(a) N2 and A1493(a) N6:G1408(a) N3 hydrogen bonds were formed. A similar configuration was observed in the X-ray structure (PDB entry 2FQN) of the eukaryotic A-site but with G1408 and A1493 forming only one hydrogen bond — A1493 N7:G1408 N2. Figures 7a and 7b present the comparison of the 2FQN X-ray structure with an MD snapshot. The A1493(a) N7:G1408(a) N2 bond was also created in the AMBER_EU simulation but, as mentioned before, A1493(a) was located outside of the RNA bulge in almost perpendicular plane to G1408(a) as shown earlier in Figure 4d.
Figure 6.
Distances between the selected atoms of A1493 and A1408 or G1408 plotted as a function of the simulation time (see text for details on the atom selection). The insets present the types of interactions.
Figure 7.
Comparison of A1492, A1493 and G/A1408 configurations observed in MD simulations and experimentally resolved structures. Only heavy atoms are shown, hydrogen bonds are denoted as dashed black lines.
In CHARMM_PRO and AMBER_PRO simulations, forming of the A1493 N6:A1408 N3 hydrogen bond was observed in both RNA parts. Moreover, in AMBER_PRO also the A1493(b) N6:A1408(b) N1 hydrogen bond was present for approximately 2.5 ns (see Figure 6). Such interaction can be also found in prokaryotic NMR structures (PDB entry 1A3M) where the adenine triplet accomplished a very similar configuration. The comparison with an MD snapshot is shown in Figures 7c and 7d. Also other conformations of A1493(b) and A1408(b) were seen in the AMBER_PRO simulation (data not shown) where a A1493 N1:A1408 N1 hydrogen bond was formed, similarly to the X-ray structure of the 30S subunit (PDB entry 1J5E). However, this hydrogen bond was not stable in our simulation.
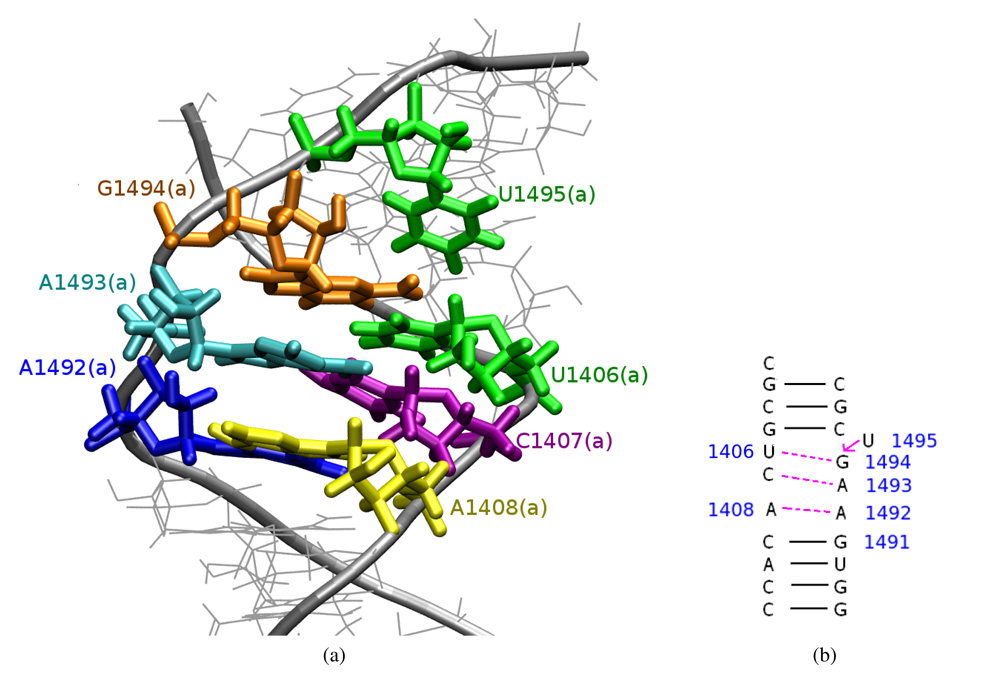
In CHARMM_PRO simulation an interesting configuration was observed and is presented in Figure 8. U1495(a) bulged out which led to the formation of U1406(a):G1494(a), C1407(a):A1493(a) and A1408(a):A1492(a) pairs. This conformation was quite stable and lasted from the 13.5 ns till the end of the simulation. Moreover, a similar shift of the pair-forming was observed in analogous simulation in complex with paromomycin (see below). The RNA is, in general, characterized by a complicated internal dynamics and this kind of pair forming with one nucleotide bulging out of the helix is often found in RNA structures50, however in the A-site this kind of shift was not yet observed.
Figure 8.
A configuration of the A-site observed in the last 7 ns of the CHARMM_PRO simulation: (a) the trajectory snapshot; (b) a scheme of the formed hydrogen bonds — compare with Figure 1b.
Extra-helical states
Our simulations also provided a wide range of the adenine flipped-out conformations. Due to the available configurational space these are more difficult to quantify than the flipped-in states, therefore, in Figure 9 we only present an ensemble of configurations derived from the MD trajectory. The conformations with the most bulged out adenines were found mainly in the simulations with the Amber force field and resembled the 1J7T and 1IBM X-ray structures. Types of flipped-out configurations where adenines interact with the minor groove of the RNA helix are similar to those observed in the 1A3M NMR models.
Figure 9.
Snapshots of the A-site model derived from the AMBER_EU simulation depicting the range of moves of A1492 and A1493 in the bare A-site.
Time scale of adenine states
In several experimental studies, concerning the prokaryotic A-site, it was suggested that A1492 can occupy the flipped-in and flipped-out states with the same probability11,12 or with a slight preference toward the intra-helical configuration18 (69% of the time A1492 was in a flipped-in state). In theoretical studies, the A1492 and A1493 flipping events were observed to occur both together and separately17,19 or only for A149220. Interestingly, in19 more flipping events were noticed for A1493, whereas in20 it was A1492 that was more dynamic. Moreover, in19 the authors estimated that the flipped-in state is slightly preferred energetically (the difference between the flipped-in and flipped-out state was 0.66 and 1.01 kcal/mol for A1492 and A1493, respectively). This result correlates with our observations because overall the adenines prefer an intra-helical configuration. In every simulation of the bare RNA adenines moved toward the inside of the RNA helix although the hydrogen bonds were not always formed and sometimes bases returned to their initial extra-helical state. This fact can be best analyzed with the time scale of measurable base pair opening angle presented in Table 3.
