Abstract
Recently, we reported that human breast cancer-associated fibroblasts show functional inactivation of the retinoblastoma (RB) tumor suppressor and down-regulation of caveolin-1 (Cav-1) protein expression. However, it remains unknown whether loss of Cav-1 is sufficient to confer functional RB inactivation in mammary fibroblasts. To establish a direct cause-and-effect relationship, mammary stromal fibroblasts (MSFs) were prepared from Cav-1−/− null mice and subjected to phenotypic analysis. Here, we provide evidence that Cav-1−/− MSFs share many characteristics with human cancer-associated fibroblasts. The Cav-1−/− MSF transcriptome significantly overlaps with human cancer-associated fibroblasts; both show a nearly identical profile of RB/E2F-regulated genes that are up-regulated, which is consistent with RB inactivation. This Cav-1−/− MSF gene signature is predictive of poor clinical outcome in breast cancer patients treated with tamoxifen. Consistent with these findings, Cav-1−/− MSFs show RB hyperphosphorylation and the up-regulation of estrogen receptor co-activator genes. We also evaluated the paracrine effects of “conditioned media” prepared from Cav-1−/− MSFs on wild-type mammary epithelia. Our results indicate that Cav-1−/− MSF “conditioned media” is sufficient to induce an epithelial-mesenchymal transition, indicative of an invasive phenotype. Proteomic analysis of this “conditioned media” reveals increased levels of proliferative/angiogenic growth factors. Consistent with these findings, Cav-1−/− MSFs are able to undergo endothelial-like transdifferentiation. Thus, these results have important implications for understanding the role of cancer-associated fibroblasts and RB inactivation in promoting tumor angiogenesis.
The tumor microenvironment plays a previously unrecognized role in human breast cancer onset and progression. Although the mammary microenvironment is composed of a host of cell types, tissue fibroblasts are an integral part of the mammary stroma and are thought to become “activated” or hyperproliferative during tumor formation (known as the desmoplastic reaction). These cancer-associated fibroblasts (CAFs) take on the characteristics of myofibroblasts often observed during the process of wound healing.1 Little is known about the molecular events that govern the conversion of mammary stromal fibroblasts to tumor-associated fibroblasts. During wound healing, this process is known to be driven by activation of the TGF-β signaling cascade.2,3 In addition, CAFs have been shown to secrete important growth factors, such as transforming growth factor (TGF)-β, platelet-derived growth factor, and hepatocyte growth factor (HGF), suggesting a role in tumor cell invasion.4,5
Recently, we isolated CAFs from human breast cancer lesions and studied their properties, as compared with normal mammary fibroblasts isolated from the same patient.6 Interestingly, we demonstrated that 8 out of 11 CAFs show dramatic down-regulation of caveolin-1 (Cav-1) protein expression; Cav-1 is a well-established marker that is normally decreased during the oncogenic transformation of fibroblasts.6 We also performed gene expression profiling studies (DNA microarray) and established a new CAF gene expression signature. Interestingly, the expression signature associated with CAFs includes a large number of genes that are regulated via the RB-pathway.6 This CAF-associated RB/E2F gene signature is also predictive of poor clinical outcome in breast cancer patients that were treated with tamoxifen monotherapy, indicating that CAFs may be useful for predicting the response to hormonal therapy.
In direct support of these findings, implantation of mammary tumor tissue in the mammary fat pads of Cav-1−/− null mice results in up to a ∼twofold increase in tumor growth, indicating that the mammary stroma of Cav-1−/− null mice has tumor-promoting properties.7 However, it remains unknown whether loss of Cav-1 is sufficient to confer RB functional inactivation in mammary stromal fibroblasts (MSFs).
Here, to establish a direct cause-effect relationship, we have now used a genetic approach using Cav-1−/− null mice. Importantly, we show that the Cav-1−/− MSF transcriptome significantly overlaps with that of human CAFs; both show a nearly identical profile of RB/E2F-regulated genes that are up-regulated, consistent with RB functional inactivation. Thus, Cav-1−/− MSFs may represent the first molecular genetic model for dissecting the activated signaling networks that govern the phenotypic behavior of human breast CAFs.
Experimental Procedures
Materials
Antibodies and their sources were as follow: phospho-Rb (pS807/811) from Cell Signaling; Rb (M-153), Cav-1 (N-20), and HGF β from Santa Cruz Biotechnology; α-smooth muscle actin and β-actin from Sigma; collagen type I from Novus Biologicals; and CAPER from BioVision. Other reagents were as follows: 4,6-diamidino-2-phenylindole (DAPI), propidium iodide, Prolong Gold Antifade mounting reagent, Slow-Fade Antifade reagent (from Molecular Probes); phalloidin-fluorescein isothiocyanate, hydrocortisone, cholera toxin, insulin, and gentamicin (from Sigma); collagenase type I (from Gibco); reduced growth factor Matrigel (from Trevigen); and Lab-TekII 8-well chamber slides (from Nalgene Nunc).
Isolation and Primary Culture of Mammary Stromal Fibroblasts
Primary mammary fibroblasts were isolated from the mammary glands of 8-week old virgin mice. Briefly, the fourth and fifth mammary glands from wild-type and Cav-1−/− mice were removed aseptically, minced with surgical blades, incubated in a shaker (for 2 to 3 hours at 37°C) in 30 to 35 ml of digestion media (DMEM [Dulbecco’s modified Eagle’s medium]/F12, 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, pen/strep) containing 2 mg/ml collagenase type I and 50 μg/ml gentamicin. Then, the cell suspensions were spun 10 minutes at 1000 rpm to eliminate floating fat cells. Cell pellets were washed twice in 10 ml of MSF growth media (DMEM, 10% fetal bovine serum, pen/strep). Then, cell pellets were disaggregated by pipetting up and down 10 to 15 times with a sterile 1-ml blue pipette tip. Mammary fibroblasts were then cultured in growth media and passaged three times. At this point, greater than 95% of the cells were mammary fibroblasts (Figure 1). At least three independent isolates of primary MSFs for each genotype were used for our experiments. Each of these cultures was derived from separate mice. These MSF cultures appeared very homogeneous, as shown in Figure 1, and failed to express adipocyte (adiponectin), epithelial (keratin 8/18), and endothelial (CD31/Pecam1) cell markers. Under our culture conditions, which dramatically favor fibroblasts, other possible contaminating cell types (such as skeletal muscle and macrophages) fail to proliferate. Finally, >90% of the cells in these MSF cultures highly express fibroblast-specific markers, such as vimentin and collagen type I.
Figure 1.
Primary cultures of wild-type and Cav-1−/− mammary stromal fibroblasts. Phase contrast images at the same magnification are shown. MSFs spread out and are enlongated, as expected. Mammary epithelia are shown for comparison. Note that the mammary epithelial cells form a densely packed monolayer, with a characteristic “cobble-stone” appearance. WT, wild-type; KO, Cav-1−/−.
