Abstract
Small noncoding microRNAs (miRNAs) have been shown to be abnormally expressed in every tumor type examined. The importance of miRNAs as potential cancer prognostic indicators is underscored by their involvement in the regulation of basic cellular processes such as cell proliferation, differentiation, and apoptosis. In this study, miRNA expression profiles of head and neck squamous cell carcinoma (HNSCC) tumor and adjacent normal tissue were examined by microarray analysis and validated by quantitative TaqMan real-time polymerase chain reaction. Using TaqMan real-time polymerase chain reaction we measured the quantitative associations between a subset of miRNAs identified on microarrays in primary tumors at diagnosis and cancer survival in a cohort of 104 HNSCC patients undergoing treatment with curative intent. The majority of miRNAs exhibiting altered expression in primary human HNSCC tumors (including miR-1, miR-133a, miR-205, and let-7d) show lower expression levels relative to normal adjacent tissue. In contrast, hsa-miR-21 is frequently overexpressed in human HNSCC tumors. Using univariate and multivariable statistical models we show that low levels of hsa-miR205 are significantly associated with loco-regional recurrence independent of disease severity at diagnosis and treatment. In addition, combined low levels of hsa-miR-205 and hsa-let-7d expression in HNSCC tumors are significantly associated with poor head and neck cancer survival Our results show that miRNA expression levels can be used as prognostic markers of head and neck cancer.
MicroRNAs (miRNAs) are a group of noncoding 22 nucleotide RNA molecules that posttranscriptionally regulate the expression of target mRNA.1 These newly discovered small RNAs regulate processes as fundamental as cellular proliferation, differentiation, and apoptosis, and a subset of miRNAs have been identified as potential diagnostic and prognostic markers in cancer.2,3,4
Among these, recent studies have shown that overexpression of miR-21 in lung cancer tumors may act as a classic oncogene, and down-regulation of let-7 miRNA family of genes, which target the Ras oncogene, may be correlated with poor survival and relapse in non-small cell lung cancer.5,6 Aberrant regulation of miRNAs has also revealed roles for specific genes as either tumor suppressors, when deleted or repressed, or as oncogenes when amplified or otherwise overexpressed.2,7,8 However, miRNA expression is highly tissue-specific; separate and distinct profiles have been described for every cancer type.9 To date there have been only two studies investigating miRNAs in head and neck squamous cell carcinoma (HNSCC) immortal cell lines, neither of which assessed the role of these noncoding RNA molecules in human tissue or attempted to correlate miRNA expression with HNSCC prognosis.9,10
HNSCC is the fifth most common malignancy in men worldwide and includes tumors of the oral cavity, oropharynx, and larynx. Survival rates for HNSCC have remained unchanged throughout the last 3 decades, and half of all cases die within 5 years of diagnosis.11 Efforts to identify prognostic biomarkers for HNSCC, including p53, EGFR, Bcl-2, MMPs, cyclins, and other markers, have proved to be primarily unsuccessful to date.12,13,14 As with mRNA expression, high-throughput genomic technologies can be used to shed new light on alterations in gene expression that are associated with HNSCC carcinogenesis.15,16,17 This is the first report to comprehensively identify and measure differentially transcribed miRNAs in HNSCC and assesses their role in cancer prognosis in humans.
Materials and Methods
Collection of Samples and Patient Data
Fresh HNSCC tumor specimens were obtained at the time of diagnosis. Adjacent normal tissue was sampled for comparison. This was taken from the same surgical field, as often done during the tumor survey, and did not put the patient at increased or additional risk. Standard biopsy techniques were performed, as were standard surgical resection when appropriate. Additional biopsies of normal tissue adjacent to the tumor mass were taken when required. Patients were excluded if their primary tumor was too small to allow both diagnostic biopsy and biopsy for the study. Patients were recruited and consented following an institutional review board-approved protocol before inclusion in the study.
Detailed clinical and demographic information was collected by physicians on all patients enrolled in the study and patients were prospectively followed-up to determine clinical outcome and disease progression. Patient information is entered on an ongoing basis into a secure clinical database system developed for the head and neck cancer program at the Albert Einstein College of Medicine. Patients undergo treatment for HNSCC, as deemed appropriate by the treating physicians blinded to miRNA results.
RNA Extraction
Fresh frozen tumor samples were prospectively collected from HNSCC patients at Montefiore Medical Center in the Bronx. Tumor tissue was snap-frozen in liquid nitrogen within 30 minutes of surgical resection or biopsy, and before treatment. Total RNA was extracted from an additional 50 to 100 mg of tissue using TRIzol by standardized protocol (Invitrogen, Carlsbad, CA). RNA was collected by alcohol precipitation and quantitated for microarray or real-time PCR analysis Selected samples were checked for integrity of RNAs and presence of microRNA peaks using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA) and RNA pico chips as described by the manufacturer.
