Abstract
We present a family of water-soluble quantum dots (QDs) that exhibit low nonspecific binding to cells, small hydrodynamic diameter, tunable surface charge, high quantum yield, and good solution stability across a wide pH range. These QDs are amenable to covalent modification via simple carbodiimide coupling chemistry, which is achieved by functionalizing the surface of QDs with a new class of heterobifunctional ligands incorporating dihydrolipoic acid, a short poly(ethylene glycol) (PEG) spacer, and an amine or carboxylate terminus. The covalent attachment of molecules is demonstrated by appending a rhodamine dye to form a QD-dye conjugate exhibiting fluorescence resonance energy transfer (FRET). High-affinity labeling is demonstrated by covalent attachment of streptavidin, thus enabling the tracking of biotinylated epidermal growth factor (EGF) bound to EGF receptor on live cells. In addition, QDs solubilized with the heterobifunctional ligands retain their metal-affinity driven conjugation chemistry with polyhistidine-tagged proteins. This dual functionality is demonstrated by simultaneous covalent attachment of a rhodamine FRET acceptor and binding of polyhistidine-tagged streptavidin on the same nanocrystal to create a targeted QD, which exhibits dual-wavelength emission. Such emission properties could serve as the basis for ratiometric sensing of the cellular receptor’s local chemical environment.
Introduction
Semiconductor nanocrystal quantum dots (QDs) are photo-stable fluorophores with narrow emission spectra tunable through visible and near-infrared wavelengths, large molar extinction coefficients, and high quantum yields.1–5 These properties make QDs powerful tools for labeling and optical imaging in biological,3,6–11 biomedical,12–18 and chem-/bio-sensing applications.19–22 Imaging cellular events at the single-molecule level is a particular area where the superior brightness and photostability of QDs excel as compared to more typical dye and fluorescent protein imaging reagents.11 Notwithstanding, the optimal design of QDs for single-molecule imaging in live cells presents a unique set of challenges. Ideally, the nanocrystal should be easily derivatized such that various secondary reporters or biomolecules can be appended to the QD to allow for sensing and/or targeting of cellular receptors. In doing so, the QD must maintain the properties of low nonspecific binding to cells, small size, high quantum yield, and good pH stability. Since the solution properties of QDs ultimately depend on the nature of the surface ligands, the foregoing criteria must be addressed through rational ligand design.
The dominant class of QDs currently used for cellular or in vivo imaging retains hydrophobic surface ligands, and thus these QDs are typically encapsulated in amphiphilic polymer micelles.6,23–28 Such encapsulated QDs benefit from high quantum yield, but the polymeric shell produces large hydrodynamic diameters (HDs) on the order of 20–30 nm for an inorganic core/shell diameter of only 4–6 nm.29 The excessive size of polymer-coated QDs, which are often much larger than the cellular receptors being labeled, presents a barrier to the widespread implementation of QDs for biological imaging. Large particles potentially interfere with the function of labeled proteins and could limit access to hindered regions such as neuronal synapses.8,30 Furthermore, amphiphilic polymer coatings are often highly charged and consequently contribute to nonspecific binding to cell membranes, thus rendering them unsuitable for single-particle imaging where low background is essential. Nonspecific adsorption can be mitigated via PEGylation of polymer-encapsulated QDs, but this further increases nanocrystal size.31 QDs coated with phospholipids or silica shells suffer from similar limitations of inherently large size and the need for a bulky PEG passivating layer.7,32
The size of water-soluble QDs can be dramatically reduced by displacing the native hydrophobic coating with small molecule coordinating thiol-based ligands such as mercaptoacetic acid (MAA).5,33–35 Nonetheless, many ligand exchange approaches face a tradeoff between size, stability, and derivatizability. MAA and various other monothiol-capped QDs are small (HD ≈ 6–8 nm) and can be derivatized using carbodiimide coupling chemistry, but they tend to aggregate rapidly due to weak ligand-QD interactions.29,33 In addition, the water solubility of MAA-capped QDs relies critically on the ionization state of the carboxylic acid group, causing solution instability under slightly acidic conditions. Peptides bearing cysteine residues have also been used to form stable aqueous QDs, but the relatively high molecular weight (~20 amino acids) of the peptides used compared to small molecule ligands still results in large QD sizes (HD ≈ 15 nm).36 Dithiol ligands, such as dihydrolipoic acid (DHLA), are much more stable with respect to ligand dissociation compared to monothiol ligands, but exhibit pH instability, poor derivatizability5 and high nonspecific binding to cells. Ligand exchange with DHLA appended to various length PEGs via ester bond formation (DHLA-PEG) yielded QDs that were highly stable in aqueous solution and suitable for live cell imaging.13,37 However, the hydroxy-terminated surface of these DHLA-PEG QDs lacks the functionality required for efficient and selective covalent derivatization under mild conditions, for example with targeting biomolecules for receptor labeling on cells, and the ester bonds are prone to hydrolysis.
Two commonly employed QD derivatization strategies are (1) direct covalent modification of QDs using common bioconjugation methods such as 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) mediated cross-coupling between amino and carboxyl functionalities, and (2) self-assembly of biomolecules onto QDs via metal-affinity interactions through a polyhistidine (His6) tag.23 QDs encapsulated in polymeric/phospholipid/silica shells are generally derivatized by covalent conjugation. QDs capped with DHLA or DHLA-PEG are amenable to conjugation using metal-affinity interactions between a His6-tagged biomolecule and metal ions at the surface of the QD, leading to stable conjugates that retain both QD luminescence and functionality of the coordinated biomolecule.38 However, covalent and His6-tag conjugation on a single nanocrystal has not heretofore been combined with the properties of small size, low nonspecific binding, and solution stability.
We now demonstrate an efficient route to compact, derivatizable aqueous CdSe(ZnCdS) core(shell) QDs. These QDs are enabled by a new suite of DHLA-PEG-derived ligands terminating in amine or carboxylic acid functional groups. The quantum yield (QY) of these DHLA-PEG derivatized QDs was enhanced to ~40% via an alloyed ZnxCd1−xS shell.39 The QDs display good pH stability, a tunable ζ-potential, very low nonspecific binding to cells, and an HD between 9 and 12 nm, which is substantially smaller than polymer encapsulated QDs. We demonstrate derivatization of these QDs via both covalent bond formation (to small molecule and protein partners) and His6-protein coupling for targeted cell labeling. The versatility of this QD system is highlighted by imaging the interaction of epidermal growth factor (EGF) with EGF receptors on live cells, and by simultaneously derivatizing QDs with streptavidin for targeting and an organic fluorophore for potential sensing applications by FRET. The versatile QD scaffold reported here facilitates a highly modular approach for tuning the composition of functional surface molecules on QDs for cellular targeting and sensing applications.
Results and Discussion
Ligand Synthesis and Design
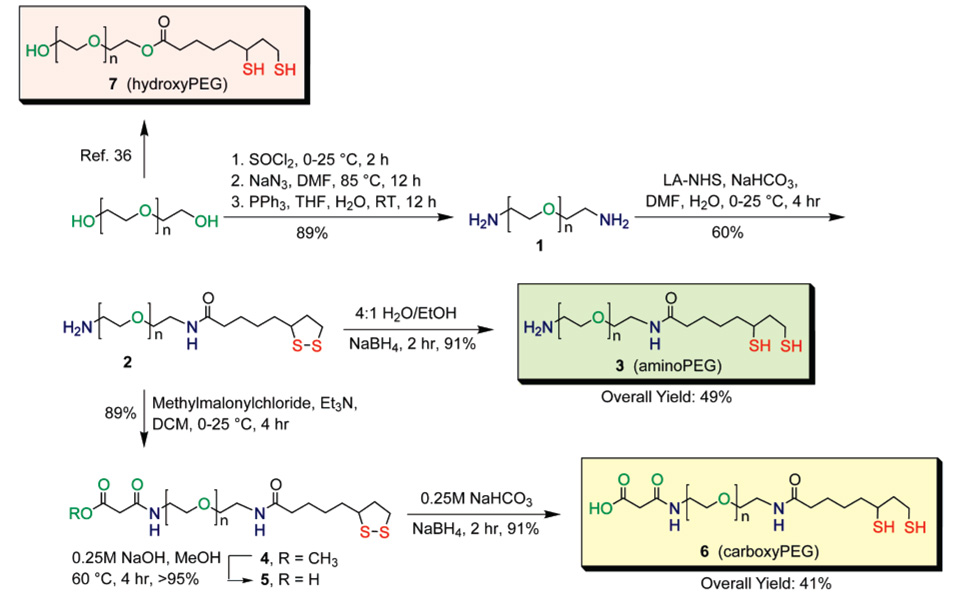
Compounds 3 (aminoPEG) and 6 (carboxyPEG) presented in Scheme 1 were designed with the following considerations in mind: (1) a DHLA moiety provides strong coordination to the QD surface, (2) a PEG spacer conveys water solubility and reduces nonspecific binding, and (3) a carboxy or amine functionality enables further covalent derivatization.
