Abstract
The osteoinductive BMP2 signal facilitates commitment to the osteoblast phenotype by inducing several classes of early response genes. Among these are bone-related HOX factors, homeodomain, RUNX and OSTERIX proteins. Here we demonstrate molecular events among BMP2-induced transcription factors that constitute a network of molecular switches on promoters of bone-related genes to coordinate their temporal expression during cellular differentiation. Our studies provide evidence for (1) selective association of HOXA10, MSX2, DLX3 and DLX5 homeodomain transcription factors on Runx2 and OC genes at stages of osteoblast maturation as well as (2) participation of these factors with RUNX2 in chromatin remodeling of bone-specific genes for repression, activation and attenuation of transcription. These findings reveal the requirement for multiple levels of control for the appropriate timing of osteoblast-related gene expression.
Key Words: Osteoblasts, Gene regulation, Homeodomain, Chromatin remodeling, Runx2, MSX2, DLX3, DLX5
Introduction
Among the principal regulatory cascades for skeletal development are Hox genes that determine the timing, position and shape of a tissue element during embryogenesis. Hox and other homeodomain (HD) transcription factors also function in the adult skeleton to support osteoblast differentiation [Depew et al., 2005; Zakany and Duboule, 2007]. The BMP2 osteogenic signal activates several Hox and HD proteins as early response genes. HOXA10 and DLX3 and DLX5 HD proteins were characterized as enhancers of osteoblast gene transcription on gene promoters [Hassan et al., 2006, 2007]. RUNX2, the transcription factor essential for bone formation, interacts with genes on mitotic chromosomes as an epigenetic mechanism for development of the osteoblast phenotype [Young et al., 2007b], attenuates global protein synthesis [Young et al., 2007a] and integrates osteogenic signals through phosphorylation of Runx2 and formation of complexes with the coregulatory proteins that transduce the response to gene targets [Lian et al., 2006]. Here, we have used two signature osteoblast-specific gene promoters (Runx2 and OC) as models to probe interactions of HOXA10, MSX2, DLX3, DLX5 and RUNX2 with chromatin of target genes for their induced and physiological expression during stages of osteoblastogenesis.
Materials and Methods
Western Blotting
Electrophoresis conditions and Western blotting have been described previously [Hassan et al., 2006].
Chromatin Immunoprecipitation Assays
The procedures for chromatin immunoprecipitation in primary rat osteoblasts are detailed elsewhere [Hassan et al., 2006, 2007]. The cross-linked rat calvarial osteoblast cells were lysed and sonicated using appropriate conditions. Precleared soluble chromatins were immunoprecipitated with specific antibodies. DNA fragments were purified from the immunocomplexes and specific promoters analyzed by PCR.
Small Interfering RNA
HOXA10 siRNA was obtained from Qiagen and protein knockdown tested against a nonspecific siRNA duplex oligo using antibodies to HOXA10 from Santa Cruz [Hassan et al., 2007].
Results
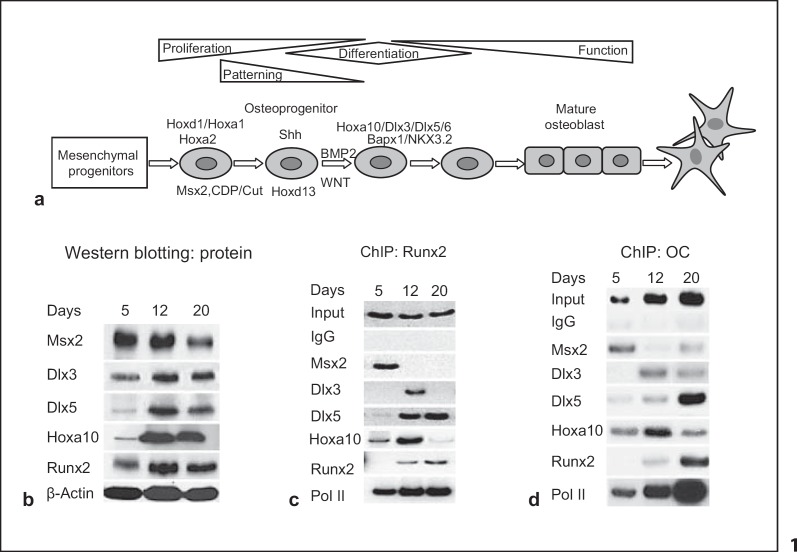