MicroRNA Microarrays
MiRNA oligonucleotide microarrays used in this study were custom printed at the Albert Einstein College of Medicine Microarray Facility. We printed a custom miRNA microarray that consisted of a set of antisense probes from Ambion Inc., Austin, TX (version 1) and the same oligonucleotide set described by Croce’s group19 that included sense strand probes, anti-sense controls, and selected miRNA precursor-specific probes representing a total 236 unique human miRNA genes. Each oligonucleotide was triple-spotted along with Ambion controls used for probe labeling and hybridization efficiency monitoring. MiRNAs were enriched from 5 μg of total RNA by fractionation using Ambion Flashpage polyacrylamide gels followed by column purification. Purified miRNAs were labeled by addition of polyA tails followed by chemical coupling of Alexa florescent dyes using the Vana miRNA labeling kit (Ambion) and hybridized using an Ambion hybridization kit recommended for these miRNA probes. Tumor sample RNA was labeled with Alexa 645 (red) and normal counterpart RNA with Alexa 555 (green). We also conducted dye flip experiments and yellow tests (comparison of identical RNA samples with both dyes on one microarray) to confirm the specificity of our microarray hybridizations.
Processing and Analysis of Microarray Gene Expression Data
Tumor versus normal signal intensities for each element on the array were calculated using the GenePix Pro 6.0 software package (Molecular Devices, Sunnyvale, CA). This software gives an integrated intensity per spot for each channel in addition to an integrated background count. In all subsequent analyses, we use the average background subtracted intensity for the two channels. For each spot, we calculated the mean intensity over the spot in the two channels and from this subtract the median of the background and log2 transformed. To correct for dye incorporation efficiency, fluorescence yield, and laser power used in scanning, we scaled the intensities from the two channels relative to each other, and computed an intensity-dependent normalization factor by first finding the rank invariant subset of the spots.20 MiRNAs with missing signal intensity microarray expression data for two or more HNSCC cases were excluded from analysis. Furthermore, to account for labeling and hybridization efficiencies among samples, intensity ratios for each miRNA element were normalized by first taking the average value of the three spot replicates on the microarray and then correcting the values for each color channel using prespiked control oligonucleotides that are fluorescently labeled and hybridized along with the miRNA sample.21
Real-Time PCR
Both semiquantitative and quantitative real time PCR methods were used to validate and extend the microarray data. Initially, Ambion mirVana qRT-PCR primer sets were used along with TAQ polymerase as recommended by the supplier to validate microarrays. Each primer set (miR-21, miR-1, miR-133a, let-7d, miR-205, and miR-206 and 5S RNA) was individually titrated and a cycle chosen such that the PCR product visualized by agarose gel analysis was in the exponential range of amplification. Tumor and normal samples were amplified and levels of products were estimated by gel electrophoresis and normalized to 5S RNA levels. Quantitative analysis of miRNA expression from large collections of tumor and normal samples was conducted by quantitative real-time PCR TaqMan protocol using a 7900 real time PCR apparatus (Applied Biosystems, Foster City, CA) exactly as recommended by the supplier. Results were expressed as a cycle threshold (CT) value, which is inversely proportional to the sample starting copy number. Normalized (Δ CT) values were computed by subtracting the CT value (averaged across three replicates) of a small noncoding RNA control gene (RNU 48) from the raw CT value of the miRNA element. To compare tumor and paired normal tissue, we generated the ΔΔCT value by subtracting the ΔCT value of the normal tissue from the ΔCT value of the tumor. We then examined the ΔΔCT distribution for each miRNA element graphically, assuming approximately equal amplification efficiencies between tumor and normal tissue.21
Statistical Analysis
We analyzed the association with cancer survival in a cohort of HNSCC patients undergoing treatment with curative intent in relation to miRNA level at diagnosis. Enrollment for the clinical study began in 2002 and patients were followed prospectively after histological confirmation of SCC. Time to event was measured from treatment start to the first instance of a local or regional recurrence, disease-free and overall survival, or to the last recorded follow-up visit date for censored patients. Survival and progression-free probability curves were estimated using Kaplan-Meier estimates. Cumulative incidence curves for loco-regional recurrence were also estimated, taking into account the competing risks of death and distant metastases. We investigated the potential effect of confounding by age, gender, ethnicity, smoking history, alcohol consumption, tumor location, disease stage, TNM status, treatment modality (primary resection +/− adjuvant therapy versus neoadjuvant therapy), and prevalence of human papillomavirus (HPV) (tested by MY09/11 PCR assay).22,23 Adjusted models based on an a priori model containing known or suspected confounders were presented; however, we also examined all possible models using an exhaustive search of the model space. The a priori model included anatomical site (oropharynx, larynx, lip/oral), T stage (T0/T1 versus T2/T3), HPV status (HPV+ versus HPV−), and treatment (chemoradiation therapy yes/no). Hazard ratios (HRs) and corresponding confidence intervals for the adjusted models were estimated using Cox proportional hazards models. To test the null hypothesis that the regression coefficient was equal to zero, P values based on the Wald χ2 test were computed and presented for each microRNA. Each event of interest (overall survival, loco-regional recurrence, distant metastases) was modeled separately, censoring at the occurrence of any competing event, to estimate the cause-specific hazard of event. To evaluate the overall impact of each covariate across all events of interest a single proportional hazards model, stratified by failure type, was estimated. This model allowed a different baseline hazard for each event type, but also allowed us to efficiently pool information across event type to generate an overall measure of variable importance in an adjusted model. Interactions between event type and covariate were examined for each covariate of interest. Robust standard errors using the sandwich estimator of Wei and colleagues24 were computed to correct for any model misspecification.