Scheme 1.
The diamine functionalized polyethylene glycol (1) was synthesized from commercially available PEG (avg MW 400, n ≈ 8) via a simple three step reaction in high yield with minimal purification steps required. The overall yield was 89% on a 30 g scale. Synthon 2, which served as the precursor to both 3 and 6, was delivered from the reaction of 1 with the N-hydroxysuccinimidyl ester of lipoic acid (LA-NHS). 3 was furnished smoothly from ring opening lipoic acid (LA) to the dihydrolipoate form by reduction with sodium borohydride.37 Alternatively, 2 could be modified by its reaction with methylmalonylchloride, followed by methylester deprotection to form an amide linkage to a malonic acid group (compound 5 ). Reduction with sodium borohydride yielded compound 6.
The synthetic strategy of Scheme 1 incorporates several advantages. Precursor 1 is obtained in large quantities without the need for extensive purification. Heterobifunctional ligands were obtained in a straightforward fashion from compound 1, with a DHLA moiety for robust coordination to the QD surface, a PEG spacer for water solubility, and a head group consisting of a primary amine (compound 2) or in principle any of a wide variety of derivative forms including carboxylic acid-terminated 6. Moreover, these ligands feature amide bonds, which are more robust toward hydrolysis than ester bonds.
Ligand Exchange and Characterization of Hydrophilic QDs
Water-soluble QDs were obtained by displacing the native hydrophobic ligands of CdSe/ZnxCd1−xS core/shell QDs with aminoPEG, carboxyPEG, hydroxyPEG (compound 7), or mixtures thereof. Ligand exchange was carried out by modifications to previous procedures37 using QD samples with selected emission wavelengths between 558 and 605 nm, synthesized according to modified literature procedures.1,40
Figure 1A presents the absorption and emission spectra of a representative QD sample with emission centered at 605 nm (QD605) before and after ligand exchange with carboxyPEG 6. Similar spectral properties are obtained for QDs exchanged with aminoPEG 3. The QY of QD605 in hexane solution was 65% and after ligand exchange was typically 30–40%. We note that the aqueous QY of alloyed shell QDs (CdSe/ZnxCd1−xS, x = 0.7) was enhanced by as much as 2-fold compared to that of QDs overcoated with a pure ZnS shell (x = 1) (Supporting Information, Figure S1). QDs bearing a pure ZnS shell, rather than the alloy ZnxCd1−xS shell, exhibited similar QYs in hexane, but experienced a loss in QY of up to 75% after ligand exchange with aminoPEG or carboxyPEG. The retention of the high QY values after water solubilization using the alloyed shell was attributed to reduced lattice mismatch between the core and shell, which provides better passivation of interfacial trap states.41–43 The samples used in the experiments reported here were overcoated with approximately five monolayers of an x ≈ 0.7 alloy. The shell was applied with a uniform mole ratio of Cd to Zn precursors throughout the overcoating process, thus we expect Zn and Cd to be present throughout the shell and also on the surface. The formation of the alloy shell in each case was accompanied by a red-shift of the absorbance maximum by approximately 30 nm from that of the CdSe cores.
Figure 1.
(A) Absorbance spectrum of CdSe(ZnxCd1−xS) core(shell), QD605, in hexane with x = 0.7 (–, red). Fluorescence spectra with absorption normalized at the excitation (λexc = 520 nm) of QD605 in hexane after one cycle of precipitation (– – –, green, QY = 65%), and in PBS buffer after ligand exchange with carboxyPEG (····, blue, QY = 43%), showing a small decrease in QY. (B) Representative dynamic light-scattering histogram of QD605 ligand exchanged with carboxyPEG in PBS, giving HD = 11.4 nm.
The HD of QDs ligand-exchanged with carboxyPEG (abbreviated carboxyQDs) was measured by dynamic light scattering and found to be ~9.9 nm and ~11.4 nm for QD565 (Figure S2A) and QD605 (Figure 1B, Figure S2B), respectively. Similar HDs were found for aminoPEG-functionalized QDs (abbreviated aminoQDs). The HD of these QDs coated with either aminoPEG or carboxyPEG is approximately a factor of 2 smaller than QDs coated with amphiphilic polymeric shells (e.g., commercial QDs), which have HDs on the order of 14–60 nm (Figure S2).44 The reduced size of these QDs relative to commonly used commercial QDs makes them attractive for cell labeling applications, as the smaller size reduces the possibility of the probe interfering with receptor function, and potentially increases access of these QDs to more hindered regions, such as the synapse. In addition, the monomodal size distribution shown by the dynamic light-scattering histogram indicates that these QDs form well-dispersed aggregate-free solutions, which is important for cell labeling and single-particle tracking applications.
Faithful imaging and single-particle tracking also depends on the ability of QDs to remain well dispersed in solution when exposed to pH fluctuations from pH 7.4 in the cell medium down to pH 5 within lysosomes. DHLA-capped QDs aggregate below pH 6, since their water solubility depends largely on the negative charge of the ionized carboxylic acid group.37 However, both carboxyQDs and aminoQDs were found to be stable in aqueous solutions between pHs 5.0 and 9.5 with no significant change in brightness for at least 3 days (Figure S3). We surmise that the PEG linker of aminoQDs and carboxyQDs further enhances water solubility, and hence one would expect greater pH stability, similar to what was previously reported for hydroxyQDs.37
Tuning Surface Charge and Functional Valency of QDs
One of the next major challenges in designing QDs suitable for cell labeling and single-particle tracking is nonspecific cell binding, which has been partly attributed to electrostatic attraction of the cell surface with charged QD ligand coatings.31 Therefore, the QD surface charge is a parameter that must be controlled. QDs ligand-exchanged with aminoPEG or carboxyPEG will have surfaces that are positively and negatively charged, respectively, in neutral buffer. Properties of QDs have been tuned using ligand mixtures.36,37 Following this lead, we employed a mixture of aminoPEG or carboxyPEG with non-charged hydroxyPEG (compound 7)37 in varying mole fractions to control the surface charge of aqueous QDs. For convenience, nomenclature of the mixed-ligand system refers to only aminoPEG or carboxyPEG mole fraction with the balance accounted for by hydroxyPEG. For instance, 20% aminoQDs refer to QDs ligand exchanged with 20% aminoPEG and 80% hydroxyPEG by mole fraction.
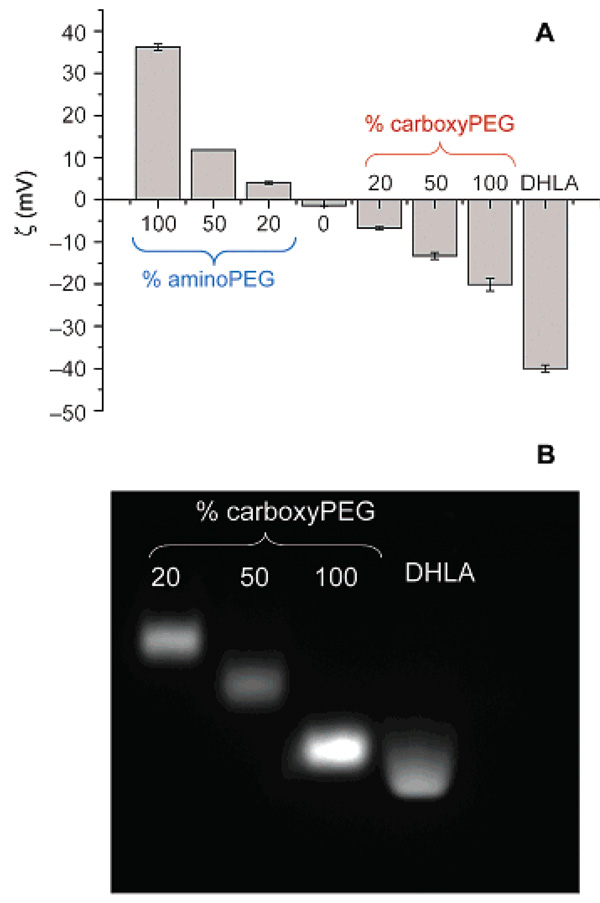
The surface charge of the resulting QD samples was analyzed by ζ-potential measurements and agarose gel electrophoresis. Figure 2A shows that aminoQDs capped with pure (100%) aminoPEG have a ζ-potential of +36.2 mV, which decreases in an approximately linear fashion with decreasing −NH2 composition in the ligand blend. A similar trend in the ζ-potential was observed with decreasing −CO2H composition in the ligand blend for carboxyQDs. The results from ζ-potential measurements were also mirrored in measurements of the electrophoretic gel mobility (Figure 2B). QDs coated with 100% DHLA were also included for comparison, revealing that DHLA-capped QDs have a higher electrophoretic mobility than carboxyQDs, which could result either from the higher ligand packing density or the smaller HD of the DHLA-capped QDs.