Gene Expression Profiling
These studies were performed essentially as we have previously described for other cell types.8 RNA was prepared from three wild-type and three Cav-1−/− MSF isolates; each of these biological replicates were derived from separate mice. Total RNA (5 μg) was reverse-transcribed using Superscript III First-Strand Synthesis System (Invitrogen) using a HPLC-purified T7-dT24 primer (Sigma Genosys), which contains the T7 polymerase promoter sequence. The single-stranded cDNA was converted to double stranded cDNA using DNA polymerase I (Promega) and purified by cDNA spin column purification using GeneChip Sample Cleanup Module (Affymetrix). The double-stranded cDNA was used as a template to generate biotinylated cRNA using Bioarray HighYield RNA Transcription Labeling Kit (Enzo) and the labeled cRNA purified by GeneChip Sample Cleanup Module (Affymetrix). Fifteen micrograms of cRNA was fractionated to produce fragments of between 35 and 200 bp using 5× fragmentation buffer provided in the Cleanup Module. The sample was hybridized to mouse 430 2.0 microarray (Affymetrix) representing over 39,000 transcripts. The hybridization and washing steps were performed in accordance with Affymetrix protocols for eukaryotic arrays. The arrays were scanned at 570 nm with a confocal scanner from Affymetrix. Analysis of the arrays was performed using the R statistics package and the limma library of the Bioconductor software package.8,9 Arrays were normalized using robust multi-array analysis, and P value of 0.05 was applied as criteria for statistically differentially expressed genes.
Gene Array Data Analysis
Gene ontology analyses were performed using the DAVID 2007 bioinformatics resource. Microarray data (series GSE1378 and GSE1379) from X.J. Ma et al10 were obtained from the National Center for Biotechnology Information Gene Expression Omnibusweb site (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GPL1223) and manipulated using GeneSpring GX software (version 7.2; Agilent Technologies). For each series, the raw data were obtained from GEO as log2 of normalized Cy5/Cy3 ratio, where tumor sample RNA and human universal reference RNA were labeled with Cy5 and Cy3, respectively. The raw data were transformed from log2 to linear values followed by per-gene median normalization in GeneSpring. The expression levels of Cav-1−/− MSF-associated genes were clustered based on standard correlation as the similarity measurement. Subsequently, a condition tree based on distance correlation was created to order the tumor specimens. The patients exhibiting the highest expression level of the Cav-1−/− MSF gene signature were used to define the impact of the Cav-1−/− MSF signature on disease outcome. For Kaplan-Meier analysis, statistical calculations were performed using GraphPad Prism 4.0 software. ES (embryonic stem) cell-specific gene sets11 and estrogen-induced gene sets12,13 were as previously described.
Statistical Analysis of Overlapping Gene Sets
For determining the statistical significance of gene set overlap, we used P values.14 We calculated the statistical significance for the transcriptome intersection by using hypergeometric probabilities for any two groups of genes. By considering the commonality between human and mouse platforms based on identical transcript identifiers, we generated a P value for the interesting sets. All comparisons were statistically significant at P < 0.009. In this case, the P value is the probability of finding the number of overlapping genes in the two sets by pure chance. This is determined by the equation:
 |
where c(n,j) is the number of combinations that one choose j objects from n objects, t is the total number of observable genes, i is the number of genes in the overlap, and m and n are the numbers of differentially expressed genes in the two sets. For the present comparisons (CAFs versus MSFs), t the number of observable genes was taken as number of common genes based on the Gene Symbol for the two chips MU74Av2 and HU133_2.0.
Other Statistical Considerations
The biological replicates demonstrate robust coherency within the phenotypic sample sets, inconsistent variation would result in an increased SD and a high P value. While we accept that based on a P value <0.05, the data set will have an inherent false positive element, we have compensated by using at least a twofold cut off. When the P value of Cav-1 MSFs 832 transcript set is examined, we observe more than 3/4 of the differentially regulated genes have a P value less than 0.005. A randomized Pearson correlation within biological replicates gave an r = 1, while between phenotypes gave an r = 0.6.
Target Validation by Real-Time PCR
We used SYBR green real-time quantitative reverse transcription (qRT)-PCR. To independently quantify gene expression, 2000 ng of total RNA was reverse-transcribed using random examers and Superscript-II reverse transcriptase (Invitrogen), according to the manufacturer’s protocol. All primers were designed using Primer Express Software (Applied Biosystems, Foster City, CA) and validated for specificity. Real-time PCR was performed using the myIQ real-time PCR (Biorad) according to the manufacturer’s instructions. Reactions were conducted in triplicate and performed in a 25-μl volume with 50 nmol/L of forward and reverse primer. The reaction cycles were: an initial incubation at 50°C for 2 minutes, denaturation at 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The SYBR green signal was continuously monitored. The amplified PCR products were analyzed in the linear range of amplification with standards. To confirm the amplification specificity, the PCR products were subjected to melting temperature dissociation curve analysis. In parallel, no amplification and no template controls were run to rule out the presence of fluorescence contaminants in the sample or in the thermal cycler heat block. Relative quantification of samples was assessed by arbitrarily setting the control cDNA value at 100, and changes in transcript levels of a sample expressed as a multiple thereof (relative expression). Differences in the number of mRNA copies in each PCR reaction were normalized using mouse 18S rRNA transcript levels.
Target Validation by Western Blot or Immunofluorescence Analysis
As mRNA levels do not necessarily reflect protein expression levels, we also performed target validation by Western blot or immunofluorescence. Using this approach, we evaluated the protein expression status of a number of important genes including RB, phospho-RB, collagen I, CAPER, and HGF. Interestingly, total RB protein levels remained unchanged and HGF protein levels increased by ∼tenfold in Cav-1−/− MSFs. However, this is in contrast to our DNA microarray results, which showed that Rb1 transcripts decreased 2.0-fold and HGF transcripts decreased 2.1-fold. Thus, in these two cases, other gene or protein regulatory mechanisms may be operating. In contrast, analysis of phospho-RB, collagen I, and CAPER protein expression levels are concordant with the increased transcriptional expression of RB/E2F target genes (96 transcripts), as well as collagen I, and CAPER transcripts, in Cav-1−/− MSFs. These results are also supported by other functional assays, such as 5-bromo-2′-deoxyuridine (BrdU) incorporation and retraction/contraction analysis.
Assay for BrdU Incorporation
Cell proliferation was determined using a standard BrdU assay (Roche). The incorporation of a pyrimidine analogue (BrdU) was measured in wild-type and Cav-1−/− MSFs, as suggested by the manufacturer. Briefly, fibroblasts were trypsinized and plated in a 96-well plate (Corning) at a density of 2000 cells/well. After 72 hours, the cells were given a BrdU pulse of 2 hours at 37°C.
Retraction/Contraction Assay
Passage 3 primary mammary fibroblasts were seeded at 50% confluency in 35-mm dishes and allowed to reach complete confluency in regular MSF growth media. Then, the cells were treated with ascorbic acid (40 μg/ml). After 4 days, Cav-1−/− MSFs showed a retraction phenotype, whereas wild-type cells remained completely attached to the plate.