Results
MicroRNA Profiling of HNSCC Tumors
We used custom-spotted oligonucleotide microarrays to make direct comparisons between HNSCC tumors and adjacent normal tissue to measure aberrant regulation of specific miRNA genes in HNSCC. We were able to generate a preliminary HNSCC miRNA signature of miRNAs over- and underexpressed in tumors relative to their matched normal counterparts (Table 1). The partial list shown includes 43 human miRNAs that were expressed, on average, twofold lower in tumors versus normal samples, and 6 miRNAs that were, on average, expressed by at least twofold higher in tumors versus normals. However, we observed some variability in miRNA expression across HNSCC patients. Assessing the frequency of HNSCC tumors over- or underexpressing miRNAs compared to their adjacent normal counterpart, we found only one miRNA with at least twofold higher expression in six of eight of patient tumors examined (represented by clones for miR-21, miR-021-prec, and mmu-miR-21_AS), whereas three human miRNAs were underexpressed in six of eight patients (miR-370, miR-199a-1-prec, miR-030b-prec).
Table 1.
List of the Highest Scoring MicroRNAs Corresponding to a Partial Expression Profile of HNSCC
| MicroRNA | Mean fold change (tumor:normal)* |
|---|---|
| Underexpressed in tumors | |
| hsa-let-7f | 0.29 |
| hsa-miR-10b_AS | 0.46 |
| hsa-miR-124a_AS | 0.34 |
| hsa-miR-142-3p | 0.43 |
| hsa-miR-324-5p | 0.23 |
| hsa-miR-368 | 0.32 |
| hsa-miR-370 | 0.26 |
| hsa-miR-373* | 0.44 |
| hsa-miR-373*_AS | 0.51 |
| hsa-miR-422a_AS | 0.43 |
| hsa-miR-422b | 0.16 |
| hsa-miR-422b_AS | 0.46 |
| hsa-miR-424 | 0.27 |
| hsa-miR-95_AS | 0.46 |
| hsa-miR-99a_AS | 0.47 |
| hsa-let-7a-2-prec-#1 | 0.54 |
| hsa-let-7a-3-prec | 0.29 |
| hsa-let-7d-prec | 0.40 |
| hsa-miR-007-1-prec | 0.30 |
| hsa-miR-007-2-prec-#1 | 0.41 |
| hsa-miR-009-1-#1 | 0.50 |
| hsa-miR-009-3-#1 | 0.47 |
| hsa-miR-016b-chr3 | 0.35 |
| hsa-miR-030a-prec-#2 | 0.45 |
| hsa-miR-030b-prec-#1 | 0.18 |
| hsa-miR-030b-prec-#2 | 0.45 |
| hsa-miR-093-prec-7.1 = 093-1 | 0.40 |
| hsa-miR-106-prec | 0.47 |
| hsa-miR-125b-2-prec-#2 | 0.41 |
| hsa-miR-128b-prec-#2 | 0.43 |
| hsa-miR-130a-prec-#1 | 0.35 |
| hsa-miR-135-1-prec | 0.33 |
| hsa-miR-140-#1 | 0.54 |
| hsa-miR-155-prec | 0.50 |
| hsa-miR-192-2/3-#2 | 0.49 |
| hsa-miR-199a-1-prec | 0.24 |
| hsa-miR-213-prec-#2 | 0.42 |
| hsa-miR-216-prec-#1 | 0.43 |
| hsa-miR-218-1-prec | 0.36 |
| hsa-miR-221-prec | 0.24 |
| hsa-miR-224-prec | 0.20 |
| miR1–2 | 0.44 |
| miR133a-1 | 0.31 |
| Overexpressed in tumors | |
| hsa-miR-021-prec-17-#2 | 4.44 |
| hsa-miR-024-1-prec-#1 | 1.99 |
| hsa-miR-151-prec | 3.66 |
| hsa-miR-199b-prec-#2 | 2.22 |
| miR21 | 3.29 |
| miR23b | 1.54 |
Tumor versus normal signal intensities for each element on the array were calculated using ratios of the mean spot intensities in the two channels over the eight HNSCC samples tested. AS, antisense probe; prec, precursor to the mature miRNA specific oligonucleotide probe.19
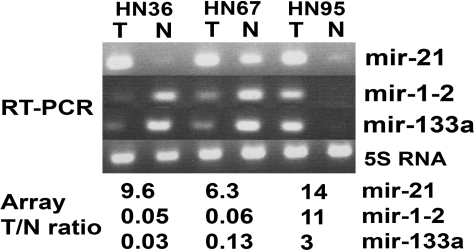
Using a semiquantitative reverse transcription (RT)-PCR assay, we independently verified the microarray data in primary tumors and matching normal tissue for six different miRNAs from our preliminary HNSCC miRNA signature (miR-21, miR-1, miR-133a, miR-205, miR-206, and let-7d). Figure 1 shows some of the RT-PCR data comparing tumor (T) and matching normal (N) RNA from three HNSCC cases. In each case examined, we observed a general agreement between microarray and RT-PCR data. The concordance of microarray and RT-PCR data demonstrated that our microarray platform could accurately identify miRNAs that are dysregulated in HNSCC and therefore candidates for biomarker analysis of individual patient samples. Given the small initial sample size, we then set out to confirm the results on a larger sample set using quantitative real-time PCR and multivariable statistical analyses.