Figure 2.
(A) ζ-Potential measurement of QDs ligand exchanged with various percentages (by mole fraction) of aminoPEG or carboxyPEG mixed with hydroxyPEG. DHLA-coated QDs shown for comparison. (B) QD mobilities in 1% agarose gel, in Tris-acetate-EDTA buffer (pH 8.3). QDs run from anode to cathode.
We probed the number of reactive functional groups per QD in order to characterize our ability to adjust the QD valency via mixtures of DHLA-PEG-derived ligands. Mixed-ligand aminoQDs with 565 nm emission were exposed to fluorescamine, which reacts rapidly with primary amines to form a fluorescent product that can be monitored at 487 nm. The QDs were purified by repeated ultrafiltration to remove free ligand and reacted with excess fluorescamine. By calibrating the fluorescence intensity signal of the fluorescamine after reaction with samples of aminoPEG at known concentration (Figure S4A), an estimate of ~140 amines/QD was obtained for 100% aminoQDs, consistent with a previously reported value from a related system.12 Furthermore, an approximately linear correlation was obtained for the amount of amine detected per QD with the percentage of amine-terminated ligands used in the ligand-exchange blend (Figure S4B).
These results suggest that the use of a mixed-ligand blend of charged and neutral DHLA-PEG-based ligands offers a simple and robust way to tune the final surface composition and charge of the resulting water-solubilized QDs. The QDs obtained from this method can be tuned to present surface amino or carboxyl groups in high enough numbers for efficient derivatization reactions, while maintaining a minimally charged particle surface to mitigate nonspecific binding for cell-labeling applications (vide infra).
Mitigation of Nonspecific Cell Binding
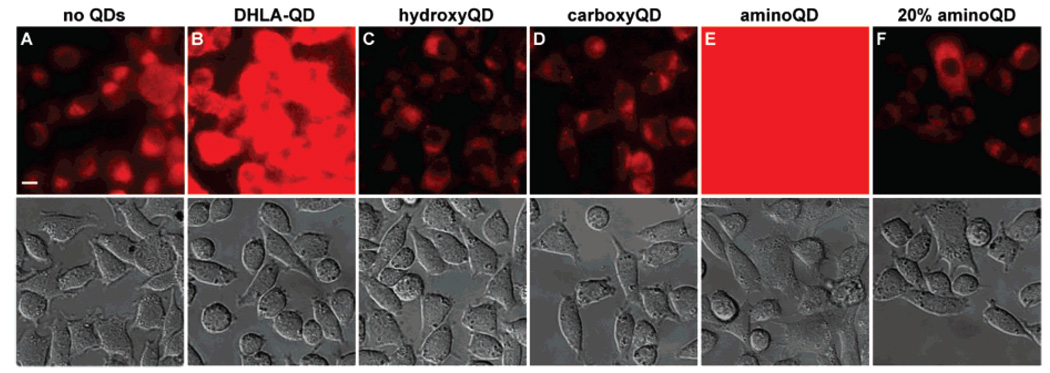
In order to directly evaluate the suitability of the charged aminoQDs and carboxyQDs for biological imaging, we investigated the nonspecific binding to a human cell line as a function of ligand composition on the QD surface. Figure 3 displays the results. The control sample (Figure 3A) shows emission from cell autofluorescence. QDs coated with negatively charged DHLA show significant nonspecific binding to both cells and glass (Figure 3B) as indicated by the increased fluorescence intensity as compared to that of the control. Neutral QDs capped with hydroxyPEG (abbreviated hydroxyQDs) show minimal nonspecific binding, as expected (Figure 3C). Surprisingly, carboxyQDs also showed minimal nonspecific cell binding, comparable to that of hydroxyQDs (Figure 3D). The lack of nonspecific binding for carboxyQDs, despite the negative charge, highlights the important role of the PEG spacer in reducing nonspecific interactions. On the other hand, aminoQDs exhibited severe nonspecific binding to cells, which at the same contrast level caused saturation of the imaging field (Figure 3E). We ascribe the high level of binding to electrostatic interactions of the highly positively charged QD with the cell surface. The problem is circumvented by coating the QDs with a mixture of 20% aminoPEG and 80% hydroxyPEG (20% aminoQDs), which do not show significantly more nonspecific binding than hydroxyQDs (Figure 3F).
Figure 3.
Nonspecific binding of QD605 to HeLa cells as a function of ligand coating. (Top row) Fluorescence image with 420 nm excitation, 605 nm emission. (Bottom row) Differential interference contrast (DIC) image. Cells were incubated with QDs for 10 min at 4 °C, and washed with buffer 4 times before imaging. (A) Control sample, showing cell autofluorescence; QDs ligand-exchanged with (B) DHLA, (C) hydroxyPEG, (D) carboxyPEG, (E) aminoPEG, (F) 20% aminoPEG (with 80% hydroxyPEG). Scale bar, 10 µm.
The results of Figure 3 indicate that both carboxyQDs and 20% aminoQDs exhibit minimal nonspecific binding to cells, making them suitable for cell labeling and single-particle imaging applications. We chose the 20% aminoPEG composition in order to provide a reasonable amount of amino groups for covalent derivatization, while sufficiently reducing the surface charge in order to mitigate nonspecific interactions with cells. This composition does not necessarily represent an optimized design for nonspecific binding, but it highlights the advantages of being able to tune the surface composition for a specific application.
Covalent Conjugation to a FRET Acceptor Dye
The suitability of 20% aminoQDs for routine derivatization was evaluated by exposing them to the amine-reactive NHS ester of carboxy-X-rhodamine (ROX), a red-emitting organic fluorescent dye (Figure 4A). Coupling to a dye offers the advantage of easily monitoring the coupling yield by following the prominent dye absorption feature. Additionally, QD–dye energy transfer systems have recently opened up new possibilities as the basis for optically reporting chemo-/bio- sensors,20,21,38,45 and the QD–ROX here serves as a model system for a dual-emission ratiometric FRET sensor platform recently developed and demonstrated for the case of pH sensing.19,20
Figure 4.
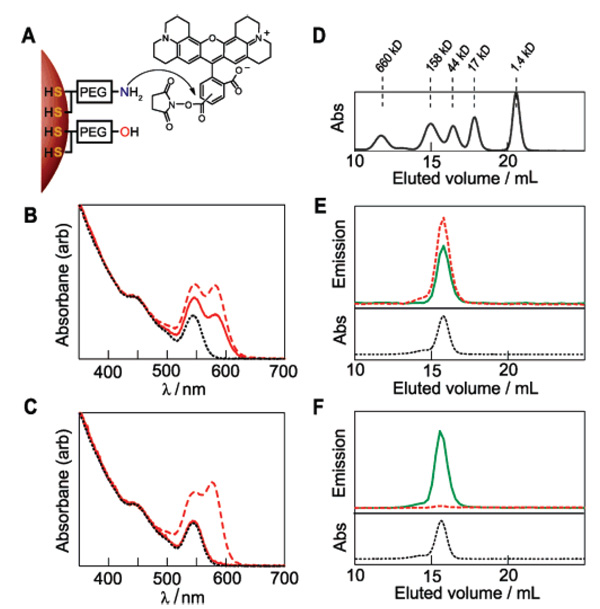
(A) Derivatization of 20% aminoQDs with the fluorescent dye ROX. (B) Absorption spectra of unconjugated QDs (····, black), QD + dye mixture (–, red) and QD + dye purified (–, red), all normalized at 400 nm. (C) Absorption spectra as in B for control experiment using the unreactive free-acid form of ROX dye: QDs (····, black), QD + dye mixture (–, red) and QD + dye purified (–, red). (D) GFC molecular weight standards. (E) GFC absorbance and fluorescence signals for the QD + dye-purified conjugate depicted in (B): QD channel (–, green), ROX channel (–, red) and absorbance at 280 nm (····, black). QD and ROX fluorescence channels are monitored at 558 and 610 nm, respectively. (F) GFC absorbance and fluorescence signals for a control sample using a mixture of QD and the free-acid form of ROX dye. Legend is same as in (E).