Western Blot Analysis
Mammary fibroblast lysates were prepared by scraping the cells into Lysis Buffer (10 mmol/L TrisHCl pH 7.5, 50 mmol/L NaCl, 1% TritonX-100, 60 mmol/L octyl glucoside with phosphatase inhibitor cocktails [Sigma] and protease inhibitor tablet). After rotation at 4°C for 40 minutes, cell lysates were spun for 10 minutes to remove insoluble material. Protein concentrations were assessed using the BCA assay kit. Cellular proteins were resolved by SDS-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes (Schleicher and Schuell, 0.2 μm). Blots were blocked for 1 hour in Tris-buffered saline(TBS)-Tween (10 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl, 0.2% Tween 20) containing 1% bovine serum albumin (BSA) and 4% nonfat dry milk (Carnation). Then, membranes were incubated for 2 hours with primary antibodies in a 1%BSA/TBS-Tween solution. Membranes were then washed with TBS-Tween, and incubated for 40 minutes with the appropriate horseradish peroxidase-conjugated secondary antibodies (Pierce, diluted 5000-fold in 1%BSA/TBS-Tween). Signal was detected with an enhanced chemiluminescence detection kit (Pierce). Nonfat dry milk was omitted from the blocking solution when we used phospho-specific antibodies.
Immunofluorescence (IF)
Cells were fixed for 30 minutes at room temperature in 2% paraformaldehyde diluted in PBS, after which they were permeabilized for 10 minutes at room temperature with IF buffer (PBS + 0.2% BSA + 0.1% TritonX-100). Then, cells were incubated with NH4Cl in PBS to quench free aldheyde groups. Primary antibodies were incubated in IF buffer overnight at room temperature. After washing with IF buffer (3×, 10 minutes each), cells were incubated for 30 minutes at room temperature with fluorocrome-conjugated secondary antibodies (Jackson Laboratories) diluted in IF buffer. Finally, slides were washed at room temperature with IF buffer (3×, 10 minutes each), and mounted with Slow-Fade Anti-fade Reagent (Molecular Probes). For collagen I staining, passage 3 primary mammary fibroblasts were allowed to reach confluency, and were treated with ascorbic acid (40 μg/ml) for 24 hours. Ascorbic acid treatment is required for collagen secretion. Then, cells were fixed and were immunostained with rabbit polyclonal antibodies against collagen I (Novus Biologicals, CO). Alternatively, a methanol-fixation protocol was used, as previously described.6 Methanol fixation was preferred for nuclear antigens, such phospho-RB and CAPER.
Preparation of Conditioned Media and 3D Mammary Culture Analysis
To prepare conditioned media, primary mammary fibroblasts were cultured until confluence. Confluent cultures were rinsed twice with serum-free medium and incubated in low-serum medium (DMEM, 10% Nu serum, glutamine, pen-strep) for 48 hours. Then, “MSF-conditioned media” was collected and incubated with 3D cultures of primary mammary epithelial cells. Primary mammary epithelial cells were purified, as previously described.15,16 Briefly, after surgical and chemical isolation, mammary gland organoids were resuspended in assay media (DMEM/F12, 2% horse serum, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, pen/strep). To wash away single cells, “organoid” pellets were subjected to repeated differential centrifugation (spun at 1000 rpm for 45 seconds, repeated for 10 cycles of pelleting and re-suspension). After the last wash, organoids were resuspended in 2 ml of growth media (DMEM/F12, 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, pen/strep), and disrupted by pipetting up and down 20 to 25 times with a sterile 1-ml blue pipette tip. Organoids were plated and allowed to attach and spread as a monolayer. Four to five days after purification, organoids attached to plastic dishes and grew as a mammary epithelial cell monolayer (Figure 1). Cell monolayers were then trypsinized. To obtain a single cell suspension, cells were passed 20 to 25× through a 1-ml blue tip. This single cell suspension was then overlaid onto Matrigel, essentially as we previously described.15,16 Briefly, wild-type mammary epithelial cells were diluted in wild-type or Cav-1−/− MSF-conditioned media supplemented with 2% Matrigel. Then, 5000 cells were overlaid on top of Matrigel (ie, 8-well chamber slides, which were pre-coated with 40 μl of Matrigel). Chambers were placed in a standard cell culture incubator at 37°C. All experiments were performed with primary mammary epithelial cells that were at passage 1.
Proteome Analysis of Secreted Proteins
Levels of up to 40 different growth factors, cytokines, and chemokines in tissue culture supernatants were measured using SearchLight Proteome Arrays (Pierce Biotechnology, Woburn, MA). The SearchLight Proteome Array is a quantitative multiplexed sandwich enzyme-linked immunosorbent assay (ELISA) containing different capture antibodies spotted on the bottom of a 96-well polystyrene microtiter plate. Each antibody captures specific protein present in the standards and samples added to the plate. The bound proteins are then detected with a biotinylated detection antibody, followed by the addition of streptavidin-horseradish peroxidase (HRP) and lastly, SuperSignal ELISA Femto chemiluminescent substrate (patent #6432,662). The luminescent signal produced from the HRP-catalyzed oxidation of the substrate is measured by imaging the plate using the SearchLight Imaging System, a cooled charge-coupled device camera. The data are then analyzed using ArrayVision customized software. The amount of luminescent signal produced is proportional to the amount of each protein present in the original standard or sample. Concentrations are extrapolated from a standard curve.
Identification of Endothelial and Pro-Angiogenic Factors by qRT-PCR
RNA was extracted from confluent mammary fibroblasts, grown in 10% fetal bovine serum and DMEM medium supplemented with 40 μg/ml ascorbic acid. RNA was extracted using RNAeasy columns (Qiagen) according to manufacturer’s instructions including Dnase treatment to eliminate genomic DNA contamination, and retro-transcribed using RT2 first strand kit (Superarray) following the manufacturer’s instructions. Mouse Angiogenesis and Cancer Pathway Finder RT2 Profiler PCR Arrays and RT2 Real-Timer SyBR Green/ROX PCR Mix were purchased from SuperArray Bioscience Corporation (Frederick, MD). For data analysis, the comparative Ct method (ΔΔCT) was used using the average of four housekeeping genes to normalize the values, according to Superarray’s pre-developed software analysis; for each gene, fold-changes were calculated as the difference in gene expression between the average of the expressions of three different assays run on two separate preparations of wild-type and Cav-1−/− MSFs.
CD31 (Pecam1) Immunostaining
Mammary glands (inguinal) from 6-month-old virgin female wild-type and Cav-1−/− mice were harvested and used to prepare frozen tissue sections. Frozen sections were subjected to fixation in acetone for 5 minutes at −20°C or 4% paraformaldehyde in PBS for 10 minutes at 4°C. For immunohistochemical detection, a 3-step biotin-streptavidin-HRP method was used after blocking with 10% rabbit serum. Tissue sections were then incubated overnight at 4°C with rat anti-mouse CD31 (BD Biosciences, San Jose, CA) at a dilution of 1:200 (0.08 μg/ml) followed by biotinylated rabbit anti-rat IgG (1:200; Vector Labs, Burlingame, CA) and streptavidin-HRP (Dako, Carpinteria, CA). Immunoreactivity was revealed with 3,3′ diaminobenzidine. For immunofluorescence detection, CD31 antibody was used at a 1:50 dilution (0.3 μg/ml) after blocking with 10% goat serum. Sections were incubated for 1.5 hours at room temperature and after washing, immunoreactivity was detected using goat anti-rat rhodamine red–X F(ab′)2 (Jackson ImmunoResearch, West Grove, PA) at a 1:200 dilution (6.7 μg/ml). Images were collected with a Zeiss LSM510 meta confocal system using a 543 nm HeNe excitation laser and a detector band pass filter range of 560 to 615 nm.