Figure 1.
RT-PCR data comparing tumor (T) and matching normal (N) microRNA from three representative HNSCC cases. Top: Ratios of T/N expression derived from the microarrays are shown below the agarose gels displaying the PCR products derived from each RT-PCR reaction. 5S RNA was used as a control to show equal amounts of tumor and normal RNAs were included in each reaction. Bottom: Microarray data (array) shown as the ratio of tumor to normal expression (T:N ratio) for the samples used for RT-PCR is provided.
Quantification of MicroRNA Expression in HNSCC
We selected five miRNA genes from the list of miRNA above (miR-21, miR-1, miR-133a, miR-205, and let-7d) that might fit a HNSCC signature based on evidence in the literature and the preliminary results from the TvN microarray comparisons. For example, miR-1 and miR-133a showed severely low expression levels relative to normal adjacent tissue (Table 1). We therefore hypothesized that these genes might discriminate between different HNSCC tumor behaviors. MiR-205 was also selected because it has been shown to be highly enriched in HNSCC cell lines relative to other tumor types,10 and therefore represents a cell-type-specific molecule that might regulate important downstream targets. Using primers for these candidate gene species, we then performed quantitative TaqMan real-time PCR analysis of an independent set of 104 HNSCC patient samples.
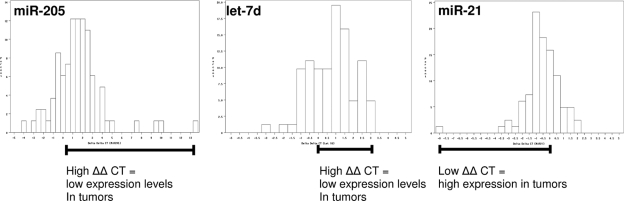
Patients were recruited from the Bronx, a high-risk area of New York City with a high incidence of head and neck cancer. Clinical characteristics of 104 patients with primary HNSCC tumors are shown in Table 2, and RNA prepared from these samples was used for our quantitative real-time PCR measurements. Matched tumor and normal adjacent tissue miRNA expression measurements were available on 95 of the 105 HNSCC patients tested. RNU 48 RNA expression levels in all samples were used as a control for the amount of RNA used in the RT-PCR reactions. Measurements of RNU 48 in tumor samples was consistent with a Gaussian distribution with a mean of 22.8 cycles, whereas normal samples were slightly skewed toward larger CT values, and thus had a higher mean (CT = 23.6). Measurements of the normalized miR-205 RNA expression levels in tumors versus normal tissue from the same patients by real-time PCR revealed a significant difference and indicated that tumors averaged ∼3.1-fold less miR-205 than normal tissue (ΔΔCT = +1.63, P value <0.001). In contrast, miR-21 expression levels were higher in tumors versus normals by ∼1.5-fold on average (ΔΔCT = −0.58, P = 0.002) (Figure 2). Furthermore, looking at the distribution in miRNA expression across patients, two–fold lower miR-205 levels were detected in 25% of tumors, whereas two-fold higher miR-205 expression was seen in only 4.5% of cases. Let-7d showed a similar distribution, with 26% and 2% of cases showing lower and higher let-7d expression in tumor versus normals, respectively. In contrast, equal proportions (15%) of tumors showed two–fold lower or higher miR-21 expression relative to their matched adjacent normal tissue. Interestingly, miR-205, miR-21, and let-7d demonstrated minor differences in expression levels with respect to tumor site within the head and neck, although these were within the limits of what may be considered biological variability (Table 3).
Table 2.
Demographic and Clinical Characteristics of HNSCC Patients
| Characteristic | N | % |
|---|---|---|
| Sex | ||
| Male | 71 | 68% |
| Female | 33 | 32% |
| Age | ||
| <60 | 41 | 40% |
| >60 | 63 | 61% |
| Ethnicity | ||
| Hispanic | 26 | 25% |
| Non-Hispanic | 77 | 74% |
| Race | ||
| White | 65 | 62% |
| African-American/Black | 30 | 29% |
| Other | 2 | 2% |
| Anatomic site | ||
| Oral cavity | 31 | 30% |
| Oropharynx | 32 | 31% |
| Hypopharynx | 9 | 9% |
| Larynx | 32 | 31% |
| Tumor Stage* | ||
| I/II | 24 | 23% |
| III/IV | 80 | 77% |
| T status | ||
| I/II | 46 | 44% |
| III/IV | 56 | 54% |
| N status | ||
| I/II | 40 | 40% |
| III/IV | 56 | 54% |
| Treatment | ||
| Primary chemo/RT | 45 | 43% |
| Primary surgery | 33 | 32% |
| Surgery -> chemo | 22 | 21% |
| Smoking history | ||
| Current | 46 | 44% |
| Former | 39 | 38% |
| Never | 17 | 16% |
| Missing | 2 | 2% |
| HPV status | ||
| HPV16− | 59 | 57% |
| HPV16+ | 37 | 36% |
| HPV status missing | 8 | 8% |
| Total | 104 | 100% |
Some cases pending histological confirmation. Totals may not add up to 104 because of missing data.
Figure 2.