QDs emitting at 558 nm (QD558) were selected to afford good spectral overlap with the ROX absorbance peak in order to form a donor/acceptor FRET couple. The ROX emission at 610 nm can then be easily monitored independent of the QD emission. Following reaction with the ROX succinimidyl ester, the QDs were separated from unbound dye and NHS byproduct via ultrafiltration. Figure 4B shows the absorption spectra of the initial QDs, the reaction mixture, and the purified product, with the dye absorption peak clearly visible. A fit of the spectrum as a sum of QD and dye contributions revealed a dye/QD molar ratio of 5.0 as-mixed, and 2.9 upon purification, indicating a coupling yield of 58%. In a control experiment (Figure 4C), a QD sample from the same batch was mixed with the free-acid form of ROX under identical reaction conditions. Upon purification, <3% of the dye remained, indicating minimal nonspecific adsorption of the dye onto the QD surface.
The FRET efficiency of the QD–dye couple was estimated to be ~90% by measuring the quenching of QD fluorescence after dye conjugation (Figure S5). The Förster distance was calculated to be R0 = 5.6 nm from the spectral overlap. From the measured FRET efficiency, the measured amount of dye per QD (m) and the calculated R0, the QD–dye separation distance was calculated to be r = 4.8 nm,46 which is in good agreement with the hydrodynamic diameter of these QDs (9.9 nm).
The purified QD–ROX conjugate was additionally characterized by gel filtration chromatography (GFC) using a size-exclusion column. Comparison with protein molecular weight (MW) standards (Figure 4D) provides an estimate of size for samples under investigation. Figure 4E shows the GFC results for the purified QD–ROX conjugate shown in Figure 4B. QD–ROX elutes at 15.7 mL volume, corresponding to a protein-equivalent MW of ~94 kD. The emission spectrum at this volume shows a peak in both QD and ROX channels. Fluorescence detection at 558 nm (QD emission) and 610 nm (ROX emission) reveals ROX emission at an elution volume corresponding to the QDs, confirming that the QD and dye are indeed bound.
In contrast, GFC of a mixture of free QDs and ROX dye at the same dye/QD ratio as that for the coupled sample shows UV absorbance and QD emission signals at the same elution volume as for the couple, but no dye fluorescence (Figure 4F). Indeed, the dye is not detected at all because of its low absorption cross section at the absorbance and excitation wavelengths used by the instrument. The ROX emission is detected in the coupled case because dye molecules bound to the QD are excited by FRET. Uncoupled QDs run under the same conditions also elute at 15.7 mL in this experiment, suggesting that attachment of a dye does not significantly alter the QD size. Together, the GFC data indicate that small-molecule dyes in NHS ester form can be covalently appended to the mixed ligand QDs without perturbing the QD size, and that control experiments with nonactivated dye show no indication of coupling/binding.
Covalent Conjugation to Streptavidin for High Affinity Cell Labeling
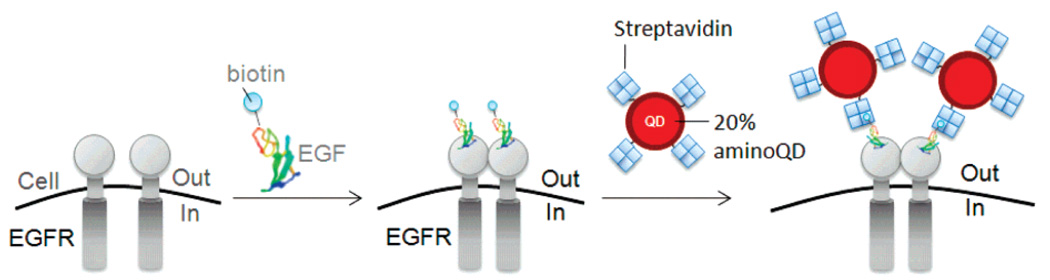
The scope of covalent derivatization using amine-functionalized QDs was extended by conjugating the protein streptavidin (SA) to provide a method for specific targeting to cellular receptors. SA was conjugated to 605 nm emitting 20% aminoQDs (aminoQD–SA) using EDC/NHS coupling chemistry. QDs modified in this manner showed strong binding to nonspecifically biotinylated cells. In control experiments in which the coupling agents or the SA were omitted, QDs showed minimal binding to biotinylated cells (Figure S6), confirming the role of covalent bond formation (as opposed to electrostatic interactions) in tethering SA to the QDs. We applied the aminoQD–SA conjugates to specific cell labeling and single-particle tracking of EGF receptors (EGFR) on live cells. EGFR is an important activator of cell division and a target for therapy of many cancers.47 There are still many questions about the mechanism of receptor association and internalization in EGFR signal transduction, which are best addressed through study at the single-molecule level.28,48
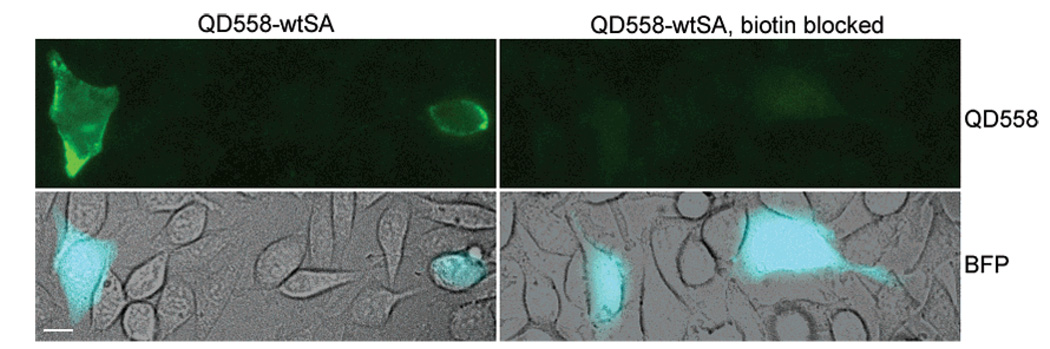
Figure 5 schematically illustrates the targeting of live cells transfected with human EGFR. COS7 cells were incubated with biotinylated EGF (bioEGF) and then stained with aminoQD–SA. We observed specific binding of the QD to the surface of EGFR transfected cells (Figure 6, left), shown by the blue fluorescent protein (BFP) cotransfection marker. Adjacent untransfected cells, indicated by the absence of the BFP marker, did not show QD staining, illustrating the specificity of labeling. Control experiments in which QDs were blocked with excess free biotin also showed no binding (Figure 6, right). Furthermore, the photostability, specificity, and high QY of these QDs enabled single-particle tracking of EGF interaction with EGFR on the cell surface (Figure 7). Individual QDs were identified by their fluorescence intensity and intermittency, or blinking, behavior and could be seen in motion on the surface of cells in a manner consistent with active transport of the labeled receptor upon receptor internalization (see movie associated with Figure 7).
Figure 5.
Targeting QDs to EGFR. EGFR labeled with biotinylated EGF (bioEGF), followed by staining with aminoQDs covalently conjugated to streptavidin (not drawn to scale).
Figure 6.
Targeting of 20% aminoQD–SA conjugates to EGFR on live cells. EGFR expressing cells are indicated by blue fluorescent protein (BFP) cotransfection marker. (Top row) QD558 channel. (Bottom row) BFP merged with DIC channel. (Left) EGFR-transfected COS7 cells treated with biotinylated EGF and stained with aminoQD–SA. (Right) Control experiment in which aminoQD–SA was blocked with excess free biotin before application to cells. Scale bar, 10 µm.
Figure 7.
Single-particle tracking of EGF via low density labeling with 20% aminoQD–SA. A single cell is shown in the imaging field (QD605 channel). Large patches of brightness represent autofluorescence, bright spots represent clusters of QDs, movie shows fluorescence intermittency of single QDs, which migrate in a manner consistent with active transport of labeled receptors. Scale bar, 5 µm.
ⓦ A movie in AVI format is available.
Cell Labeling with a Targeted Dual-Emission QD
Metal-affinity coordination and covalent attachment to QDs each has its advantages and limitations. For small molecules such as dyes, a His6-tagged form is not readily available, and so covalent conjugation is preferable. For recombinant proteins, expression with a His6 tag is routine, and metal-affinity conjugation to QDs is convenient due to the high binding affinity, fast kinetics of binding, controlled orientation of the protein on the QD surface, and ease of preparation with little or no purification steps necessary.49 Here we combine the advantages from both methods of bioconjugation to achieve a versatile nanocrystal that still maintains the desired properties for cell labeling, forming a basis for the realization of QDs with both targeting specificity and environmental sensing capabilities.