Results
Transcriptional Gene Profiling of Cav-1−/− MSFs Reveals Striking Similarities with Human CAFs and RB Tumor Suppressor Functional Inactivation
Human breast CAFs show functional inactivation of the RB tumor suppressor and down-regulation of Cav-1 protein expression.6 However, it remains unknown whether loss of Cav-1 is sufficient to confer RB functional inactivation in mammary fibroblasts. Here, to establish a direct cause-effect relationship, we have now used a genetic approach using Cav-1−/− null mice. MSFs were prepared from wild-type and Cav-1−/− null mice (Figure 1) and subjected to genome-wide transcriptional profiling.
The expression of 832 known genes and transcripts was changed in Cav-1−/− MSFs, as compared with Cav-1+/+ MSFs; 380 transcripts were up-regulated and 452 transcripts were down-regulated (Figure 2). All of these genes and transcripts changed by >twofold and achieved statistical significance (P < 0.05). Gene ontology analysis revealed that the 380 up-regulated transcripts exhibit a strong enrichment for genes involved in cell cycle control (supplemental Table S1 and supplemental Figure S1, see http://ajp.amjpathol.org). Interestingly, the Cav-1−/− MSF transcriptome significantly overlaps with that of human breast CAFs. We previously reported a CAF gene signature consisting of 118 up-regulated known gene transcripts.6 Table 1 shows a list of 55 genes that were commonly up-regulated in both human breast CAFs and Cav-1−/− MSFs. Many of these genes are RB/E2F-regulated genes.17
Figure 2.
CAF versus Cav-1−/− MSF gene signatures. Venn diagrams summarizing the similarities and differences between gene transcript changes in human breast CAFs and Cav-1−/− MSFs.
Table 1.
Common Gene Expression Changes in Human Breast CAFs and Cav-1−/− MSFs
| A. Genes up-regulated in both human breast CAFs and Cav-1−/− MSFs (55 genes)
| ||
|---|---|---|
| Symbol | Gene name | Fold-upregulation in Cav-1 KO |
| Anln | Anillin, actin binding protein (scraps homolog, Drosophila) | 2.5 |
| Aurka | Aurora kinase A | 3.3 |
| Birc5 | Baculoviral IAP repeat-containing 5 (survivin) | 3.8 |
| Blm | Bloom syndrome | 2.0 |
| Brca1 | Breast cancer 1, early onset | 2.3 |
| Bub1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | 3.6 |
| Bub1b | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 2.5 |
| Casc5 | Cancer susceptibility candidate 5 | 2.5 |
| Ccnb2 | Cyclin B2 | 2.3 |
| Ccnf | Cyclin F | 2.1 |
| Cdc20 | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 2.4 |
| Cdc45l | CDC45 cell division cycle 45-like (S. cerevisiae) | 2.0 |
| Cdca1 | Cell division cycle associated 1 | 2.4 |
| Cdca3 | Cell division cycle associated 3 | 2.7 |
| Cdca5 | Cell division cycle associated 5 | 2.1 |
| Cdca8 | Cell division cycle associated 8 | 3.0 |
| Cenpa | Centromere protein A | 3.9 |
| Cenpf | Centromere protein F, 350/400ka (mitosin) | 2.8 |
| Cenpk | Centromere protein K | 2.0 |
| Cep55 | Centrosomal protein 55kDa | 3.2 |
| Ckap2l | Cytoskeleton associated protein 2-like | 2.5 |
| Dlg7 | Discs, large homolog 7 (Drosophila) | 2.6 |
| E2f7 | E2F transcription factor 7 | 2.7 |
| Fabp5 | Fatty acid binding protein 5 (psoriasis-associated) | 3.9 |
| Foxm1 | Forkhead box M1 | 2.4 |
| Hmmr | Hyaluronan-mediated motility receptor (RHAMM) | 3.3 |
| Kif11 | Kinesin family member 11 | 2.7 |
| Kif20a | Kinesin family member 20A | 2.8 |
| Kif23 | Kinesin family member 23 | 2.5 |
| Kif2c | Kinesin family member 2C | 3.3 |
| Kntc2 | Kinetochore associated 2 | 2.6 |
| Mad2l1 | MAD2 mitotic arrest deficient-like 1 (yeast) | 2.3 |
| Mcm10 | MCM10 minichromosome maintenance deficient 10 (S. cerevisiae) | 2.4 |
| Mcm5 | MCM5 minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) | 3.8 |
| Melk | Maternal embryonic leucine zipper kinase | 2.3 |
| Mki67 | Antigen identified by monoclonal antibody Ki-67 | 3.1 |
| Nek2 | NIMA (never in mitosis gene a)-related kinase 2 | 2.0 |
| Nusap1 | Nucleolar and spindle associated protein 1 | 3.1 |
| Oip5 | Opa interacting protein 5 | 2.0 |
| Pbk | PDZ binding kinase | 2.7 |
| Plk1 | Polo-like kinase 1 (Drosophila) | 3.2 |
| Prc1 | Protein regulator of cytokinesis 1 | 4.0 |
| Prim1 | Primase, polypeptide 1, 49kDa | 3.7 |
| Pttg1 | Pituitary tumor-transforming 1 | 2.4 |
| Racgap1 | Rac GTPase activating protein 1 | 2.1 |
| Rad51ap1 | RAD51 associated protein 1 | 2.7 |
| Shcbp1 | SHC SH2-domain binding protein 1 | 2.9 |
| Spag5 | Sperm associated antigen 5 | 2.8 |
| Spbc24 | Spindle pole body component 24 homolog (S. cerevisiae) | 2.5 |
| Tk1 | Thymidine kinase 1, soluble | 3.3 |
| Top2a | Topoisomerase (DNA) II alpha 170kDa | 2.8 |
| Tpx2 | TPX2, microtubule-associated, homolog (Xenopus laevis) | 2.8 |
| Trip13 | Thyroid hormone receptor interactor 13 | 2.3 |
| Ttk | TTK protein kinase | 2.5 |
| Tyms | Thymidylate synthetase | 3.5 |
| B. Genes down-regulated in both human breast CAFs and Cav-1−/− MSFs (8 genes)
| ||
|---|---|---|
| Symbol | Gene name | Fold-down-regulation in Cav-1 KO |
| Casp1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | −4.2 |
| Dscr1l1 | Down syndrome critical region gene 1-like 1 | −3.2 |
| Kctd12 | Potassium channel tetramerisation domain containing 12 | −2.1 |
| Mid1 | Midline 1 (Opitz/BBB syndrome) | −2.3 |
| Rspo3 | R-spondin 3 homolog (Xenopus laevis) | −2.3 |
| Stmn2 | Stathmin-like 2 | −2.2 |
| Tnxb | Tenascin XB | −3.2 |
| Zfp36l2 | Zinc finger protein 36, C3H type-like 2 | −2.1 |
We also independently scanned the list of 380 transcripts that are up-regulated in Cav-1−/− MSFs and identified 96 RB/E2F target genes (supplemental Table S2, see http://ajp.amjpathol.org). Thus, fully one-fourth of the up-regulated genes, due to loss of Cav-1, are related to RB functional inactivation.