Distribution of tumor versus normal differences in normalized microRNA levels for miR-205, miR-21, and let-7d. Matched tumor and normal fold differences in microRNA TaqMan ΔΔCT levels normalized to the RNU48 control gene shown.
Table 3.
Difference in Tumor MicroRNA Levels by Anatomic Site
| Anatomic site | miR-205 (n = 104) | Effect (SE)* | miR-21 (n = 104) | Effect (SE)* | let-7d (n = 104) | Effect (SE)* | miR-133 (n = 41) | Effect (SE)* | miR-1 (n = 41) | Effect (SE)* |
|---|---|---|---|---|---|---|---|---|---|---|
| Oropharynx | −0.86 | 1.0 (ref) | −2.11 | 1.0 (ref) | 3.72 | 1.0 (ref) | 7.56 | 1.0 (ref) | 6.70 | 1.0 (ref) |
| Larynx/hypopharynx | −0.53 | 0.33 (0.54) | −2.27 | −0.15 (0.26) | 3.73 | 0.003 (0.23) | 6.64 | −0.92 (3.97) | 6.33 | −0.37 (3.78) |
| Lip/oral cavity | −0.08 | 0.78 (0.58) | −2.74 | −0.62 (0.28) | 3.80 | 0.08 (0.24) | 5.00 | −2.57 (1.38) | 4.67 | −2.04 (1.31) |
Effect estimates and standard errors (SEs) for relative differences in microRNA expression are shown by β coefficient based on a linear regression model showing the difference in normalized ΔCT tumor levels using oropharynx as the reference, adjusting for tumor site.
MicroRNA Expression and Clinical Outcome in HNSCC
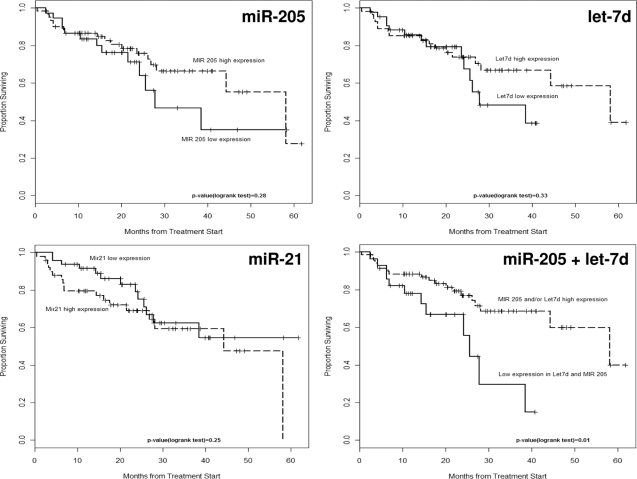
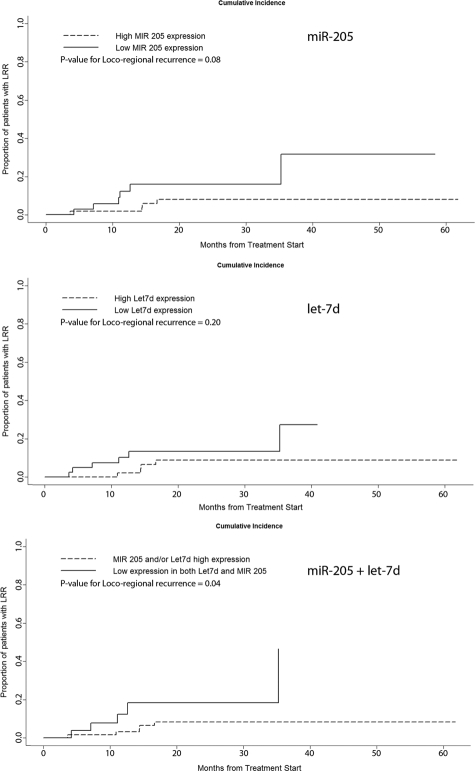
Using the normalized real-time PCR results from 104 primary tumor samples, we also assessed the prognostic association with absolute levels of expression of each of the miRNA genes tested using prospective follow-up data collected on study patients. We observed a significantly shorter time to death (Figure 3) and loco-regional recurrence (Figure 4) in HNSCC patients showing combined lower absolute levels (higher ΔCT) for miR-205 and let-7d, indicative of increased risk with reduced expression in tumors. The prognostic impact of low miR-205 and let-7d levels in tumors also appeared to be independent of anatomical site, tumor size (T-stage), treatment (chemo-radiotherapy), and HPV status (Table 4). The final model presented was based on a priori considerations, and takes into account many known or relevant predictors of progression and survival. Adjustment for anatomical site did not alter the parameter estimates substantively for any of the miRNAs, consistent with the univariate comparisons observed. More parsimonious models based on an exhaustive search across the model space did not significantly alter the results for miR-205, or miR-205, and let-7d combined (data not shown). To gain efficiency, we examined all relevant clinical outcomes simultaneously, estimating the overall effect of each variable stratified by event type. We did not observe any statistically significant interactions between event type and covariates, therefore the estimates presented are based on a summarized effect across all event types. We observed a significantly shorter time to cancer progression (loco-regional recurrence, distant metastasis) or death in HNSCC patients showing lower absolute levels (higher ΔCT) for miR-205 and let-7d, indicative of increased risk with reduced expression in tumors (Table 4). As illustrated by the unadjusted Kaplan-Meier and cumulative incidence curves, reduced expression of let-7d and miR-205 combined remained a significant predictor of cancer progression (HR = 4.61, P value <0.0001), independent of anatomical site, T-stage, treatment, or HPV.