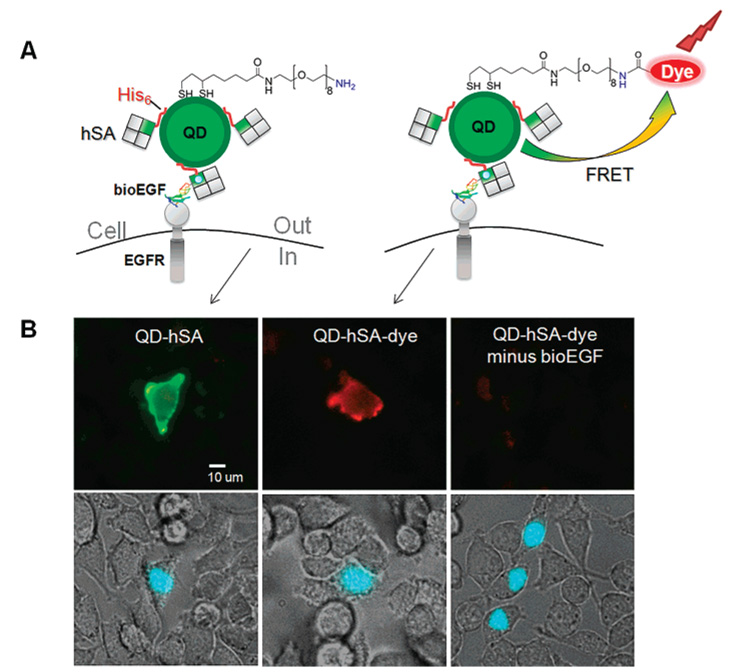
To show that aminoQDs bearing alloyed ZnxCd1−xS shells are amenable to metal-affinity driven coordination of His6-tagged proteins, we incubated 20% aminoQDs with a streptavidin bearing a His6-tag on one subunit (hSA). Polyhistidine has been shown to associate with CdSe/ZnS core/shell QDs via metal-affinity interactions with Zn ions present at the nanocrystal surface.49 In the present case, Cd ions present on the surface due to the alloyed shell could also participate in binding.50 We used hSA-tagged QDs in conjunction with biotinylated EGF for specific labeling of EGFR on live cells (Figure 8A, left). The resulting QD–hSA conjugates showed specific binding to biotinylated EGF on EGFR-expressing cells, indicated by the YFP transfection marker (Figure 8B, left). We note that previously, an analogue of compound 7 with n ≈ 12 was found to inhibit the binding of a polyhistidine-tagged protein to the metal-rich QD surface due to steric interference.37 However, we found that the PEG-based ligands described here, with n ≈ 8, enabled efficient binding to hSA.
Figure 8.
Targeting the EGF receptor with a QD–dye conjugate. (A) Schematic showing conjugation of His6-tagged streptavidin (hSA) to 20% aminoQDs (left) or combined hSA conjugation with covalent conjugation of dye to 20% aminoQDs (right) (only one surface ligand shown for clarity). (B) Labeling of EGFR transfected into HeLa cells with biotinylated EGF, followed by staining with QD–hSA (left), QD–hSA–dye (middle), or a control with QD–hSA–dye minus biotinylated EGF (right). (Top) Fluorescence pseudocolor images showing overlay of 558 nm (QD emission, green) and 605 nm (emission from QD–dye FRET, red). (Bottom) Phase contrast images are overlaid with YFP (nuclear transfection marker, shown in blue).
To generate aminoQDs bearing both a dye and streptavidin, we conjugated 20% aminoQDs to the photostable AlexaFluor 568 dye using NHS chemistry, giving ~2.3 dye molecules per QD and ~75% FRET efficiency (Figure S7). The QD–dye conjugate was then incubated with hSA and applied to HeLa cells displaying EGFR after incubation with biotinylated EGF (Figure 8A, right). QD–hSA–dye conjugates bound specifically to EGFR-transfected cells and excitation at 405 nm led to emission principally from the dye, as a result of FRET (Figure 8B, middle). Analysis of the individual channels (Figure S8) shows highly quenched QD emission in the green channel for the QD–hSA–dye conjugate compared to that for the QD–hSA conjugate. Also, upon intense irradiation of the QD–hSA–dye conjugate at 405 nm, bleaching of the dye was observed in the red channel, along with a recovery of the QD emission in the green channel, further supporting the occurrence of FRET within the targeted QD–dye conjugate on the cell surface (data not shown).
By appending an appropriate environmentally responsive chromophore, QD–dye conjugates can be made to behave as FRET-based sensors of their environment (e.g., pH),19 while the His6-tag conjugation of hSA provides a means for targeting QDs to cellular proteins with high affinity. Thus, QDs which exclusively sense a targeted receptor’s local environment can potentially be realized using this method of dual conjugation.
Conclusions
We have synthesized a series of DHLA-PEG functionalized ligands that produce aqueous derivatizable QDs with small hydrodynamic size, low nonspecific binding, high quantum yield, and good solution stability across a wide pH range for cell-labeling applications. We demonstrated the ability to tune the QD surface charge and functional valency without sacrificing pH stability by ligand exchange using a mixture of aminoPEG or carboxyPEG with hydroxyPEG. Using 20% aminoQDs, we showed efficient covalent conjugation of a dye using mild carbodiimide-coupling chemistry. Furthermore, covalent conjugation of streptavidin to 20% aminoQDs enabled high affinity labeling and single-particle tracking of EGF interaction with EGFR on live cells. We further demonstrated the use of both covalent conjugation of a dye and metal-affinity driven conjugation of a His6-tagged streptavidin to create multifunctional nanocrystals with possible application as targeted, optically reporting nanosensors. In principle, any combination of proteins, peptides, or small-molecule reporters can be conjugated to these QDs covalently and/or via His6-tag to provide the desired targeting/sensing capabilities, while maintaining the favorable properties for cell-labeling applications.
Experimental Section
Materials and Analysis
All chemicals unless indicated were obtained from Sigma Aldrich and used as received. Air-sensitive materials were handled in an Omni-Lab VAC glove box under dry nitrogen atmosphere with oxygen levels <0.2 ppm. All solvents were spectrophotometric grade and purchased from EMD Biosciences. Amine-bearing compounds were visualized on thin layer chromatography (TLC) plates using a ninhydrin solution. All other TLC plates were visualized by iodine staining. NMR spectra were recorded on a Bruker DRX 401 NMR Spectrometer. ESI-MS was measured on an Applied Biosystems QTrap mass spectrometer. Samples were dissolved in a solution of acetonitrile, water, and acetic acid (50:50:0.01 v/v) at a concentration of 2.5 pmol/µL and introduced via syringe pump at a flow rate of 10 µL/min. UV-vis absorbance spectra were taken using an HP 8453 diode array spectrophotometer. Photoluminescence spectra were recorded with a SPEX FluoroMax-3 spectrofluorimeter. The absorbance of all solutions was kept below 0.1 OD to avoid inner-filter effects. Commercial QD605’s were obtained from Invitrogen (cat. no. Q22041, component A).
Compound 1 (Diamino-PEG)
Neat PEG8 (average MW 400 g/mol) (20.0 g, 48.3 mmol) was degassed at 80 °C for 1 h with stirring to remove traces of water. The flask was back-filled with N2 and cooled on an ice bath before thionyl chloride (10.5 mL, 145.0 mmol) was slowly added. The solution was warmed to 25 °C and stirred for 2 h. The conversion was monitored by the disappearance of the broad O–H stretch at 3500 cm−1 and the appearance of a C–Cl stretch at 730 cm−1 in the IR spectrum. The product was diluted with DMF (20 mL) and the solvent removed under reduced pressure. This was repeated three times to remove all residual traces of thionyl chloride. The sample was dissolved in a solution of sodium azide (9.42 g, 145.0 mmol) in 250 mL of DMF and stirred overnight at 85 °C. The solvent was removed under reduced pressure, and 200 mL of dichloromethane was added. The precipitate was removed by vacuum filtration and the solvent evaporated in vacuo to yield the intermediate diazide. The conversion was confirmed by the appearance of a sharp azide stretch at 2100 cm−1 and the disappearance of the C-Cl stretch at 730 cm−1 in the IR spectrum. The sample was dissolved in 300 mL of tetrahydrofuran (THF), and triphenylphosphine (27.9 g, 106 mmol) was added. The solution was stirred at 25 °C for 4 h before adding 4 mL of water and stirring overnight. The THF was removed in vacuo, and 100 mL of water was added. The precipitate was removed by vacuum filtration and the filtrate washed with toluene (3 × 50 mL). The water was removed in vacuo to yield the pure product as light-yellow oil (17.8 g, 89%). ESI-MS: m/z 457 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) 3.53 (m, 28 H), 3.39 (t, J = 5.2 Hz, 4H), 2.74 (t, J = 5.2 Hz, 4H), 1.27 (s, 4H).