Table 3.
Percentage of time a pair is formed, and thus an opening angle is measurable, in various types of MD simulations (see Table 1).
| bases forming the pair: | ||||
|---|---|---|---|---|
| A/G1408(a) and | A/G1408(b) and | |||
| A1492(a) | A1493(a) | A1492(b) | A1493(b) | |
| AMBER_PRO | — | 49.8% | 9.0% | 25.0% |
| AMBER_EU | — | 2.7% | — | — |
| CHARMM_PRO | 40.8% | 24.9% | — | 27.6% |
| CHARMM_EU | 1.5% | 88.8% | — | — |
Complex with paromomycin
In order to analyze the influence of the ligand on the mobility of adenines, we performed two MD simulations of the complex with paromomycin, denoted CHARMM_PRO_PAR and CHARMM_EU_PAR. The number of hydrogen bonds between the RNA strands is presented in Figure 10. The overall structure of the A-site was more stable than in the corresponding Charmm simulations without the ligand, however, still not as stable as in no-ligand Amber force field simulations (compare Figure 10 with Figures 5d and 5c).
Figure 10.
The number of hydrogen bonds (hb) formed between base pairs in the RNA helix derived from MD simulations with paromomycin. Calculations were performed with X3DNA45.
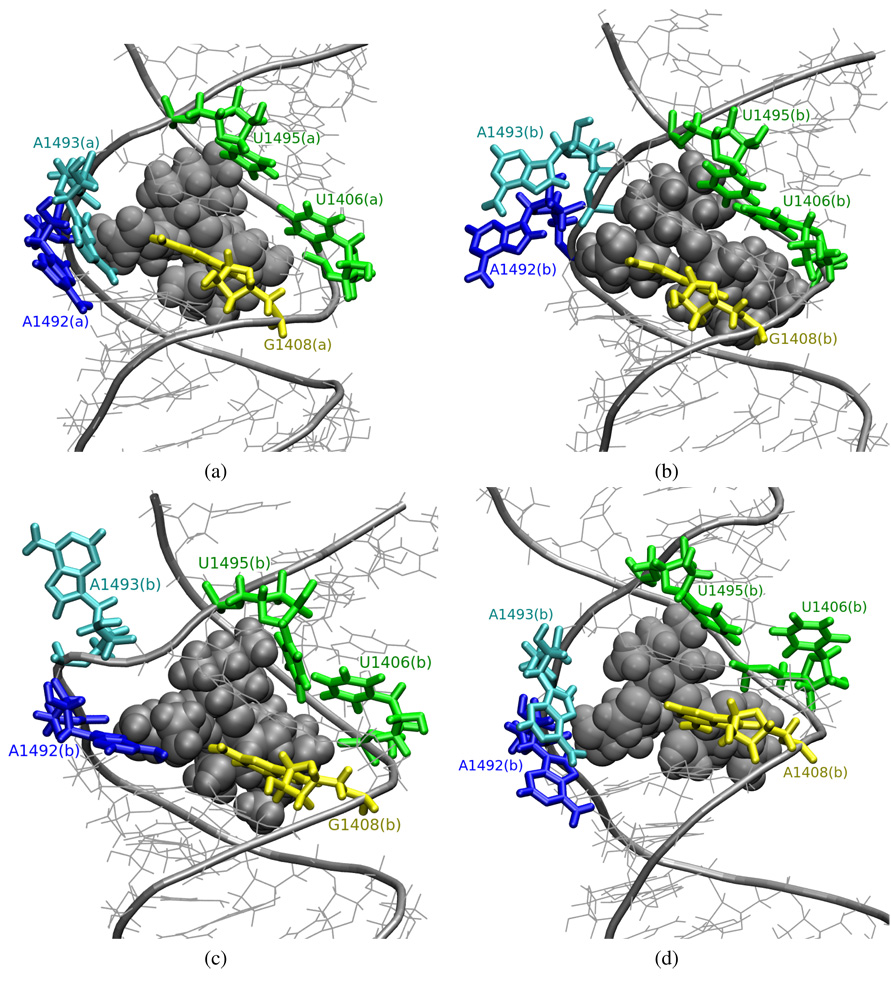
Visualization of the trajectory shows that in CHARMM_EU_PAR simulation, part (a) adenines moved toward the minor groove of the RNA helix at about 5th ns and they stayed in the conformation presented in Figure 11a until the end of the simulation. Adenines in a similar position were reported in the NMR structure of the prokaryotic A-site model complexed with paromomycin (PDB entry 1PBR). In part (b) of the RNA fragment, both adenines stayed in the flipped-out state shown in Figure 11b for about 7 ns. Then A1492(b) moved inward, and A1493(b) bulged out even more into the solvent to a conformation shown in Figure 11c. Next, between 12thand 16th ns of the simulation, A1492(b) formed a hydrogen bond with G1408(b) (see Figure 10b). A similar conformation was found in a prokaryotic structure, 1T0E, where A1493 is bulged out, and A1492 and A1408 form a pair.
Figure 11.
Snapshots from the trajectories of the RNA-paromomycin complexes depicting conformations of A1492 and A1493; van derWaals spheres represent paromomycin; (a)–(c) CHARMM_EU_PAR, (d) CHARMM_PRO_PAR.
In CHARMM_PRO_PAR, A1492(a) and A1493(a) acquired a flipped-out conformation throughout the entire simulation, resembling the configuration from the starting structure. In this part of the RNA we also noticed a slipped configuration of G1494(a), U1406(a) and U1495(a), similar to the one observed in CHARMM_PRO simulation presented in Figure 8, but here C1407(a) was not paired with A1493(a). This configuration lasted for about 5 final ns of the MD simulation. In part (b) of the RNA, both A1492(b) and A1493(b) moved toward the RNA groove in the beginning and formed a stable configuration similar to the one presented in Figure 11a and to the one found in the 1PBR structure. These adenines possessed, however, great mobility and they sometimes moved back out to the configuration presented in Figure 11d.