Figure 3.
Kaplan-Meier plots for overall survival by miR-205, miR-21, and let-7d expression individually and combined.
Figure 4.
Cumulative-incidence plots for loco-regional recurrence by miR-205 and let-7d expression individually and combined. Cumulative incidence curves for loco-regional recurrence were also estimated, taking into account the competing risks of death and distant metastases. P values for the comparison of cumulative incidence curves were computed based on the approach of Gray.41
Table 4.
Hazard Ratios (HRs) for Each MicroRNA Expression in Tumors with Survival and Loco-Regional Recurrence
| MicroRNA | Univariate HR* | P value | Adjusted HR† | P value |
|---|---|---|---|---|
| Overall survival | ||||
| miR-205 (low versus high) | 1.49 | 0.28 | 2.51 | 0.025 |
| miR-21 (high versus low) | 0.67 | 0.25 | 1.00 | 0.995 |
| miR-let7d (low versus high) | 1.44 | 0.33 | 1.73 | 0.166 |
| Loco-regional recurrence‡ | ||||
| miR-205 (low versus high) | 3.19 | 0.07 | 4.19 | 0.049 |
| miR-21 (high versus low) | 1.46 | 0.55 | 1.94 | 0.281 |
| miR-let7d (low versus high) | 2.42 | 0.17 | 2.34 | 0.132 |
| Survival or disease progression§ | ||||
| miR-205 (low versus high) | NA | 2.93 | 0.008 | |
| miR-21 (high versus low) | NA | 1.81 | 0.658 | |
| miR-let7d (low versus high) | NA | 2.30 | 0.017 |
Categorized into a binary variable using a mean ΔCTcutoff.
Adjusted for t-stage, anatomic site, treatment, and HPV.
Restricting to patients with no evidence of disease after treatment.
Local, regional recurrence, distant metastases, or death. Univariate models not estimated.
Discussion
One of the most promising aspects of molecular classification is the possibility of isolating smaller subsets of genes whose expression (or lack thereof) correlates significantly with clinical parameters, and the ability to construct a disease-specific assay that can be used to classify cancers and predict clinical outcome.25 In one of the early studies to link gene expression profiling to treatment response, Hanna and colleagues16 identified 60 tumor-related genes from a cDNA microarray containing 1187 genes that could successfully predict the radiation response of tumor samples. In oral cancer, Warner and colleagues17 identified 23 differentially expressed genes that correlated with tumor stage (III to IV) and metastasis. Gene expression signatures have also been identified that are associated with recurrence of HNSCC disease.17 This study represents the initial use of miRNAs to classify and predict behavior of HNSCC.
Most of the existing miRNA data for HNSCC comes from work with cell lines in culture. A real-time PCR study of 222 miRNAs in 32 cancer cell lines that included 4 head and neck lines confirmed that miRNA expression patterns in HNSCC cells could be distinguished from the cell-type specificity of the other 28 lines that included lung, pancreas, prostate, breast, colorectal, and blood cell types.9 This result is consistent with the prevailing idea that miRNAs are differentially expressed in tissues and during altered growth.26,27 In addition, miR-205 is a relatively enriched species in head and neck, having higher expression in the HNSCC lines than any of the other cell lines derived from different cancers.24 Comparison of miRNA expression profiles in nine HNSCC cell lines revealed 33 highly expressed and 22 lowly expressed miRNAs.10 Among the highly expressed miRNAs were miR-212 and miR-205. These data comparing cell lines from different tumor origins revealed that miR-205 is abundant in its expression in squamous cells in head and neck tissues (relative to other cell types in other tissues) but they obviously could not address the issue of miR-205 expression in tumors compared with normal tissue or of HNSCC tumors compared among themselves for correlation with clinical data. Our data are novel in that we observed that it is the repression of this cell-type-enriched miRNA in patient tumors that is important as a prognostic predictor independent of anatomical site, treatment, or disease stage at presentation.
MiR-205 therefore, seems to act as a tumor suppressor in HNSCC, and reduced expression of this miRNA should result in increased expression of target oncogenes. To demonstrate this, we attempted to identify potential miR-205 targets by looking for mRNAs whose expression levels that were inversely correlated with miR-205 levels in the same HNSCC tumors. We then used TargetRank to predict which among the highly correlated genes were also theoretical miRNA targets, representing potential direct miR-205 targets.28 One gene that met these two selection criteria was dihydrofolate reductase (DHFR). In 16 (N = 16) HNSCCs in which primary tumors and adjacent normal tissue measurements by both cDNA microarray and TaqMan RT-PCR data were available for comparison, we observed a high correlation (corr = 0.89) between miR-205 (low expression) and DHFR (high expression). TargetScan predicts a score of 0.422 (on a scale of 0 to 1) with four conserved seed matches in the DHFR gene. DHFR plays a critical role in folate metabolism, and represents the target of methotrexate, a drug commonly used in preventing tumor growth. DHFR activity is also associated with the p53 tumor suppressor protein because both are targeted for degradation through the MDM2 ubiquitin ligase pathway. In addition, it was recently shown that a specific MDM2 polymorphism, SNP309, is correlated with early tumor onset in HNSCC and poor prognosis in other tumors.29 These pathways are additionally linked through p14ARF which is frequently lost or mutated in HNSCC. The activity of p14ARF results in increased degradation of DHFR and resistance to folate antagonists in cells with nonfunctional p53.30 Our results suggest that these pathways may intersect and that miR-205 may be involved in the regulation of these events that determine these cancer-related phenotypes. Further dissection of these pathways and direct experimental determination of miR-205 target genes in HNSCC cell lines and confirmation of these targets in specific head and neck clinical samples should help to understand mechanisms involved in these responses.