Lipoic Acid NHS-Ester (LA-NHS)
To a solution of lipoic acid (LA, 5.00 g, 24.23 mmol) and N-hydroxysuccinimide (NHS, 3.35 g, 29.1 mmol) in 150 mL of tetrahydrofuran at 4 °C was added slowly a solution of dicyclohexylcarbodiimide (6.00 g, 29.1 mmol) in 10 mL of tetrahydrofuran. The mixture was warmed to room temperature and stirred for 5 h. The precipitate was removed by vacuum filtration and the solvent evaporated in vacuo. The crude product was redissolved in 100 mL of ethyl acetate and filtered once more by vacuum filtration. The product was recrystallized from a solution of hot ethyl acetate/ hexane (1:1 v/v) as a pale-yellow solid (5.88 g, 80%). 1H NMR (400 MHz, CDCl3): δ (ppm) 3.58 (m, 1H), 3.13 (m, 2H), 2.84 (s, 4H), 2.63 (t, J = 7.1 Hz, 2H)), 2.50 (m, 1H), 1.99−1.46 (m, 7H).
Compound 2 (LA-PEG-Amine)
To a solution of compound 1 (12 g, 29.1 mmol) and sodium bicarbonate (2.44 g, 29.1 mmol) in dimethylformamide/water (100 mL, 50:50 v/v) at 4 °C was added dropwise a solution of LA-NHS (1.60 g, 5.27 mmol) in 10 mL of dimethylformamide over 1 h. The solution was warmed to RT, stirred overnight, and extracted with chloroform (3 × 30 mL). The combined organic extracts were washed with water (3 × 30 mL), dried over sodium sulfate, and filtered, and the solvent was evaporated. The crude product was purified by alumina column (dichloromethane/methanol, 95:5) to give the final product as a yellow oil (1.90 g, 60%). ESI-MS: m/z 601 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) 3.63 (m, 26H), 3.52 (t, J = 5.2 Hz, 2H), 3.47 (t, J = 5.2 Hz, 2H) 3.10 (m, 2H), 2.86 (t, J = 5.2 Hz, 2H), 2.40 (m, 1H), 2.17 (t, J = 6.5 Hz, 2H), 1.99−1.46 (m, 7H).
Compound 3 (aminoPEG)
To a solution of 2 (1.50 g, 2.50 mmol) in 4:1 water/ethanol (20 mL) at 4 °C was slowly added 4 equiv of sodium borohydride over a 30 min period. The solution was stirred for 2 h at 4 °C, acidified to pH 2 with 3 M HCl, and extracted with chloroform (3 × 15 mL). The combined organics were dried over MgSO4 and filtered. The solvent was removed in vacuo to yield the product as a colorless oil (1.37 g, 91%). ESI-MS: m/z 603 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) 3.84 (m, 2H), 3.61 (m, 36H), 3.52 (m, 4H), 3.13 (m, 2H), 2.84 (m, 2H), 2.62 (m, 2H), 2.19 (m, 2H), 1.99−1.46 (m, 9H).
Compound 5 (LA-PEG-CO2H)
Into a solution of 2 (1.90 g, 3.16 mmol) and triethylamine (0.320 g, 3.16 mmol) in dichloromethane (30 mL) was dripped slowly a solution of methylmalonylchloride (0.475 g, 3.48 mmol) in dichloromethane (10 mL) at 4 °C. The solution was stirred at RT for 4 h and the solvent removed in vacuo. The crude product was purified by silica column (dichloromethane/methanol, 95:5) and the solvent evaporated to give the pure product as a yellow oil (1.97 g, 89%). Methylester deprotection was achieved by stirring with 3.5 equiv of NaOH in methanol for 5 h at 60 °C. The solvent was removed in vacuo after neutralizing to pH 7 with 3 M HCl. The product was dissolved in water, acidified to pH 2, and extracted with chloroform (3 × 20 mL) to yield the pure product in quantitative yield. ESI-MS: m/z 731 [M + H]+. 1H NMR (400 MHz, CDCl3): δ (ppm) 3.70−3.52 (m, 36 H), 3.51−3.35 (m, 6H), 3.14 (m, 2H), 2.45 (m, 1H), 2.20 (t, J = 7.3 Hz, 2H), 1.96−1.36 (m, 7H).
Compound 6 (carboxyPEG)
To a solution of 2 (1.50 g, 2.06 mmol) in 0.25 M sodium bicarbonate buffer (20 mL) at 4 °C was slowly added 4 equiv of sodium borohydride over a 30 min period. The solution was stirred for 2 h at 4 °C, acidified to pH 2 with 3 M HCl, and extracted with chloroform (3 × 15 mL). The combined organics were dried over MgSO4 and filtered. The solvent was removed in vacuo to yield the product as a colorless oil. ESI-MS: m/z 733 [M + H]+. 1H NMR (400 MHz, CDCl3):δ (ppm) 3.70−3.52 (m, 36 H), 3.51−3.35 (m, 6H), 2.87 (m, 1H), 2.65 (m, 2H), 2.18 (t,J = 7.3 Hz, 2H), 1.96−1.36 (m, 9H).
Synthesis of CdSe(ZnxCd1−xS) core(shell) QDs
CdSe cores were synthesized according to previously reported procedures.39 Overcoating with an alloyed shell was carried out via modifications to previously reported procedures. Briefly, CdSe cores precipitated from growth solution by the addition of methanol were redispersed in hexane and injected into a degassed solution of 10 g of 99% trioctylphosphine oxide (TOPO) and 0.4 g n-hexylphosphonic acid. After removing the hexane under reduced pressure at 80 °C, the flask was back-filled with dry N2and the temperature increased to 130 °C before adding 0.25 mL of decylamine and stirring for 30 min. Precursor solutions of diethylzinc (ZnEt2), dimethylcadmium (CdMe2), and hexamethyldisilathiane [(TMS)2S] were prepared by dissolving the appropriate amounts of each in 4 mL of TOP and loading them into two separate syringes for metal and sulfur under inert atmosphere. The molar quantity of ZnEt2 required to achieve the desired shell thickness (typically 5 monolayers) was calculated according to the methods of Leatherdale.51 For an alloyed shell, an appropriate mole fraction ZnEt2 was replaced by CdMe2. A 2-fold molar excess of (TMS)2S was used. The precursor solutions were injected simultaneously into the 130 °C bath at a rate of 4 mL/h. The sample was annealed overnight at 80 °C, and 4 mL of butanol was added. The QDs were stored in growth solution under ambient conditions and centrifuged once more before use. Typical fluorescence quantum yields were 68% for QD605 in hexane.
Water Solubilization of CdSe(ZnCdS) Core(Shell) QDs
Exchange of the native tri-n-octylphosphine/tri-n-octylphosphine oxide (TOP/TOPO) surface ligands on QDs for the PEG-derivatized ligand was carried out according to previously reported procedures, with modifications to masses of reagents used and the incubation times.37 To 0.2 mL of QDs in growth solution (optical density > 50 at 350 nm) was added acetone to the point of turbidity. Centrifugation and decantation yielded ~10 mg of dry pellet, to which 50 µL of neat DHLA-PEG derivatized ligand and 10 µL of methanol were added. The mixture was stirred gently under N2 at 60 °C for 2.5 h and precipitated by adding 0.3 mL of ethanol, 0.05 mL of chloroform, and 0.5 mL of hexane in succession. Centrifugation at 3000g for 2 min yielded a clear supernatant, which was discarded. The pellet was dispersed in 0.5 mL of PBS and filtered through a 0.2 µm filter. The sample was purified using gel filtration chromatography in order to remove aggregated QDs, and the fractions were concentrated at 3500g using a Vivaspin-6 10 000 MW cutoff spin concentrator. Typical concentrations of QD preparations were 8 µM. The QY in water was ~40%.
Quantum Yield Measurement
QYs of QD565 and QD605 were measured relative to rhodamine 590 (λexc = 490 nm) and rhodamine 640 (λexc = 520 nm), respectively. Solutions of QDs in PBS and dye in ethanol were optically matched at the excitation wavelength. Fluorescence spectra of QD and dye were taken under identical spectrometer conditions in triplicate and averaged. The optical density was kept below 0.1 between 300 and 800 nm, and the integrated intensities of the emission spectra, corrected for differences in index of refraction and concentration, were used to calculate the quantum yields using the expression QYQD = QYDye × (absorbancedye/absorbanceQD) × (peak areaQD/peak areaDye) × (nQD solvent)2/(nDye solvent)2.52
Gel Filtration Apparatus
GFC was performed using an ÄK-TAprime Plus chromatography system from Amersham Biosciences equipped with a Superose 6 10/300 GL column. PBS (pH 7.4) was used as the mobile phase with a flow rate of 0.5 mL/min. Typical injection volumes were 50 µL. Detection was achieved by measuring the absorption at 280 nm, and the fluorescence spectrum in 30 s intervals was simultaneously recorded using an Ocean Optics SD2000 fiber optic spectrometer (λexc = 460 nm) from an Ocean Optics LS-450 LED light source. The column was calibrated using gel filtration protein standards from Bio-Rad (cat. no. 151–1901) ranging in MW from 1.3 to 158 kDa.