We next validated these results by immunofluorescence and Western blot analysis. Figure 3, A and B shows that RB is indeed hyperphosphorylated in Cav-1−/− MSFs. However, total RB levels remain unchanged in Cav-1−/− MSFs, as seen by immunoblotting (Figure 3B). Thus, loss of Cav-1 expression is sufficient to confer functional inactivation of the RB tumor suppressor protein. Consistent with RB hyperphosphorylation and cell cycle progression, Cav-1−/− MSFs showed a ∼2.7-fold increase in BrdU incorporation (Figure 3C).
Figure 3.
Cav-1−/− MSFs show increased levels of phosphorylated RB (p-RB). A: Cav-1−/− MSF have increased p-RB, as compared with normal wild-type fibroblasts, as shown by immunofluorescence (upper panels), using a phopho-specific antibody that recognizes endogenous RB only when phosphorylated at serine 807/811 (p-RB). DAPI staining shows the nuclei of the cells imaged (lower panels). Images were taken at ×20. B: Virtually identical results were obtained by Western blot analysis. Total RB levels and β-actin levels are shown for comparison. C: Note also that Cav-1−/− MSFs show a significant increase in BrdU incorporation, consistent with cell cycle progression (*P < 0.01). WT, wild-type; KO, Cav-1−/−.
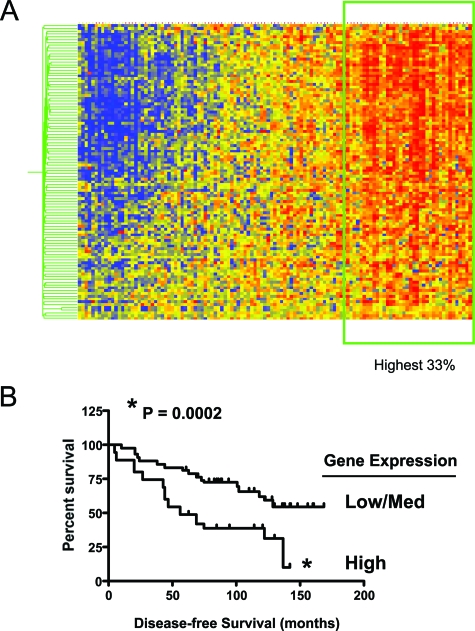
Genes that were consistently up-regulated in Cav-1−/− MSFs were used to cluster a breast cancer data set to determine their impact on disease outcome. Specifically, we observed that the Cav-1−/− MSF gene expression signature correlated with an increased risk of recurrence after tamoxifen monotherapy (Figure 4A and B). Note that this 103-member gene signature consists mainly of RB/E2F-regulated genes (supplemental Table S3, see http://ajp.amjpathol.org). Thus, high expression of the Cav-1−/− MSFs gene signature was associated with a greater than twofold decrease in disease-free survival, in breast cancer patients treated with tamoxifen monotherapy.
Figure 4.
High expression of the Cav-1−/− MSF gene signature is associated with poor clinical outcome in breast cancer patients treated with tamoxifen monotherapy. A: Gene expression data from 60 ER-positive human breast tumors that were both micro- and macrodissected were analyzed for the expression pattern of proliferative genes up-regulated in Cav-1−/− MSFs. A core of proliferation associated genes that are regulated by the RB/E2F pathway strongly co-segregated in this analysis. B: A Kaplan-Meier survival analysis was conducted, wherein the recurrence of those tumors in the highest one-third of overall expression was compared against the remainder of the cohort (*P < 0.0002). Patients in the high Cav-1−/− MSFs gene expression group had a poor prognosis on tamoxifen monotherapy, with greater than a ∼2.6-fold reduction in disease-free survival.
Cav-1−/− MSFs Behave Like Myofibroblasts and Show Evidence of Activated TGF-β Signaling
Human breast CAFs possess many of the characteristics of activated myofibroblasts,2,3 and this activation process is thought to be controlled by TGF-β signaling. Thus, we examined the expression of muscle-related genes in Cav-1−/− MSFs. Table 2 shows a list of these relevant gene changes observed in Cav-1−/− MSFs. Some of the muscle-specific gene transcripts that are up-regulated include smooth muscle actin (SMA), anillin, merosin, myosin (heavy and light chains), and tropomyosin. Similarly, genes transcripts related to activated TGF-β signaling and fibrosis were up-regulated, including collagen I and interleukin-11, and the TGF-β ligand itself. Transcripts of proteins associated with the nucleolus (site of ribosome biogenesis), ribosomal proteins, and genes associated with RNA splicing, were also up-regulated (Table 2). Interestingly, there is an emerging relationship between the nucleolus, ribosome biogenesis, and cell transformation.18
Table 2.