In our study, we find repression of miRNAs to be more common than overexpression in human tumors compared to normal tissue. This is consistent with other studies that looked at human oral specimens, and with the hypothesis that a subset of miRNA genes may be epigenetically silenced by hypermethylation of CpG island sequences within promoter or exon regions.31 Alternative mechanisms proposed for altered levels of miRNA genes in cancer include alterations in copy number, chromatin modifications, and regulation by transcription factors. One example includes the let-7 family of genes. It has been recently reported that the c-myc oncogene contributes to widespread repression of at least 21 distinct miRNA transcription units including let-7 family members and let-7d specifically.32 The let-7 gene family consists of at least nine genes within eight transcription units. C-myc is frequently overexpressed in HNSCC tumors and it is possible that one important consequence of this for the oncogenic effects of myc in HNSCC is the repression of let-7 miRNA. Low levels of expression of various members of this multigene family in tumors relative to normal tissue have been shown to predict poor prognosis in lung cancer.5,6 Our results add HNSCC as another cancer type in which pathways regulated by let-7 are critical for tumor behavior.
It will also be important to identify and experimentally confirm downstream targets of let-7d because these genes could prove to become new potential therapeutic targets in the future. One experimentally verified target of let-7 miRNAs is the KRAS oncogene.33 HMGA2 is another let-7 target gene and this gene is also regulated in part in hypoxic conditions by miR-98 in the HNSCC cell lines SCC-4 and SCC-9.34 HMGA2 activity is associated with enhanced sensitivity to the topoisomerase II doxorubicin and high levels in lung tumors is correlated with poor prognosis and metastasis.35 We also looked for mRNAs whose expression levels were inversely correlated with let-7d levels in the same HNSCC tumors in an attempt to identify additional target genes of possible interest. Many genes show highly significant inverse correlations with let-7d expression, although many of the identified target genes represented unidentified ESTs, and no known genes contained seed regions that matched let-7d binding sites. Further studies are needed to identify target let-7d target genes in HNSCC. Again, experimental determination of let-7 targets in HNSCC cells will be necessary to make clearer the mechanisms responsible for the critical role of let-7d in HNSCC tumor phenotypes.
Expression levels of miR-21 RNA were not correlated with any of the clinical parameters that we measured, however, this miRNA has experimentally verified target genes that have been shown to be altered in tumors, one of which is PDCD4, a tumor suppressor protein involved in invasion and metastasis.36,37 We have derived extensive cDNA microarray data comparing primary HNSCC tumors to their normal counterparts in the same patient (data not shown). Examination of these tumor/normal datasets revealed that PDCD4 mRNA is down-regulated twofold or more in 15 of 26 cases examined. Thus, we imply that this miR-21 target gene identified during manipulation of tissue culture lines is also relevant to actual patient samples. Additional miR-21 targets identified along with PDCD4 include the actin protein ACTA2, and the anti-proliferative protein encoded by the BTG2 gene.37 Examination of our cDNA microarray data again showed that ACTA2 expression was twofold lower or less in 7 of 26 HNSCC cases, whereas BTG-2 was lower in 9 of 26. These correlations between miRNA expression levels and potential mRNA targets in clinical samples would seem to support the relevance of at least some targets identified in tissue culture models.
Another experimentally confirmed miR-21 target gene is the PTEN tumor suppressor.38 PTEN tumor suppressor is frequently altered in HNSCC39,40 and loss of heterozygosity caused by homozygous deletion or allelic loss and point mutation have been documented. However, in HNSCC neither of these mechanisms could account for a majority of the tumors with reduced PTEN protein expression.40 PTEN, however, is a confirmed target of miR-21, the only miRNA overexpressed in all solid tumors examined as well as some additional cancers.3 PTEN encodes a phosphatase that can inhibit growth and/or survival pathways including AKT/PI3K, and its function is altered in advanced tumors of various types, including breast, lung, gastric, and prostate. As in HNSCC, the frequency of loss of heterozygosity at the PTEN locus in these tumors is relatively small (less than 10%). Using tissue microarrays to measure PTEN protein levels in (n = 46) primary HNSCC tumors used for miRNA measurements, we observed that miR-21 was up-regulated in tumors where PTEN expression was absent, albeit not significantly (P = 0.10). Looking at differences in miR-21 RNA levels in tumor versus normal tissue by case, the majority of HNSCC (79%) that showed no evidence of PTEN expression had higher miR-21 tumor levels, whereas in cases where PTEN was present, only 46% of HNSCC showed an absolute increase in miR-21 levels (P = 0.03), and no cases showed a greater than twofold difference in tumor versus adjacent normal. We did not observe these same correlations with PTEN mRNA as measured by microarray analysis. These observations would suggest that translational inhibition of PTEN mRNA via miR-21 is an alternative mechanism to loss of heterozygosity and mutation for the silencing of the PTEN tumor suppressor. This miRNA may therefore play a role in HNSCC risk or tumor development, although we do not find any association with cancer progression in the head and neck.