Dynamic Light Scattering
Light-scattering analysis was performed using a DynaPro Dynamic Light Scatterer. All QD samples were between 0.5 and 2 µM and filtered through a 0.02 µm filter before analysis. Typical count rates were between 85 and 150 kHz. Each autocorrelation function (ACF) was acquired for 10 s, and averaged for 10 min per measurement. A software filter was employed to discard all ACF fits with sum of square errors >15. The resulting ACF was fitted using the Dynamics V6 software employing a non-negative least-squares fitting algorithm. Hydrodynamic radii were obtained from a mass-weighted size distribution analysis and reported as the mean of triplicate measurements.
Agarose Gel Electrophoresis
Electrophoresis of QDs was performed using a Minicell Primo (Thermo) with 1% Omnipur agarose (EMD) in TAE (40 mM Tris-acetate, 1 mM EDTA, pH 8.3) at 7.9 V/cm for 15 min. QDs were diluted to 150 nM in TAE and mixed with 6 × loading buffer (16% sucrose) before loading onto the gel. Gels were visualized under 305 nm UV with a ChemiImager 5500 (Alpha Innotech Corporation).
ζ-Potential Measurement
ζ-potential was measured on a Zeta PALS Zeta Potential Analyzer from Brookhaven Instruments Corp. QDs coated with aminoPEG and carboxyPEG (5 µM) were measured in PBS buffer at pHs 6.0 and 7.4, respectively. AminoQDs were measured at pH 6.0 because the samples were insufficiently ionized at pH 7.4 to yield well-resolved data at the different aminoPEG/hydroxyPEG ligand compositions. Values are reported as the average of triplicate runs consisting of 100 scans each.
pH Stability Measurement
Twenty percent aminoQDs and carboxyQDs (554 nm emission) were incubated in PBS buffer, pHs ranging from 5.0 to 7.4, and borate buffer ranging from pH 8.5 to pH 9.5. at room temperature in the dark at 2 µM concentration. At various time intervals, an aliquot was diluted in PBS for fluorescence analysis in a 96-well plate. Fluorescence intensity measurements of QDs were performed on a Tecan XFluor4 plate reader, with excitation at 490 nm and emission detection at 554 nm. The emission intensities were normalized to that from a stock solution of Rhodamine 590.
Covalent Conjugation of Dye Molecules to Ligand-Exchanged QDs
Purified 20% aminoQDs were brought to ~10 µM in sodium bicarbonate buffer (30 mM, pH 8.5)), and then an aliquot of ROX NHS ester (Molecular Probes), prepared at 5 mM in anhydrous dimethylformamide, was introduced. After 1 h reaction time at room temperature, the QDs were then separated from unbound material via ultrafiltration. In control experiments, the amine-reactive dye was replaced with the nonactivated (free acid) form, or by pure DMF, and the same workup procedure was applied after the reaction time had elapsed.
Analysis of FRET in Covalently-Linked QD–Dye Conjugates
Förster theory predicts an overall efficiency E = mR06/(mR06 + r6) for the transfer of excitation from a donor to a set of m acceptors separated by a constant distance r, where R0 is the characteristic distance at which the transfer to a single acceptor would proceed at 50% efficiency. The characteristic distance (in cm) is given by R0 = (8.79 × 10−25 κ2n−4JQD)1/6 for donor quantum yieldQD and spectral overlap integral J, with orientation factor κ2 = 2/3 and refractive index n = 1.33. For a given sample, E can be obtained from the quench of the QD donor emission with respect to a control prepared with no dye, m is determined from the absorption spectrum with an appropriate estimate of the QD molar extinction coefficient, and R0 can be calculated from the spectral overlap and measured QD quantum yield. These parameters allow the typical separation r to be estimated and compared with the radius measured by other means, e.g., DLS.
Conjugation of 20% AminoQDs to Streptavidin
SA (50 µL, 10 mg/mL) was first activated using 1000 equiv of EDC and NHS in MES buffer (100 mM, pH 6.5) for 30 min. Excess coupling reagent was removed by ultrafiltration through a 10 000 MW cutoff filter (Vivaspin 2). Activated SA (16 µL) was mixed with 20% aminoQDs (50 µL, 7 µM) in sodium bicarbonate buffer (50 mM, pH 8.4) and allowed to react for 1 h. The QD–SA conjugate was purified by ultrafiltration through a 10 000 MW cutoff filter (Vivaspin 6) into PBS (pH 7.4).
Conjugation of 20% AminoQD–wtSA to Nonspecifically Biotinylated Cells
HeLa cells were incubated with 1 mg/mL of EZ-Link Biotin-Sulfo-NHS (Pierce) in PBS + 1% casein for 15 min at RT. The cells were washed 4× and incubated with 20% aminoQD–wtSA samples (100 µL, 200 nM) at 4 °C for 10 min. The cells were then washed 4× with ice-cold PBS before imaging.
Conjugation of His6-Tagged Streptavidin (hSA) to Dye-Functionalized QDs
To conjugate His6-tagged streptavidin (hSA) (A1D3, as previously described53) to the QDs, 5 µL of 3 µM control or dyeconjugated QDs in 10 mM sodium borate pH 7.4 were incubated with 5 µL of 27 µM hSA in PBS for 1 h at RT.
Cell Culture, Labeling, and Imaging
HeLa (human carcinoma) and COS7 (African green monkey kidney) cells were grown in DMEM with 5% fetal calf serum, 50 U/mL penicillin and 50 µg/mL streptomycin. Transfections were performed with lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions, and cells were imaged the day after transfection. pEYFP-H2B54 was a kind gift of A. K. Leung (MIT). This encodes histone H2B fused to enhanced yellow fluorescent protein (YFP), as a nuclear-localized cotransfection marker. The cytoplasmic cotransfection marker BFP was a gift from R. Y. Tsien (UCSD). The human EGFR gene in pcDNA3 (Invitrogen) was a gift from K. D. Wittrup (MIT).
Nonspecific Binding of QDs
HeLa cells, cooled to 4 °C in PBS for 5 min to minimize endocytosis, were incubated with 40 nM QDs in PBS with 0.5% dialyzed bovine N,N-dimethyl casein (Calbiochem) for 10 min at 4 °C. Cells were then washed 4× with ice-cold PBS and imaged in PBS.
Fluorescence and Phase Contrast Microscopy
Cells were imaged live using a Zeiss Axiovert 200M inverted epifluorescence microscope with a 40× oil-immersion lens and a Cascade II camera (Photometrics) with intensification set at maximum. Bright field images were collected using differential interference contrast and 30 ms exposure. BFP (420DF20 excitation, 550DRLP dichroic, 575DF40 emission), YFP (495DF10 excitation, 515DRLP dichroic, 530DF30 emission), Alexa Fluor 568 or ROX (560DF20 excitation, 585DRLP dichroic, 605DF30 emission), QD558 (405DF20 excitation, 515DRLP dichroic, 565DF20 emission), and QD605 or QD558/ROX FRET (405DF20 excitation, 585DRLP dichroic, 605DF30 emission) images were collected and analyzed using Slidebook software (Intelligent Imaging Innovations). Typical exposure times were 0.1–0.5 s. AlexaFluor 568 fluorescence was bleached by 30 s of intense 405DF20 excitation. Fluorescence images were background corrected. All fluorescence images were captured under identical camera conditions and are displayed at the same contrast level such that they can be directly compared. Movies were acquired with 0.2 s exposure and no delay between frames, using an additional 2.5× Optovar lens.
EGF Receptor Labeling with Streptavidin Linked to QDs by EDC Coupling
COS7 cells were transfected with 0.2 µg pcDNA3 EGFR and 7.5 ng pcDNA3 BFP per well of a 48-well plate. The next day cells were incubated in PBS with 5 mM MgCl2, 0.5% dialyzed casein, and 90 nM biotinylated EGF (biotin-XX-EGF from Invitrogen: human EGF conjugated at a single site to biotin via a long spacer arm) for 5 min at RT. Cells were washed four times with PBS and incubated with PBS, 0.5% dialyzed casein, and 70 nM 20% aminoQD605-wtSA for 5 min at RT, before washing four times in PBS and imaging in PBS. As a negative control, QD605-wtSA was incubated with a 500-fold excess of free biotin (Tanabe, U.S.A.) for 5 min at RT, before adding to cells. For movie imaging, COS7 cells were labeled as above but with 20 nM 20% aminoQD605-wtSA and were maintained in the microscope at 37 °C using an environmental control system (Solent Scientific).