In summary, the presence of paromomycin makes A1492 and A1493 acquire more often the flipped-out conformation. The two simulations of the complexes display very similar configurations of the adenines, however, in general in the simulation of the A1408G mutated structure the extreme configurations with adenines bulged out into the solvent were less frequent than in the simulation of the original, non-mutated RNA helix.
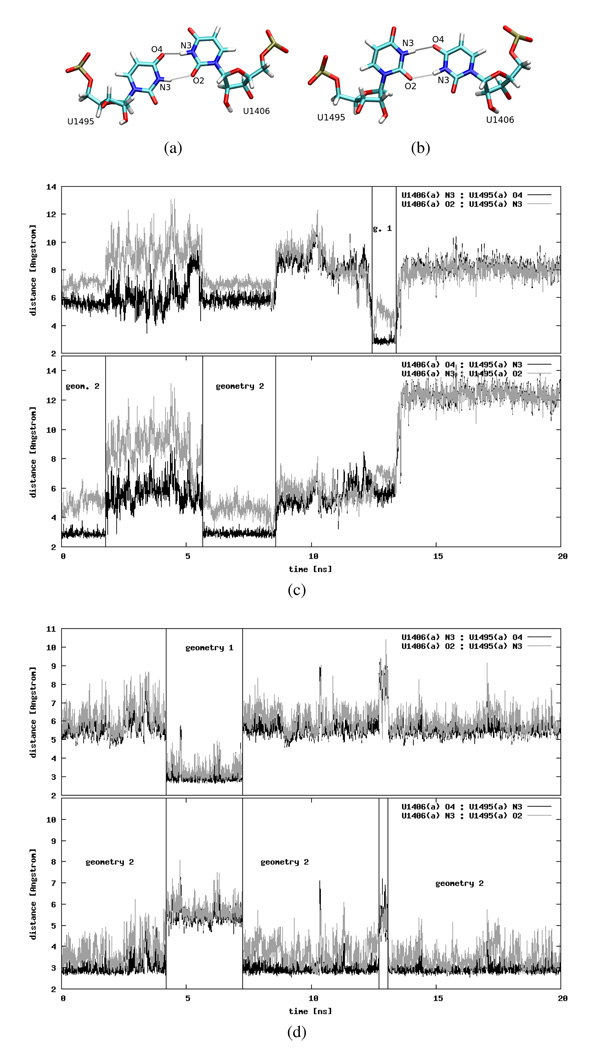
The U1406○U1495 pair
The highly conserved U1406○U1495 pseudo pair is expected to be of fundamental importance for the ribosome function7. The stability of the U1406○U1495 pseudo pair has been recently reported as an important factor for the stability of the whole A-site RNA fragment21. This pair is most often found in two distinct geometries presented in Figures 12a and 12b. In the first geometry the hydrogen bonds are formed between U1406 N3 and U1495 O4, and between U1406 O2 and U1495 N3, while in the second geometry — between U1406 O4 and U1495 N3, and between U1406 N3 and U1495 O2. According to the NMR structures14, the first geometry is the preferred one, whereas in the crystal structures the uracil pair is found in the second geometry2,22. In general, the hydrogen bond between N3 and O2 is mediated by a water molecule. However, in the initial simulation structure, the water molecule is visible only in one part of the RNA between the N3 atom of U1406(a) and the O2 atom of U1495(a). The lack of electron density from a water oxygen in part (b) may be correlated with the fact that this half was resolved with lower accuracy. Still, in the reports of recent MD simulations of the A-site RNA complexed with paromomycin, it was suggested that a water molecule should also be present in part (b)21.
Figure 12.
Two possible geometries of the uridine pair: (a) first geometry, (b) second geometry. Distances between the selected atoms of part (a) uridines plotted as a function of the simulation time: (c) CHARMM_PRO and (d) AMBER_PRO (see text for details on the atom selection). The time intervals when the uridines adopt a certain geometry are marked with horizontal lines and labels identifying the geometry.
Uridine pseudo pair in the bare RNA fragment
According to our MD simulations the aforementioned hydrogen bond distances differ between the Amber and Charmm force fields, as well as between the native and A1408G mutated trajectories. Figures 12c and 12d present exemplary data. In the CHARMM_PRO simulation the uridine pair in part (a) was formed three times and only for a few nanoseconds: twice in the second and once in the first geometry with only one of the expected hydrogen bonds truly formed (Figure 12c). In the first geometry it was U1406(a) N3:U1495(a) O4, and in the second — U1406(a) O4:U1495(a) N3. The distances between other atoms in each geometry suggest that those contacts might be mediated by a water molecule (see discussion in the Hydration density Section). In the same simulation but in part (b) (data not shown), the uracil pseudo pair as in the second geometry formed in the beginning and lasted for approximately 1.2 ns, then it switched to the first geometry for another 2 ns before the uridines drifted away from each other for the remaining simulation time.
In the CHARMM_EU simulation, in part (b) of the RNA, the uridine pseudo pair was not well defined and the four monitored distances between atoms of U1406(b) and U1495(b) showed that these bases were apart throughout the entire simulation (data not shown). On the other hand, in part (a) the uridine pair attained the second geometry for almost all of the simulation time and only a short opening event was observed between 6.12 and 6.66 ns.
In the simulations with the Amber force field the uridine pseudo pair in parts (a) and (b) was well preserved and appeared mainly in the second geometry. However, in the AMBER_PRO simulation the pair in part (a) switched its conformation to the first geometry for several nanoseconds from 4th to 7th ns (Figure 12d) and short opening events were observed in both parts – around 13th ns for 450 ps in part (a) and around 2nd ns for 465 ps in part (b). In AMBER_EU around 4.5 ns only one ~200 ps long opening occurred in part (b) of the molecule (data not shown). Also in the Amber force field we noticed that the distance between U1406 N3 and U1495 O2 fluctuated more than the one between U1406 O4 and U1495 N3, which suggests that a water-mediated hydrogen bond was formed (see Hydration density Section).