A Selection of Relevant Gene Changes in Cav-1−/− MSFs
| Symbol | Gene name | Fold-change in Cav-1 KO |
|---|---|---|
| Caveolins | ||
| Cav1 | Caveolin, caveolae protein 1 | −116.4 |
| Muscle-related genes | ||
| Acta2 | Actin, alpha 2, smooth muscle, aorta | 2.5 |
| Anln | Anillin, actin binding protein (scraps homolog, Drosophila) | 2.5 |
| Flnb | Filamin, beta | 2.0 |
| Lama2 | Laminin, alpha 2 (merosin) | 3.2 |
| Lmod1 | Leiomodin 1 (smooth muscle) | 2.1 |
| Myh11 | Myosin, heavy polypeptide 11, smooth muscle | 2.0 |
| Myl9 | Myosin, light polypeptide 9, regulatory | 2.0 |
| Myo6 | Myosin VI | −2.0 |
| Tpm1 | Tropomyosin 1, alpha | 2.6 |
| Tpm2 | Tropomyosin 2, beta | 2.6 |
| TGF-beta- and fibrosis-related genes | ||
| Bmp6 | Bone morphogenetic protein 6 | −2.1 |
| Col1a2 | Procollagen, type I, alpha 2 | 2.6 |
| Col18a1 | Procollagen, type XVIII, alpha 1 | −2.8 |
| Smad7 | MAD homolog 7 (Drosophila) | 2.4 |
| Tgfb2 | Transforming growth factor, beta 2 | 2.1 |
| Tgfb3 | Transforming growth factor, beta 3 | 3.2 |
| Cytokine/chemokine signaling | ||
| Ccl2 | Chemokine (C-C motif) ligand 2 | −2.3 |
| Ccl8 | Chemokine (C-C motif) ligand 8 | −2.6 |
| Ccl11 | Chemokine (C-C motif) ligand 11 | −12.2 |
| Cxcl7 | Chemokine (C-X-C motif) ligand 7 | 2.7 |
| Cxcl16 | Chemokine (C-X-C motif) ligand 16 | −3.0 |
| Il11 | Interleukin 11 | 2.4 |
| Socs2 | Suppressor of cytokine signaling 2 | −2.2 |
| Socs3 | Suppressor of cytokine signaling 3 | −2.0 |
| Estrogen receptor co-activator genes | ||
| Greb1 | Gene regulated by estrogen in breast cancer protein | 2.0 |
| Ncoa7 | Nuclear receptor coactivator 7 | 4.0 |
| Rbm39 | Rbm39; CAPER ((coactivator of AP-1 and estrogen receptors); EST C79248) | 8.1 |
| Stem cell-related genes | ||
| Aldh18a1 | Aldehyde dehydrogenase 18 family, member A1 | 2.0 |
| Cyr61 | Cysteine rich protein 61 | 2.5 |
| Hells | Helicase, lymphoid specific | 2.7 |
| Mphosph1 | M-phase phosphoprotein 1 | 2.1 |
| Rspo2 | R-spondin 2 homolog (Xenopus laevis) | 2.2 |
| Rspo3 | R-spondin 3 homolog (Xenopus laevis) | −2.3 |
| Sprr1a | Small proline-rich protein 1A | 2.6 |
| Nucleolar components/ribosomal proteins/RNA-binding proteins/RNA splicing | ||
| Hnrpa1 | Heterogeneous nuclear ribonucleoprotein A1 | 2.1 |
| Lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 2.0 |
| Nol5 | Nucleolar protein 5 | 2.2 |
| Npm3 | Nucleoplasmin 3 | 2.1 |
| Nusap1 | Nucleolar and spindle associated protein 1 | 3.1 |
| Pop4 | Processing of precursor 4, ribonuclease P/MRP family, (S. cerevisiae) | 3.0 |
| Raly | HnRNP-associated with lethal yellow | 2.2 |
| Rbm16 | RNA binding motif protein 16 | 2.3 |
| Rbm39 | RNA binding motif protein 39; CAPER; EST C79248 | 8.1 |
| Rpl17 | Ribosomal protein L17 | 8.8 |
| Rpl18 | Ribosomal protein L18 | 2.0 |
| Rps24 | Ribosomal protein S24 | 2.0 |
| Rrm1 | Ribonucleotide reductase M1 | 2.8 |
| Snhg3 | Small nucleolar RNA host gene (non-protein coding) 3 | 2.0 |
| Snord22 | Small nucleolar RNA, C/D box 22 | 2.1 |
| Snrpa1 | Small nuclear ribonucleoprotein polypeptide A′ | 2.1 |
| Snrpd3 | Small nuclear ribonucleoprotein D3 | 2.0 |
| Txnl4 | Thioredoxin-like 4 | 2.1 |
| U2af1 | U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 1 | 2.2 |
Next, we took several different approaches to functionally validate the potential myofibroblastic phenotype of Cav-1−/− MSFs. We first visualized the cytoskeletal organization of Cav-1−/− MSFs using fluorescein isothiocyanate-phalloidin. As predicted, Cav-1−/− MSFs showed more intense fluorescein isothiocyanate-phalloidin staining, with thicker F-actin-based stress fibers, consistent with a more myofibroblastic phenotype (Figure 5). The distribution of Cav-1 immunostaining is shown for comparison.
Figure 5.
Status of the cytoskeleton in Cav-1−/− MSFs. Mammary fibroblasts were isolated from the mammary glands of wild-type and Cav-1−/− mice. Cells were then fixed and immunostained with antibodies against Cav-1 (N-20, Santa Cruz). In parallel, cells were directly labeled with fluorescein isothiocyanate-conjugated phalloidin, to stain actin filaments. Note that Cav-1−/− MSFs exhibit thicker actin filaments. As expected, Cav-1+/+ wild-type MSFs display a strong Cav-1 membrane staining. However, Cav-1 is absent in Cav-1−/− cells. WT, wild-type; KO, Cav-1−/−.
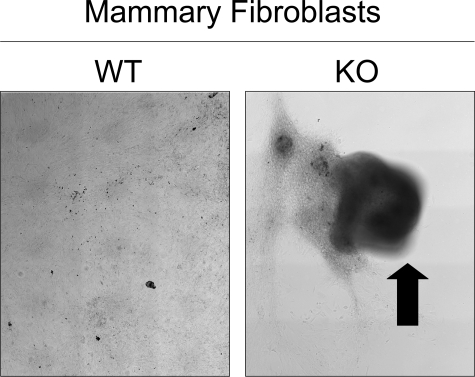
Remarkably, extended culture of confluent Cav-1−/− MSFs (for 4 days) with ascorbic acid consistently resulted in retraction/contraction, indicative a more myofibrolastic phenotype. Wild-type mammary fibroblasts did not undergo retraction/contraction. For Cav-1−/− MSFs, eight out of eight 35-mm dishes plated showed this retraction phenotype. In contrast, for Cav-1+/+ MSFs, none of eight 35-mm dishes showed detachment/retraction. Figure 6 shows representative low-power phase images of wild-type and Cav-1 knockout (KO; Cav-1−/−) fibroblasts. An arrow points at the area of retraction/contraction.
Figure 6.
Cav-1−/− MSFs undergo detachment and retraction/contraction. Primary mammary fibroblasts were isolated from the mammary glands of wild-type and Cav-1−/− mice. At passage 3, cells were allowed to reach confluency and were treated with ascorbic acid (40 μg/ml) for 24 hours. Ascorbic acid treatment facilitates collagen secretion. Then, cells were cultured for an additional 4 days. Remarkably, Cav-1−/− MSFs underwent monolayer detachment and retraction/contraction (arrow), consistent with a more myo-fibroblastic phenotype. Both panels are montages of low-power phase images that were assembled. For Cav-1−/− MSFs, eight of eight 35-mm dishes plated showed this retraction phenotype. In contrast, for wild-type fibroblasts, none of eight 35-mm dishes showed detachment/retraction. WT, wild-type; KO, Cav-1−/−.
Activation of TGF-β/Smad signaling is thought to be one of the major cell signaling mechanisms that confers a myofibroblastic phenotype. Interestingly, we have previously demonstrated that Cav-1 functions as a kinase inhibitor of the TGF-β type I receptor.19 Thus, loss of Cav-1 would be predicted to result in constitutive TGF-β/Smad signaling. Consistent with this hypothesis, our microarray analysis showed upregulation of SMA and interleukin (IL)-11 (Table 2), as well as other TGF-β/Smad responsive genes.