Transcriptional mechanisms that program osteoblast growth and differentiation include DNA binding of regulatory complexes and chromatin modifications for activation and repression of a gene during stages of differentiation (fig. 1a). Western blot analyses reveal changing protein levels of the transcription factors during osteoblast maturation (fig. 1b). Although MSX2 decreases, RUNX2, DLX3 and DLX5 increase to maximal levels with formation of bone nodules (day 12 in primary rat calvarial cells). The chromatin immunoprecipitation studies define the in vivo occupancy of the factors on the respective gene promoters (fig. 1c, d). We found a temporal recruitment of HD proteins in relation to Runx2 (fig. 1c) and OC (fig. 1d) transcription. MSX2 is associated with both promoters only in the proliferation period (day 4) and is no longer bound to both of them by day 12 despite significant cellular MSX2 levels (fig. 1b). On day 12 both DLX3 and DLX5, together with RUNX2, are recruited to the Runx2 and OC promoters. A concomitant increase in RNA polymerase II occurs at this time reflecting induced transcription. However, DLX3 binding to Runx2 is transient in the matrix maturation stage of differentiation with preferential binding of DLX5 at later stages. This suggests an important role for DLX5 in the HD network to regulate terminal differentiation. In the mineralization stage, an increased association of RUNX2 to OC is accompanied by maximal expression of the gene. However, the increased binding of RUNX2 to its own promoter results in a slight decrease in cellular protein; yet, transcription is maintained as indicated by RNA polymerase II binding and likely sustained by DLX5 binding. Thus Runx2 and OC are examples of a coordinated temporal regulation by an HD network, and with RUNX2, these factors support the timing of expression and physiological levels of target genes during osteoblast differentiation.
Fig. 1.
Regulation of stage-specific expression of HD proteins (MSX2, DLX3, DLX5), HOXA10 and RUNX2 transcription factors during osteoblast differentiation. a Schematic of osteogenic lineage cells and expression of regulatory and signaling factors at different stages of osteogenesis. b HD proteins exhibit different temporal expression profiles during development of the osteoblastphenotype. c, d Molecular switches and temporal recruitment of HD (MSX2, DLX3 and DLX5), HOXA10 and RUNX2 and coordinate changes in binding of RNA polymerase II (Pol II) to the Runx2 (c) and OC (d)promoters. Stages shown are growth (day 5), matrix formation (day 12) and mineralization (day 20) of primary fetal rat calvarial cell cultures. ChIP = Chromatin immunoprecipitation.
The early association of HOXA10, prior to RUNX2, on bone-related gene promoters when expression is very low (Runx2) or not detected (BSP, OC), indicates a specialized function (fig. 1c, d). We previously reported the effects of HOXA10 depletion (by siRNA) on histone acetylation and H3K4 methylation of gene promoters, which reflect transcriptionally active chromatin [Hassan et al., 2007]. Our findings showed a significant decrease in acetylation of Runx2, BSP and OC chromatin in the absence of HOXA10 in osteoblasts. From these studies, a pattern emerges that defines functional activities of HOXA10, HD proteins and RUNX2 and a mechanism for activating gene transcription and regulating their temporal expression profile during stages of osteoblast maturation.