In summary, in the simulations with the Charmm force field the uridines were not as stable as in the Amber force field. Furthermore, the A1408G mutation possibly reduced the flexibility of the system because in both force fields the U1406○U1495 pair in the mutated structure formed the second geometry almost throughout the entire simulation, similar as in experimental studies2.
Uridine pseudo pair in complex with paromomycin
In MD simulations of the RNA-paromomycin complex, the uridine pair was formed, however for a limited time. In the CHARMM_PRO_PAR simulation, only part (a) uridines acquired any of the previously described geometries. Figure 13a shows that the first geometry with both hydrogen bonds formed around 4th ns for approximately 200 ps. Later around 6th ns only one interaction i.e., U1406(a) N3:U1495(a) O4 was created for about 7 ns, with an opening event of 0.5 ns. After-wards, U1495(a) bulged out and stayed in that configuration until the end of the dynamics.
Figure 13.
Distances between the selected atoms of uridines during MD simulations (a) CHARMM_PRO_PAR (part (a) of the RNA helix) and (b) CHARMM_EU_PAR (part (b) of the RNA helix); (see text for details on the atom selection, and Figures 12a and 12b for the definition of the first and the second geometry, respectively). The time intervals when the uridines adopt a certain geometry are marked with horizontal lines and labels identifying the geometry.
In the simulation of the mutated RNA complexed with paromomycin (CHARMM_EU_PAR) the uridine pair was formed only in part (b) of the RNA helix (see Figure 13b). The U1406(b)○U1495(b) pair formed two hydrogen bonds in the first geometry only once from ~3th ns to ~9th ns. Still, several times one contact per geometry was created. Part (a) uridines often formed one hydrogen bond per each geometry for a very limited time (the longest event lasted for about 0.5 ns, data not shown).
Our data suggest that paromomycin does not necessarily stabilize the uridine pseudo pair. In both simulations with the ligand, this pair was formed only for a limited time and U1495 often bulged out into the solvent. In the experimentally resolved structures of the model A-site complexed with neamine and its derivatives, the uridines also do not form a pair although it is U1406 that bulges out2. Interestingly, even when no hydrogen bonds were formed, only U1495 flipped out and its movements were more dynamic than U1406, probably because of the interactions between U1406 and paromomycin, which prevented this uridine from flipping out.
Ion densities
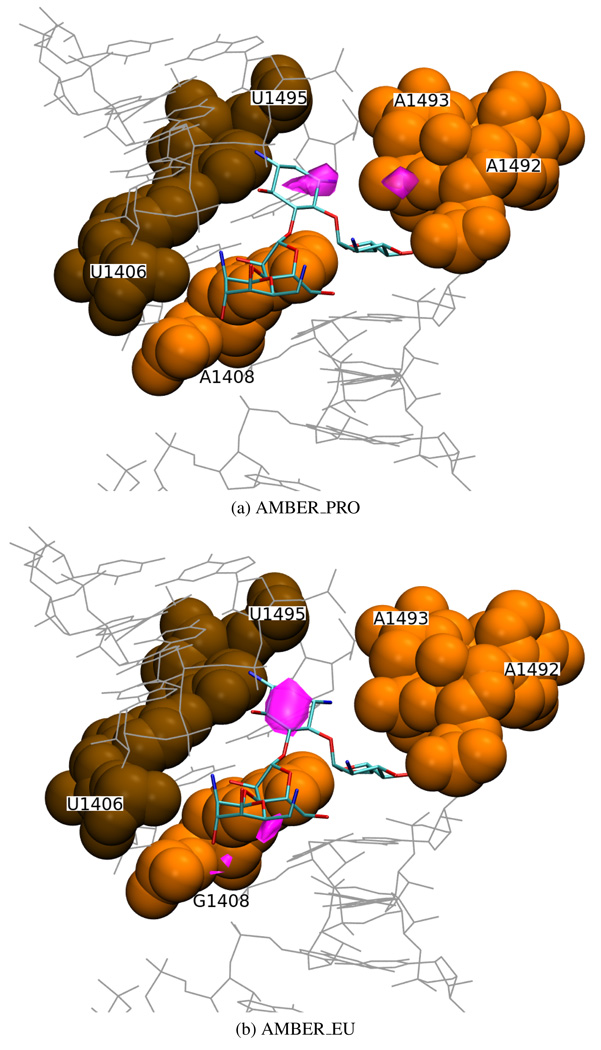
In physiologic pH, all five paromomycin amino groups (Figure 1a) are positively charged3. Their electrostatic interactions with the negatively charged RNA provide a major contribution to the binding free energy26,51–53. It was suggested that, upon binding, aminoglycosides expel positive ions that natively occupy the RNA bulge12,54,55. In order to check if there is any localized ion density in the binding site in the absence of paromomycin, we analyzed an average distribution of sodium ions in the simulations without the antibiotic. We wanted to detect any differences introduced by the choice of force field and by the A1408G mutation.
One of the most dense areas (more than 0.053 sodium ions per Å3) was found at similar positions in parts (a) and (b) in every simulation of the bare RNA (see Figure 14, pink areas). It corresponds to the binding location of paromomycin ring II, that carries two amino groups, indicating a particular complementarity of the receptor and ligand electrostatic fields in this area. This stands in agreement with other experimental1 and MD21 studies which show that rings I and II, common to most aminoglycosides, are the “anchor” for their binding to the A-site.
Figure 14.
Areas of sodium ion density (in pink) of ≥ 0.053 ions per Å3 derived from MD simulations in the Amber force field (part (b) of the molecule) and the superimposed crystal structure of the complex with paromomycin (PDB entry 1J7T); van der Waals spheres denote the U1406(b)○U1495(b) pair (in brown) and A/G1408(b), A1492(b), A1493(b) (in orange).