To independently validate the increased expression of SMA, we subjected Cav-1+/+ and Cav-1−/− MSFs to qRT-PCR analysis. Consistent with TGF-β/Smad activation, qRT-PCR analysis of Cav-1−/− MSFs showed quantitative increases in SMA (fourfold), collagen I (1.9-fold), and connective tissue growth factor (CTGF; 2.1-fold)—all TGF-β/Smad responsive genes (Figure 7). Interestingly, the co-up-regulation of IL-11 and CTGF in human breast cancers has been shown to be associated with a poor prognosis, and an increased risk of metastatic disease.20 Similarly, the increased expression of collagen I in Cav-1−/− MSFs was independently validated by immunofluorescence analysis and is shown in Figure 8. Overall, these data suggest that Cav-1−/− MSFs are more myofibroblastic, likely due to constitutive TGF-β/Smad signaling.
Figure 7.
Cav-1−/− MSFs show increased levels of TGF-β/Smad-responsive genes. Relative quantification of samples was assessed by arbitrarily setting the control cDNA value at 100, and changes in transcript levels of a sample were expressed as a multiple thereof (relative expression). The differences in the number of mRNA copies in each PCR reaction was corrected for using mouse 18S rRNA endogenous control transcript levels. (P < 0.05). WT, wild-type; KO, Cav-1−/−.
Figure 8.
Cav-1−/− MSFs show increased collagen I staining. Primary mammary fibroblasts were isolated from the mammary glands of wild-type and Cav-1−/− mice. At passage 3, cells were allowed to reach confluency and were treated with ascorbic acid (40 μg/ml) for 24 hours. Ascorbic acid treatment facilitates collagen secretion. Then, cells were fixed and immunostained with rabbit polyclonal antibodies against collagen I (Novus Biologicals, CO). Note that Cav-1−/− MSFs exhibit a higher collagen I content as compared with wild-type cells. Both panels are the same exposure time. WT, wild-type; KO, Cav-1−/−.
Estrogen Receptor Signaling, HGF/Scatter Factor Expression, and the Epithelial-to- Mesenchymal Transition
We noticed that several estrogen-receptor (ER) co-activator genes were up-regulated in Cav-1−/− MSFs, suggesting that ER signaling may also be activated. These included Rbm39 (a.k.a., CAPER),21 Ncoa7,22 and Greb123 (Table 2). Since CAPER showed the highest level of up-regulation, we chose to validate its expression by immunofluorescence analysis. Figure 9 shows that CAPER protein expression is dramatically elevated in Cav-1−/− MSFs. Since CAPER functions as an ER-co-activator gene at the level of the nucleus, the nuclear distribution of CAPER in Cav-1−/− MSFs may reflect its constitutive activation. Consistent with this hypothesis, a number of estrogen-regulated genes are appropriately up-regulated or down-regulated in Cav-1−/− MSFs (supplemental Table S4, see http://ajp.amjpathol.org). Many of these genes are known RB/E2F-regulated genes, as estrogen also drives proliferation in certain cell types.
Figure 9.
Cav-1−/− MSFs show increased levels of CAPER, an ER-co-activator gene. Cav-1−/− MSF have increased levels of CAPER, as compared with normal wild-type fibroblasts, as shown by immunofluorescence (upper panels). DAPI staining shows the nuclei of the cells imaged (lower panels). Images were taken at ×20.
Estrogen/ER signaling normally controls HGF expression and secretion in mammary stromal fibroblasts.24 Thus, we next assessed HGF expression in Cav-1−/− MSFs. Figure 10 shows that the levels of HGF expression in Cav-1−/− MSFs are significantly increased by ∼tenfold.
Figure 10.
Cav-1−/− MSFs express higher levels of HGF, a key epithelial morphogen. Lysates from wild-type and Cav-1−/− MSFs were subjected to immunoblot analysis with an antibody directed against the β-chain of HGF. Immunoblotting with β-actin is shown as an equal loading control.
The secretion of certain known stromal cell factors (HGF/scatter factor) from mammary fibroblasts is thought to profoundly regulate the phenotypic behavior of mammary epithelial cells. Stromally derived HGF normally induces an epithelial-mesenchymal transition (EMT) in mammary epithelia, driving their conversion from a mammary epithelial phenotype to a more invasive myo-epithelial/myo-fibroblastic phenotype.24 Thus, we hypothesized that Cav-1−/− MSFs may secrete increased levels of growth-promoting and EMT-promoting factors.
To test this hypothesis directly, we overlaid single-cell suspensions of wild-type mammary epithelial cells onto a 3D Matrigel culture, and stimulated them with “conditioned media” from Cav-1+/+ and Cav-1−/− MSFs for a period of 4 days. Then, we subjected these cultures to immunostaining with α-SMA to visualize the onset of an EMT, and co-staining with propidium iodide to visualize the distribution of cell nuclei.
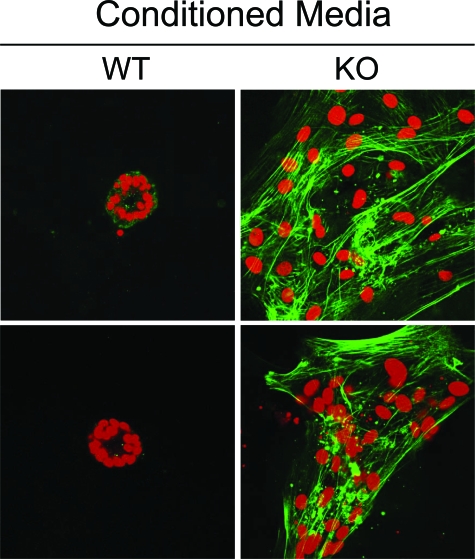
Interestingly, our results directly show that “conditioned media” derived from Cav-1−/− MSFs has a profound effect on the phenotypic behavior and morphology of wild-type mammary epithelial cells. Notably, we observed that Cav-1−/− MSFs “conditioned media” drives the onset of an EMT in normal wild-type mammary epithelial cells. Under these conditions, wild-type mammary epithelia fail to form normal 3D acinar structures, but instead appear as clusters of flattened fibroblastic cells that are positive for immunostaining with SMA, an EMT-marker (Figure 11).
Figure 11.
Conditioned media from Cav-1−/− MSFs drives wild-type mammary epithelial cells to undergo an EMT. Conditioned media was prepared from wild-type and Cav-1−/− MSFs. This conditioned media was then applied to 3D cultures of isolated wild-type mammary epithelial cells shortly after plating on day 0. After 4 days of treatment, cells were fixed with paraformaldehyde and immunostained with antibodies directed against α-smooth muscle actin (α-SMA; shown in green). Cells were counterstained with propidium iodide (shown in red) to visualize cell nuclei. Note that wild-type epithelial cells exposed to Cav-1−/− MSF-conditioned media appear fibroblastic, and express SMA, consistent with an EMT. In contrast, wild-type epithelial cells exposed to wild-type fibroblast conditioned media appear epitheloid and form typical 3D acinar structures with a central lumen, as expected. WT, wild-type MSF-conditioned media; KO, Cav-1−/− MSF-conditioned media.