Discussion
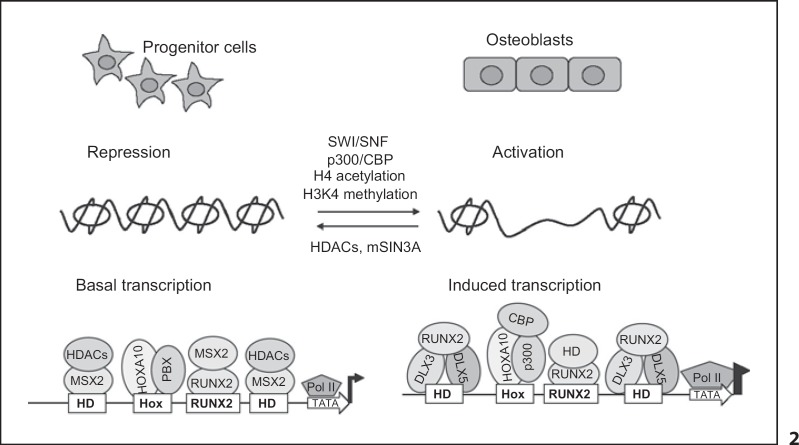
There is a requirement for chromatin remodeling of genes silenced by condensed chromatin for BMP2 induction of osteogenesis [Young et al., 2005]. The signals for induction of tissue-specific genes occur at multiple levels of control as indicated by the presented studies (fig. 2). RUNX2 functions in bone-specific activation by supporting chromatin modification. This was established by mutation of the Runx2 sites in the OC gene [Javed et al., 1999]. We propose that the recruitment of HOXA10 to gene promoters prior to RUNX2 binding may fulfill several roles. HOXA10 complexes with the corepressors PBX and MEIS as well as coactivators such as p300 [Saleh et al., 2000; Capellini et al., 2006]. Its binding to bone promoters prior to Runx2 suggests an epigenetic modification without transcription that later contributes to activation. Another level of gene regulation is needed for physiological control of transcription driven by cellular representation of the factors and formation of heterodimeric complexes with coregulatory factors (fig. 2) that may alter DNA-binding affinity. We postulate that the MSX and DLX HD proteins function to regulate the timing of stage-specific gene expression coordinated with requirement for RUNX2 and HOXA10. In summary, combinatorial mechanisms are operative for regulated transcription of osteoblast genes through diversification of both sequence-specific activators/repressors that contribute to patterns of gene expression and the multistep process of programming involved in bone formation.
Fig. 2.
Mechanisms for coordinated control of transcription factors and chromatin modifiers for regulating the program of osteoblast differentiation. A general model is proposed for the sequential expression of osteoblastic genes. In osteoprogenitor cells, the initial transcription complexes (such as MSX2 and HOXA10) are composed of transcription factors partnering with repressor coregulatory proteins. RNA polymerase II (Pol II) binding is presented as a result of epigenetic modifications to poise the genes for activated transcription in osteoblasts. For induced transcription, HOXA10 and RUNX2 recruit different coregulatory factors to become activation complexes and DLX3 and DLX5 become bound to the genes. This unique mechanism of switching occupancy of transcription factor complexes regulates gene-specific expression during osteoblast differentiation.
Acknowledgements
This study was supported by National Institutes of Health grants DE12528 and AR39588. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Abbreviations used in this paper
- BMP
bone morphogenetic protein
- BSP
bone sialoprotein
- HD
homeodomain
- HDAC
histone deacetylases
- OC
osteocalcin
References
- Capellini T.D., Di G.G., Salsi V., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:133–2263. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Depew M.J., Simpson C.A., Morasso M., Rubenstein J.L. Reassessing the Dlx code the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:207–501. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M.Q., Tare R.S., Lee S., Mandeville M., Morasso M.I., Javed A., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by Dlx3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:281–40515. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- Hassan M.Q., Tare R.S., Lee S., Mandeville M., Weiner B., Montecino M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:27–3337. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A., Gutierrez S., Montecino M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:19–7491. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J.B., Stein G.S., Javed A., van Wijnen A.J., Stein J.L., Montecino M., Hassan M.Q., Gaur T., Lengner C.J., Young D.W. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:7–1. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Saleh M., Rambaldi I., Yang X.J., Featherstone M.S. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol Cell Biol. 2000;20:20–8623. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.W., Hassan M.Q., Pratap J., Galindo M., Zaidi S.K., Lee S., Yang X., Xie R., Underwood J., Furcinitti P., Imbalzano A.N., Penman S., Nickerson J.A., Montecino M.A., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:445–442. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young D.W., Hassan M.Q., Yang X.-Q., Galindo M., Javed A., Zaidi S.K., Furcinitti P., Lapointe D., Montecino M., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:104–3189. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.W., Pratap J., Javed A., Weiner B., Ohkawa Y., van Wijnen A., Montecino M., Stein G.S., Stein J.L., Imbalzano A.N., Lian J.B. SWI/SNF chromatin remodeling complex is obligatory for BMP2-induced, Runx2-dependent skeletal gene expression that controls osteoblast differentiation. J Cell Biochem. 2005;94:94–720. doi: 10.1002/jcb.20332. [DOI] [PubMed] [Google Scholar]
- Zakany J., Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:17–359. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]