Interestingly, another area of high sodium ion density (more than 0.053 per Å3) was found in the vicinity of the O6 atom of G1408 in all the simulations of the mutated RNA (see Figure 14b), including the one with paromomycin. No such elevated ion density in the corresponding location in the simulations of the prokaryotic RNA was observed because in those structures the O6 oxygen is exchanged to an amino nitrogen (A1408 N6). A well localized positive ion density around the O6 oxygen of G1408 indicates that it is an important source of negative potential in the eukaryotic binding site.
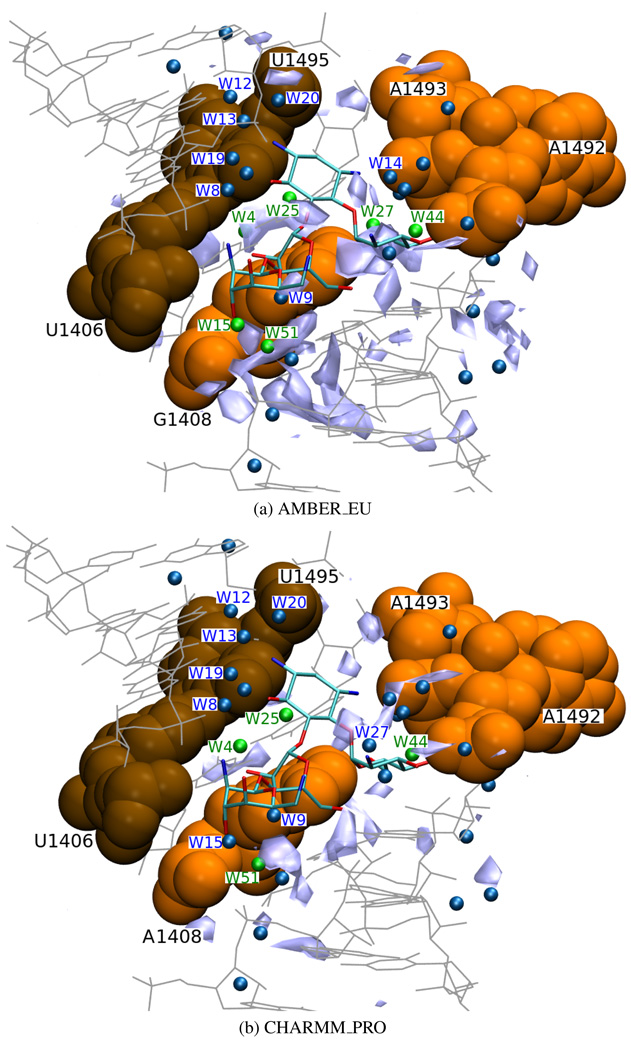
Hydration density
Several contacts between paromomycin and RNA are accomplished indirectly, through water molecules2,22. In the 1J7T structure the positions of numerous crystal water molecules were resolved. To determine the distribution of water molecules within the binding cleft in the absence and presence of the antibiotic, we analyzed the MD hydration patterns. After superimposing the crystal water molecules of the initial structure of the complex onto the MD derived water densities, some areas of high water density (more than 0.23 water oxygens per Å3) were identified. Table 4 and Figure 15 present those areas and compare them with the positions of some of the crystallo-graphically resolved waters (see Figure 1a for the location of the water molecules situated less than 4 Å from paromomycin and reference22 for the information on the remaining ones).
Table 4.
Selected water molecules of the 1J7T crystal structure and their MD corresponding water density areas. “+” denotes water density areas higher than 0.22 water molecules per Å3 observed in the position of the corresponding crystallographic water molecule; “−” denotes lack of high water density in this position. Brackets denote water molecules located in part (b) of the RNA duplex.
| water molecule | AMBER_PRO | AMBER_EU | CHARMM_PRO | CHARMM_EU | CHARMM_PRO_PAR | CHARMM_EU_PAR |
|---|---|---|---|---|---|---|
| W1 | + | + | − | − | − | + |
| W4 | + | + | + | + | − | + |
| (W7) | (−) | (−) | (−) | (−) | (−) | (−) |
| W8 (W54) | −(−) | −(−) | −(−) | −(−) | −(+) | −(−) |
| W9 (W2) | − (−) | −(+) | −(−) | −(+) | +(+) | +(+) |
| W12 | − | − | − | − | − | − |
| W13 | − | − | − | − | − | − |
| W14 | − | − | − | − | + | + |
| W15 | − | + | − | − | + | + |
| W19 | − | − | − | − | + | − |
| W20 | − | − | − | − | + | + |
| W25 | + | + | + | + | − | + |
| W27 | − | + | − | − | − | + |
| W28 | − | + | − | − | − | + |
| W32 | − | − | − | − | + | + |
| W44 | − | close | + | + | − | + |
| (W45) | (+) | (+) | (+) | (+) | (+) | (+) |
| W49 | + | + | + | − | − | − |
| W51 | − | + | + | + | + | + |
Figure 15.
Areas of high water density (light blue, ≥ 0.23 water oxygens per Å3) located in part (a) of the simulated structure, superimposed on the crystal structure of the complex with paromomycin (PDB entry 1J7T); van der Waals spheres denote the U1406(b)○U1495(b) pair (in brown) and A/G1408(b), A1492(b), A1493(b) (in orange); smaller spheres symbolize crystal water oxygen atoms, the ones which were identified in the simulation are marked in green and the ones discussed in the text are numbered.
Water molecules number W8, W9, W14, W32 and W44 link the paromomycin with nucleotides A1408, A1492, A1493, U1406 and U1495 (Figure 1a). Therefore, we checked if there is an increased water density in their positions prior to the ligand binding (i.e., in the bare RNA) or whether it is the ligand that stabilizes those water molecules, possibly contributing to an unfavorable entropic effect. Table 4 shows that the high water density for W32 was not reproduced in any of the simulations without the antibiotic. This can be explained by the fact that W32 mediates between the O6′ oxygen of paromomycin ring I and O2P atom of A1493 that displayed higher mobility than other atoms in the binding site due to A1492 and A1493 flipping. Similarly, the position of water W14, connecting ring I of paromomycin with the O1P atom of A1493, was not reproduced in any simulation of the bare RNA. However, in proximity of W44, which is also located near the bulge, the areas of high water density were observed in most of the simulations. The reason may be that W44 is close to and stabilized by G1494.