Imaging of FRET between QD and Dye while Bound to EGF Receptor
A 5-fold molar excess of hSA in PBS was incubated with 20% aminoQD558 or 20% aminoQD558-Alexa Fluor 568 for 30 min at RT, allowing stable binding of the His6-tag of hSA to the QD shell. HeLa were transfected with 0.2 µg of pcDNA3 EGFR and 5 ng of H2B-YFP per well of a 48-well plate. The next day cells were incubated in PBS with 5 mM MgCl2, 0.5% dialyzed casein, and 60 nM biotinylated EGF (biotin-XX-EGF from Invitrogen: human EGF conjugated at a single site to biotin via a long spacer arm) for 5 min at 4 °C, to stop receptor internalization. Cells were washed four times with cold PBS and incubated with PBS, 0.5% dialyzed casein, and 40 nM QD558-hSA or QD558-hSA-Alexa Fluor 568 for 5 min at 4 °C, before washing four times in cold PBS and imaging.
Acknowledgment
This research was supported by the NSFMRSEC program (DMR-0117795) and made use of its shared user facilities, the Harrison Spectroscopy Laboratory and the MIT-Harvard NIH Center for Cancer Nanotechnology Excellence (1U54-CA119349) (M.G.B.); National Institutes of Health (1 P20GM072029-01) (A.Y.T.); and the Army Research Office (W911NF-06-1-0101) (D.G.N.). W.L. was supported by a National Science Foundation Graduate Research Fellowship. M.H. was supported by a Computational and Systems Biology Initiative MIT-Merck postdoctoral fellowship. A.B.G. is a Novartis fellow of the Life Sciences Research Foundation. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH GM68762) is gratefully acknowledged.
Footnotes
Supporting Information Available: Supporting material contains dynamic light-scattering data, fluorescamine-aminoQD emission data, spectra for 20% aminoQDs-Alexa Fluor 568, channel images of QD dye conjugate targeting EGF receptor, and 1H NMR and ESI-MS spectra of 1, 2, 3, 5, 6, and LA-NHS.
ⓦ This paper contains enhanced objects available on the Internet at http://pubs.acs.org/journals/jacsat.
References
- 1.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J. Phys. Chem. B. 1997;101:9463–9475. [Google Scholar]
- 2.Chan WCW, Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 3.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 4.Peng ZA, Peng X. J. Am. Chem. Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 5.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J. Am. Chem. Soc. 2000;122:12142–12150. [Google Scholar]
- 6.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Nat. Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 7.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 8.Howarth M, Takao K, Hayashi Y, Ting AY. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X, Chan WCW, Nie S. J. Biomed. Opt. 2002;7:532–537. doi: 10.1117/1.1506706. [DOI] [PubMed] [Google Scholar]
- 10.Anikeeva N, Lebedeva T, Clapp AR, Goldman ER, Dustin ML, Mattoussi H, Sykulev Y. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16846–16851. doi: 10.1073/pnas.0607771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Bioconjugate Chem. 2004;15:79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 13.Zimmer JP, Kim S-W, Ohnishi S, Tanaka E, Frangioni JV, Bawendi MG. J. Am. Chem. Soc. 2006;128:2526–2527. doi: 10.1021/ja0579816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 15.So M-K, Xu C, Loening AM, Gambhir SS, Rao J. Nat. Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 16.Soltesz EG, Kim S, Laurence RG, DeGrand AM, Parungo CP, Dor DM, Cohn LH, Bawendi MG, Frangioni JV, Mihaljevic T. Ann. Thorac. Surg. 2005;79:269–277. doi: 10.1016/j.athoracsur.2004.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Nat. Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka E, Choi HS, Fujii H, Bawendi MG, Frangioni JV. Ann. Surg. Oncol. 2006;13:1671–1681. doi: 10.1245/s10434-006-9194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snee PT, Somers RC, Nair G, Zimmer JP, Bawendi MG, Nocera DG. J. Am. Chem. Soc. 2006;128:13320–13321. doi: 10.1021/ja0618999. [DOI] [PubMed] [Google Scholar]
- 20.Somers RC, Bawendi MG, Nocera DG. Chem. Soc. Rev. 2007;36:579–591. doi: 10.1039/b517613c. [DOI] [PubMed] [Google Scholar]
- 21.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Tetsuo Uyeda H, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Nat. Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 22.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 23.Medintz I, Uyeda H, Goldman E, Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Nakatani E, Gronenberg LS, Tokimoto T, Wirth MJ, Hruby VJ, Roberts A, Lynch RM, Ghosh I. Bioconjug. Chem. 2007;18:323–332. doi: 10.1021/bc0601929. [DOI] [PubMed] [Google Scholar]
- 25.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 26.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nanolett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 27.Groc L, Heine M, Cognet L, Brickley K, Stephenson FA, Lounis B, Choquet D. Nat. Neurosci. 2004;7:695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- 28.Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. J. Cell. Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AM, Duan H, Rhyner MN, Ruan G, Nie S. Phys. Chem. Chem. Phys. 2006;8:3895–3903. doi: 10.1039/b606572b. [DOI] [PubMed] [Google Scholar]
- 30.Groc L, Heine M, Cognet L, Brickley K, Stephenson FA, Lounis B, Choquet D. Nat. Neurosci. 2004;7:695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- 31.Bentzen EL, Tomlinson ID, Mason J, Gresch P, Warnement MR, Wright D, Sanders-Bush E, Blakely R, Rosenthal SJ. Bioconjugate Chem. 2005;16:1488–1494. doi: 10.1021/bc0502006. [DOI] [PubMed] [Google Scholar]
- 32.Parak WJ, Gerion D, Zanchet D, Woerz AS, Pellegrino T, Micheel C, Williams SC, Seitz M, Bruehl RE, Bryant Z, Bustamante C, Bertozzi CR, Alivisatos AP. Chem. Mater. 2002;14:2113–2119. [Google Scholar]
- 33.Aldana J, Wang YA, Peng X. J. Am. Chem. Soc. 2001;123:8844–8850. doi: 10.1021/ja016424q. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Bawendi MG. J. Am. Chem. Soc. 2003;125:14652–14653. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 35.Algar WR, Krull UJ. Langmuir. 2006;22:11346–11352. doi: 10.1021/la062217y. [DOI] [PubMed] [Google Scholar]
- 36.Pinaud F, King D, Moore HP, Weiss S. J. Am. Chem. Soc. 2004;126:6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. J. Am. Chem. Soc. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- 38.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat. Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 39.Snee PT, Chan Y, Nocera DG, Bawendi MG. Adv. Mater. 2005;17:1131–1136. [Google Scholar]
- 40.Murray CB, Norris DJ, Bawendi MG. J. Am. Chem. Soc. 1993;115:8706–8715. [Google Scholar]
- 41.Manna L, Scher EC, Li LS, Alivisatos AP. J. Am. Chem. Soc. 2002;124:7136–7145. doi: 10.1021/ja025946i. [DOI] [PubMed] [Google Scholar]
- 42.Yu Z, Guo L, Du H, Krauss T, Silcox J. Nano Lett. 2005;5:565–570. doi: 10.1021/nl048245n. [DOI] [PubMed] [Google Scholar]
- 43.Xie R, Kolb U, Li J, Basche T, Mews A. J. Am. Chem. Soc. 2005;127:7480–7488. doi: 10.1021/ja042939g. [DOI] [PubMed] [Google Scholar]
- 44.Pons T, Uyeda HT, Medintz IL, Mattoussi H. J. Phys. Chem. B. 2006;110:20308–20316. doi: 10.1021/jp065041h. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, DePaoli V, Rosenzweig N, Rosenzweig Z. J. Am. Chem. Soc. 2006;128:10378–10379. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- 46.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. J. Am. Chem. Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 47.Moasser MM. Oncogene. 2007;26:6577–6592. doi: 10.1038/sj.onc.1210478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teramura Y, Ichinose J, Takagi H, Nishida K, Yanagida T, Sako Y. EMBO J. 2006;25:4215–4422. doi: 10.1038/sj.emboj.7601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. J. Phys. Chem. C. 2007;111:11528–11538. [Google Scholar]
- 50.Prabhukumar G, Matsumoto M, Mulchandani A, Chen W. Environ. Sci. Technol. 2004;38:3148–3152. doi: 10.1021/es035150z. [DOI] [PubMed] [Google Scholar]
- 51.Leatherdale CA, Woo WK, Mikulec FV, Bawendi MG. J. Phys. Chem. B. 2002;106:7619–7622. [Google Scholar]
- 52.Eaton D. Pure Appl. Chem. 1988;60:1107–1114. [Google Scholar]
- 53.Howarth M, Chinnapen DJF, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. Nat. Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Platani M, Goldberg I, Lamond AI, Swedlow JR. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]