In conclusion, microRNAs miR-205 and let-7 expression levels can be used as prognostic markers of head and neck cancer survival and recurrence. These observations may have diagnostic and therapeutic implications for the management and treatment of HNSCC patients. Our results also expand the known pathways and genes that are important for the behavior of this disease and represent an important source of new avenues of research in head and neck cancer.
Acknowledgments
We thank Catherine Sarta for management of the patients and specimen collection, Xin Zheng and Mark Yin for database management, and Anne Dunne for assistance with the HPV analyses protocols.
Footnotes
Address reprint requests to Dr. Geoffrey Childs, Albert Einstein College of Medicine, Department of Pathology, 1300 Morris Park Ave., Bronx, NY 10461. E-mail: childs@aecom.yu.edu.
Supported by the National Cancer Institute (grants CA115243 to N.F.S., CA103547 to M.B.P., and CA104402 to T.J.B.).
References
- Ambros V. MicroRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- American Cancer Society Atlanta: American Cancer Society,; Cancer Facts and Figures 2007. 2007 [Google Scholar]
- Kyzas PA, Stefanou D, Batistatou A, Agnantis NJ. Prognostic significance of VEGF immunohistochemical expression and tumor angiogenesis in head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2005;131:624–630. doi: 10.1007/s00432-005-0003-6. [DOI] [PubMed] [Google Scholar]
- Lothaire P, de Azambuja E, Dequanter D, Lalami Y, Sotiriou C, Andry G, Castro G, Jr, Awada A. Molecular markers of head and neck squamous cell carcinoma: promising signs in need of prospective evaluation. Head Neck. 2006;28:256–269. doi: 10.1002/hed.20326. [DOI] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- Belbin TJ, Singh B, Barber I, Socci N, Wenig B, Smith R, Prystowsky MB, Childs G. Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res. 2002;62:1184–1190. [PubMed] [Google Scholar]
- Hanna E, Shrieve DC, Ratanatharathorn V, Xia X, Breau R, Suen J, Li S. A novel alternative approach for prediction of radiation response of squamous cell carcinoma of head and neck. Cancer Res. 2001;61:2376–2380. [PubMed] [Google Scholar]
- Warner GC, Reis PP, Jurisica I, Sultan M, Arora S, Macmillan C, Makitie AA, Grenman R, Reid N, Sukhai M, Freeman J, Gullane P, Irish J, Kamel-Reid S. Molecular classification of oral cancer by cDNA microarrays identifies overexpressed genes correlated with nodal metastasis. Int J Cancer. 2004;110:857–868. doi: 10.1002/ijc.20197. [DOI] [PubMed] [Google Scholar]
- Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng GC, Oh MK, Rohlin L, Liao JC, Wong WH. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 2001;29:2549–2557. doi: 10.1093/nar/29.12.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Bauer HM, Ting Y, Greer CE, Chambers JC, Tashiro CJ, Chimera J, Reingold A, Manos MM. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–477. [PubMed] [Google Scholar]
- Hildesheim A, Schiffman MH, Gravitt PE, Glass AG, Greer CE, Zhang T, Scott DR, Rush BB, Lawler P, Sherman ME, Kurman RJ, Manos MM. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- Wei LJ, Lin DY, Weissfeld L. Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. J Am Stat Assoc. 1989;84:1065–1073. [Google Scholar]
- Leethanakul C, Knezevic V, Patel V, Amornphimoltham P, Gillespie J, Shillitoe EJ, Emko P, Park MH, Emmert-Buck MR, Strausberg RL, Krizman DB, Gutkind JS. Gene discovery in oral squamous cell carcinoma through the Head and Neck Cancer Genome Anatomy Project: confirmation by microarray analysis. Oral Oncol. 2003;39:248–258. doi: 10.1016/s1368-8375(02)00107-0. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- Nakashima M, Kondo S, Shimizu Y, Wakisaka N, Murono S, Furukawa M, Yoshizaki T. Impact of MDM2 single nucleotide polymorphism on tumor onset in head and neck squamous cell carcinoma. Acta Otolaryngol. 2008;128:808–813. doi: 10.1080/00016480701724904. [DOI] [PubMed] [Google Scholar]
- Magro PG, Russo AJ, Li WW, Banerjee D, Bertino JR. p14ARF expression increases dihydrofolate reductase degradation and paradoxically results in resistance to folate antagonists in cells with nonfunctional p53. Cancer Res. 2004;64:4338–4345. doi: 10.1158/0008-5472.CAN-03-1045. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, Karjalainen A, Knuutila S, Anttila S. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Lorenz G, Kleist B. Detection of new PTEN/MMAC1 mutations in head and neck squamous cell carcinomas with loss of chromosome 10. Cancer Genet Cytogenet. 2002;132:20–24. doi: 10.1016/s0165-4608(01)00509-x. [DOI] [PubMed] [Google Scholar]
- Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, Gonzalez MV. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. [Google Scholar]