The position of the water molecule W9, which mediates between paromomycin and base no. 1408, was reproduced only in the simulations of the complex, in contrast to molecule W51 in its vicinity which was reproduced in most of the simulations. Interestingly, near W2 in part (b), which corresponds to W9 in part (a), areas of high water density were observed in the simulations of the mutated structure and with the antibiotic. In both AMBER_EU and CHARMM_EU, A1492(b) and A1493(b) did not flip inside the bulge what could explain the presence of water molecule W2. Molecules W1, W4, W25, W27 and W28, located at the extra-helical side of nucleotide 1408, were generally reproduced in the simulations of the bare RNA. This probably means that it is the paromomycin that stabilizes the water molecules inside the cleft during the binding process.
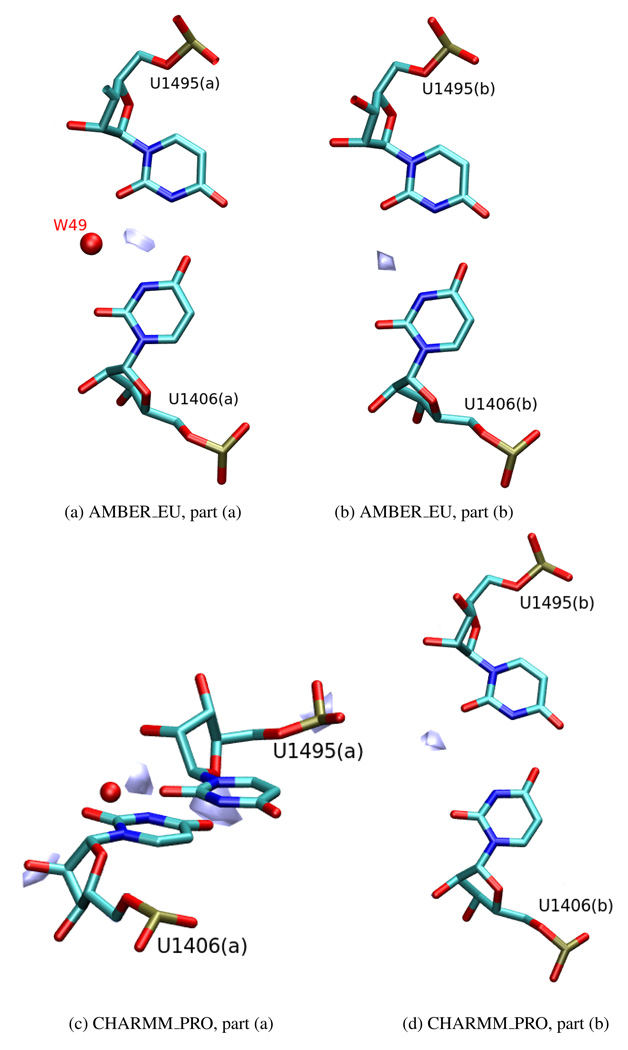
The W8 water molecule in part (a) and the corresponding W54 in part (b) bridge the interactions between the paromomycin and the uridines U1406 and U1495. An increased water density, corresponding to their crystallographic positions, was found only in CHARMM_PRO_PAR simulation and only in part (b) of the system. On the other hand, the water molecule W49, which is localized at the outer side of the helix and contributes to the stability of the U1406(a)○U1495(a) pair1,21 was reproduced in both Amber simulations (AMBER_PRO and AMBER_EU) and in both parts of the system (Figures 16a and 16b) even though in the crystal structure it is present only in the (a) part. Likewise, in the CHARMM_PRO simulation we noticed an area of increased water density in the vicinity of both uridine pairs, outside of the RNA helix (Figures 16c and 16d), however in part (a) this density was situated above the plane of the base pair and in part (b) it was further away from the pair than in the Amber force field simulations. Moreover, as reported in The U1406○U1495 pair Section, in the CHARMM_PRO simulation these uridines formed a pair only for a limited time. Therefore, one cannot be sure whether these areas indicate a water-bridged hydrogen bond or if they correlate with the water simply occupying a gap created when the pair was destroyed.
Figure 16.
Areas of high water density ((a) 0.28, (b) 0.35, (c) 0.22 and (d) 0.24 water oxygen per Å3) located near the U1406○U1495 pseudo pair. For the sake of clarity, only one area or several areas of high water density nearest the uridine pair are shown; the red sphere denotes the water oxygen (W49) in the position as in the 1J7T crystal structure.
Apart from these identified areas of high water density, in all of the simulations of the bare RNA, we observed water dense areas located very near or even in the same position as atoms of rings I and II of paromomycin. Therefore, the antibiotic would have to expel these water molecules out of the cleft in order to bind.
Conclusions
To investigate the dynamics of the ribosomal A-site we performed six 20-ns long MD simulations, which differed in the applied force field (Amber or Charmm) and initial structure and sequence (original or A1408G mutated A-site RNA with or without paromomycin). We analyzed the differences in the dynamics between the two mirrored parts of the RNA fragment, as well as between trajectories corresponding to prokaryotic and eukaryotic-like structures.
A1492 and A1493 intra- and extra-helical states
We observed multiple intra- and extra-helical states of A1492 and A1493 whose mobility was also reported in17,19 but with the use of the targeted or replica-exchange MD. In all simulations, the adenines in part (a) behaved differently than those in part (b) of the RNA helix. In general, adenines moved separately and only one adenine pair adopted a stable flipped-in configuration. The observed adenine configurations correspond to available experimental structures both, NMR and X-ray. Interestingly, in the simulations of the bare RNA, configurations with adenines bulged out into the solvent to the extent as in the starting 1J7T X-ray structure were found only in the simulations with the Amber force field. Similar studies but on a different RNA sequence38 indicate that Charmm force field may favor configurations observed in the NMR, whereas Amber – those in the X-ray experiments. However, in our case, the positions of A1492 and A1493 in all six simulations correlated uniformly with the ones from NMR and X-ray structures. We must note that not all of the experimentally seen configurations were observed in our simulations, probably due to not long enough sampling or force field limitations.
Based on the simulations with the Charmm force field, the hydrogen bond network between A1492 or A1493 and A1408 or G1408 suggests that in the eukaryotic-like structure the interactions are more stable and thus more difficult to break. This fact may explain the aminoglycosidic resistance gained by the bacteria upon the A1408G mutation7–9.
U1406○U1495 pairing affects the flexibility of the A-site
The U1406○U1495 pseudo pair is an important factor for the stability of the A-site. Our studies show that even a small difference in the RMSF of this pair can lead to an increased mobility of other A-site nucleotides. The number of hydrogen bonds formed within this uridine pair and their duration suggest that this pair is more stable in Amber than in Charmm force field. In the Amber force field simulations, the uridine pair appeared in the same configuration as in the starting crystal structure22 almost throughout the entire simulation.
On the other hand, in the simulations of the RNA-paromomycin complex the stability of the uridine pair does not influence that much the flexibility of the A-site. It seems that binding of the antibiotic provides enough interactions to stabilize this site on its own. In several X-ray structures of the RNA complexed with neamine and its derivatives2, the uridine pair was broken and U1406 was found in a flipped-out state. In our simulations, however, it was the U1495 that bulged out.
Na+ and water molecules gather in the binding cleft
It was proposed that upon binding aminoglycosides expel water molecules and ions from the binding site12,54. The crystal structure of the A-site complexed with the antibiotic reveals some indirect bonds formed via water molecules.
In the bare RNA we observed that the sodium ion density has its maximum in the location of paromomycin ring II, which suggests complementarity of the receptor and ligand electrostatic fields. We also found that the A1408G mutation causes slight changes in the positions of the sodium ions within the binding cleft because they tend to gather near the O6 atom of G1408 and are absent around N6 in the presence of A1408. It seems that this additional possible cation location in the mutated RNA may also contribute to hindering the binding of aminoglycosides that would need to expel more ions from the binding cleft.
Some of the water-mediated contacts between paromomycin and RNA may be easily formed because simulations show that water molecules are already close to their locations as in the crystal structure of the complex. Also one of the hydrogen bonds formed between U1406 and U1495 is probably accomplished through a water molecule because in the Amber force field simulations, where the uridine pseudo pairs were well-defined, we found high water density areas close to and between the uridines.
In conclusion, while approaching the binding site, paromomycin has to displace both, water molecules and ions, which are located in positions of rings I and II. Interestingly, the A1408G mutation alters the sodium ions’ density distribution in the paromomycin binding site.
Performance of the force fields
MD studies of RNA are in general more problematic than simulations of proteins or even DNA. Apart from the fact that RNA is highly charged and adding many counterions to the simulated environment is required, RNA, in contrast to a typical DNA helix, can form complicated tertiary structures leaving the well defined equilibrium states that the force fields were primarily designed to describe. Moreover, force field parameters designed for nucleic acids did not undergo so many tests as the ones for proteins because they were introduced later. In our study, we compared the simulations with two popular force fields Amber and Charmm. RMSD and RMSF values, as well as the observed stability of the selected base pairs, suggest that in the Amber force field the RNA double helix is more stable than in the Charmm force field.
One must note that the applied force fields were not initially designed for long, 20-ns simulations. Amber force field may perhaps restrain the system too much. Moreover, with these force fields we were not able to include divalent ions, such as Mg2+, although they seem significant in certain processes, because these ions are poorly represented (e.g. the force fields do not provide for polarization effects). Therefore, more MD studies on RNA simulations are desirable to assess force field limitations which would help MD become more reliable.
Supplementary Material
The movie presents the flipping event observed in the AM-BER PRO simulation (see Table 1). First, A1493 (see Figure 1b; cyan) moves inside the helix and pairs with A1408 (yellow), then A1492 (blue) follows and they both stay inside the helix. Additional file provides Charmm force field parameters for paromomycin and some exemplary data comparing the MD simulations of paromomycin with the NMR data.
This information is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The authors acknowledge support from University ofWarsaw (115/30/E-343-/S/2007/ICM BST 1255 and 115/30/E-343/BST1345/ICM2008), Polish Ministry of Science and Higher Education (3 T11F 005 30, 2006–2008), Fogarty International Center (National Institutes of Health Research Grant no R03 TW07318) and Foundation for Polish Science. Simulations were performed at ICM University of Warsaw grant no G31-4.
References
- 1.Vicens Q, Westhof E. Biopolymers. 2003;70:42–57. doi: 10.1002/bip.10414. [DOI] [PubMed] [Google Scholar]
- 2.François B, Russell RJM, Murray J, Aboul-ela F, Masquida B, Vicens Q, Westhof E. Nucleic Acid Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jana S, Deb J. Appl. Microbiol. Biotechnol. 2006;70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 4.Kaul M, Barbieri C, Kerrigan J, Pilch D. J. Mol. Biol. 2003;326:1373–1387. doi: 10.1016/s0022-2836(02)01452-3. [DOI] [PubMed] [Google Scholar]
- 5.Pilch DS, Kaul M, Barbieri CM. Top. Curr. Chem. 2005;253:179–204. [Google Scholar]
- 6.Lynch SR, Puglisi JD. J. Mol. Biol. 2001;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- 7.Hobbie S, Bruell C, Kalapala S, Akshay S, Schmidt S, Pfister P, Boettger E. Biochimie. 2006;88:1033–1043. doi: 10.1016/j.biochi.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Hobbie S, Pfister P, Bruell C, Sander P, François B, Westhof E, Boettger E. Antimic. Agents Chem. 2006;50:1489–1496. doi: 10.1128/AAC.50.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobbie SN, Pfister P, Brüll C, Westhof E, Böttger EC. Antimicrob. Agents Chemother. 2005;49:5112–5118. doi: 10.1128/AAC.49.12.5112-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Careter A, Clemons W, Brodersen D, Morgan-Warren R, Wimberly B, Ramakrishnan V. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 11.Shandrick S, Zhao Q, Han Q, Ayida BK, Takahashi M, Winters GC, Simonsen KB, Vourloumis B, Hermann T. Angew. Chem. Int. Ed. Engl. 2004;43:3177–3182. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 12.Chao P, Chow C. Bioorgan. Med. Chem. 2007;15:3825–3831. doi: 10.1016/j.bmc.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fourmy D, Yoshizawa S, Puglisi J. J. Mol. Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- 14.Lynch S, Puglisi J. J. Mol. Biol. 2001;306:1023–1035. doi: 10.1006/jmbi.2000.4419. [DOI] [PubMed] [Google Scholar]
- 15.Arya D, editor. Aminoglycoside antibiotics. Wiley & Sons, Inc.; 2007. [Google Scholar]
- 16.Fan-Minogue H, Bedwell D. RNA. 2008;14:148–157. doi: 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meroueh S, Mobashery S. Chem. Biol. Drug Des. 2007;69:291–297. doi: 10.1111/j.1747-0285.2007.00505.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaul M, Barbieri C, Pilch D. J. Am. Chem. Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- 19.Sanbonmatsu KY. Biochimie. 2006;88:1053–1059. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Réblova K, Lankaš F, Rázga F, Krasovska MV, Koča J, Šponer J. Biopolymers. 2006;82:504–520. doi: 10.1002/bip.20503. [DOI] [PubMed] [Google Scholar]
- 21.Vaiana AC, Westhof E, Brodersen D. Biochimie. 2006;88:1061–1073. doi: 10.1016/j.biochi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Vicens Q, Westhof E. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 23.Foloppe N, MacKerell A. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- 24.Banavali N, MacKerell A. J. Comput. Chem. 2000;21:105–120. [Google Scholar]
- 25.Wang J, Cieplak P, Kollman P. J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 26.Pilch D, Kaul M. Biochemistry. 2002;41:7695–7705. doi: 10.1021/bi020130f. [DOI] [PubMed] [Google Scholar]
- 27.Brooks BR, Karplus M, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S. J. Comput. Chem. 1983;4:187–217. http://www.charmm.org. [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Klein ML, Impey RW. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Frisch MJ, Trucks GW, Schlegel HB, Scuseriax GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Wallingford, CT: Gaussian, Inc; 2004. [Google Scholar]
- 30.Asensio JL, Hidalgo A, Cuesta I, González C, Cañada J, Vicent C, Chiara JL, Cuevas G, Jiménez-Barbero J. Chem. Eur. J. 2002;8:5228–5240. doi: 10.1002/1521-3765(20021115)8:22<5228::AID-CHEM5228>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 31.Phillips JC, Schulten K, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. http://www.ks.uiuc.edu/Research/namd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feller SE, Zhang Y, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103 [Google Scholar]
- 33.Schlick T. Molecular Modeling and Simulation. Springer: 2002. [Google Scholar]
- 34.Darden T, Perera L, Li L, Pedersen L. Structure. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 35.Ryckaert J, Ciccotti G, Berendsen H. J. Comput. Phys. 1977;32:327–341. [Google Scholar]
- 36.Mura C, McCammon J. Nucleic Acids Res. 2008;36:4941–4955. doi: 10.1093/nar/gkn473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDowell SE, Špačková N, Šponer J, Walter NG. Biopolymers. 2006;85:169–183. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarzyńska J, Réblová K, Šponer J, Kuliński T. Biopolymers. 2008;89:732–746. doi: 10.1002/bip.21001. [DOI] [PubMed] [Google Scholar]
- 39.Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, van Drunen R, van der Spoel D, Sijbers K, Keegstra H, Reitsma B, Renardus MKR. Physics Computing 92, World Scientific, chapter Gromacs: A parallel computer for molecular dynamics simulations. 1993 [Google Scholar]
- 40.Berendsen HJC, van Drunen R, van der Spoel D. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 41.Lindahl E, Hess B, van der Spoel D. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 42.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 43.van der Spoel D, Lindahl E, Hess B. GROMACS, User Manual, version 3.3. http://www.gromacs.org.
- 44.Bloomfield V, Crothers D, Tinoco I. Nucleic Acids: Structures, Properties, and Functions. University Science Books; 2000. http://books.google.com/books?id=Qfg028GkovEC. [Google Scholar]
- 45.Lu X-J, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. http://www.ks.uiuc.edu/Research/vmd. [DOI] [PubMed] [Google Scholar]
- 47.Pantano S, Tyagi M, Giacca M, Carloni P. Eur. Biophys. J. 2004;33:344–351. doi: 10.1007/s00249-003-0358-z. [DOI] [PubMed] [Google Scholar]
- 48.Eaton JW. GNU Octave. www.octave.org.
- 49.MolDyAna. http://moldyana.icm.edu.pl/moldyana/
- 50.Batey R, Rambo R, Doudna J. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Tor Y. J. Am. Chem. Soc. 1997;119:8734–8735. [Google Scholar]
- 52.Hermann T, Westhof E. J. Med. Chem. 1999;42:1250–1261. doi: 10.1021/jm981108g. [DOI] [PubMed] [Google Scholar]
- 53.Yang G, Trylska J, Tor Y, McCammon JA. J. Med. Chem. 2006;49:5478–5490. doi: 10.1021/jm060288o. [DOI] [PubMed] [Google Scholar]
- 54.Hermann T, Westhof E. J. Mol. Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]
- 55.Dlugosz M, Antosiewicz JM, Trylska J. J. Chem. Theory Comput. 2008;4:549–559. doi: 10.1021/ct700210n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie presents the flipping event observed in the AM-BER PRO simulation (see Table 1). First, A1493 (see Figure 1b; cyan) moves inside the helix and pairs with A1408 (yellow), then A1492 (blue) follows and they both stay inside the helix. Additional file provides Charmm force field parameters for paromomycin and some exemplary data comparing the MD simulations of paromomycin with the NMR data.
This information is available free of charge via the Internet at http://pubs.acs.org.