Proteome Analysis of Cav-1−/− MSF-Secreted Factors
We also subjected conditioned media derived from Cav-1+/+ and Cav-1−/− MSFs to broad-spectrum ELISA analysis to detect potential differences in their patterns of secreted factors. The concentrations of approximately 40 secreted factors (growth factors, cytokines, and chemokines) were quantitatively evaluated by ELISA (Pierce SearchLight Multiplexed Proteome Arrays) and expressed as pg/ml. Our results are summarized in Table 3. Interestingly, the secretion of several pro-angiogenic or pro-tumorigenic factors was significantly increased in Cav-1−/− MSFs, such as platelet-derived growth factor, vascular endothelial growth factor, macrophage inflammatory proteins 2 and 1alpha, granulocyte-macrophage colony-stimulating factor, among others (thymus and activation-regulated chemokine, L-Selectin, IL-6, IL-10, and IL-13).
Table 3.
Proteome Analysis of Cav-1−/− MSF Secreted Proteins
| Name | KO (pg/ml) | WT (pg/ml) | Fold change |
|---|---|---|---|
| PDGF-BB | 2820 ± 359.5 | ND | ∞ |
| VEGF | 139.64 ± 7.72 | 53.5 ± 2.1 | 2.6** |
| MIP2 | 47.74 ± 10.36 | 18.5 ± 3.26 | 2.6* |
| MIP1-alpha | 3.12 ± 0.04 | 1.42 ± 0.24 | 2.2** |
| GM-CSF | 13.92 ± 0.96 | 7.38 ± 0.34 | 1.9** |
| TARC | 234.22 ± 17.98 | 147.62 ± 4.48 | 1.6* |
| L-Selectin | 100.2 ± 4.2 | 65.9 ± 0.2 | 1.5* |
| IL-6 | 7675.76 ± 137.46 | 3903.12 ± 140.72 | 2.0*** |
| IL-10 | 74.46 ± 8.44 | 45.86 ± 0.7 | 1.6* |
| IL-13 | 170.78 ± 12.3 | 124.54 ± 10.74 | 1.4* |
NOTE: No changes (NC) were observed in the levels of TGFbeta1, Leptin, IL-18, IFN-gamma, MIP1beta, KC, OPN, RANTES, P-Selectin, SDF1beta, CRP, JE, E-Selectin, MMP9, and sVCAM1.
P < 0.05;
P < 0.001;
P < 0.0001. ND, not detectable.
Cav-1−/− MSFs have the Capacity to Undergo Endothelial-Like Transdifferentiation
As Cav-1−/− MSF-conditioned media showed the up-regulation of a number of pro-angiogenic growth factors (Table 3), we next assessed their potential to undergo endothelial cell differentiation. Since collagen I is important for endothelial cell differentiation and endothelial “tube formation,” we optimized the expression and secretion of collagen I by treating Cav-1+/+ and Cav-1−/− MSFs with ascorbic acid at confluency.
Twenty-four hours later, we assayed these MSFs for the expression of a number of endothelial-specific markers by qRT-PCR. Interestingly, our results directly demonstrate that Cav-1−/− MSFs show the clear up-regulation of a number of endothelial-specific marker genes and pro-angiogenic factors, as compared with Cav-1+/+ MSFs treated identically (Table 4). These endothelial and pro-angiogenic markers included: Pecam1, Tek, Pgf, Plau, Il-6, Tbx4, Tgfb3, Col18a1, platelet-derived growth factor-A, Timp1, and vascular endothelial growth factor-C. Notably, Pecam1 gene expression was increased in Cav-1−/− MSFs by ∼17.5-fold.
Table 4.
Endothelial and Pro-Angiogenic Genes Are Up-Regulated in Cav-1−/− MSFs, as Seen by qRT-PCR Analysis
| Gene symbol | Gene name | Fold KO increase | P value |
|---|---|---|---|
| Pecam1 | Platelet/endothelial cell adhesion molecule 1 | 17.5 | 0.020 |
| Tek | TEK Tyrosine Kinase, endothelial | 4.5 | 0.003 |
| Pgf | Placental growth factor | 4.3 | 0.005 |
| Plau | Plasminogen activator, urokinase | 3.8 | 0.04 |
| Il-6 | Interleukin 6 | 3.5 | 0.007 |
| Tbx4 | T Box 4 | 3.0 | 0.005 |
| Tgfb3 | Transforming growth factor, beta 3 | 2.1 | 0.010 |
| Col18a1 | Procollagen, type XVIII, alpha 1 | 2.0 | 0.015 |
| PDGF-A | Platelet derived growth factor, alpha | 1.9 | 0.011 |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 1.9 | 0.033 |
| Vegf-C | Vascular endothelial growth factor C | 1.6 | 0.001 |
It is important to note that most of these markers were not up-regulated when subconfluent Cav-1−/− MSFs were examined by gene expression profiling (DNA microarray). Thus, these findings appear to be specific for confluent Cav-1−/− MSFs monolayers treated with ascorbic acid.
Furthermore, when Cav-1−/− MSF confluent monolayers were cultured for extended periods of time (30 days), they underwent spontaneous endothelial “tube formation” (Figure 12). Thus, under certain culture conditions that optimize collagen I production, Cav-1−/− MSFs biochemically and functionally undergo endothelial-like transdifferentiation.
Figure 12.
Cav-1−/− MSFs spontaneously undergo endothelial-like tube formation. Primary mammary fibroblasts were isolated from the mammary glands of wild-type and Cav-1−/− mice. The cells were allowed to reach confluency in 10-cm dishes, and were treated with ascorbic acid (40 μg/ml). Ascorbic acid treatment facilitates collagen secretion. Then, the cells were subjected to extended culture (30 days) without changing their media. Interestingly, Cav-1−/− MSFs spontaneously underwent endothelial-like tube formation (right panels). Four representative phase images are shown for each genotype. WT, wild-type; KO, Cav-1−/−.
To determine the in vivo relevance of these findings, we next assessed the vascularization of Cav-1−/− mammary fat pads. For this purpose, we immunostained frozen sections derived from age-matched wild-type and Cav-1−/− virgin female mammary glands with a well-established endothelial cell marker protein, namely CD31 (Pecam1). Figure 13 directly demonstrates that Cav-1−/− mammary fat pads show dramatically increased vascularization, as compared with wild-type mice. These findings are consistent with the idea that Cav-1−/− MSFs can promote mammary stromal angiogenesis and/or undergo endothelial cell transdifferentiation in vivo.
Figure 13.
Increased vascularization of Cav-1−/− mammary fat pads, as visualized by CD31 immunostaining. Mammary glands (inguinal) from 6-month-old virgin female wild-type and Cav-1−/− mice were harvested and used to prepare frozen tissue sections. Samples were then subjected to immunostaining with a rat mAb directed against CD31 (Pecam1). Bound primary antibodies were visualized with HRP-conjugated (A) or rhodamine-conjugated (B) secondary antibodies. In A, samples were also counterstained with hematoxylin to visualize overall tissue morphology, and boxed areas are shown at higher magnification. Note that CD31 staining is dramatically increased in Cav-1−/− mammary fat pads, as compared with wild-type mice processed in parallel. This is consistent with the idea that Cav-1−/− mammary fat pads are highly vascularized. Original magnification = ×40 (A)×20 (B) WT, wild-type; KO, Cav-1−/−.
Transcriptional Comparison of Cav-1−/− MSFs with CAFs Isolated from Individual Patients
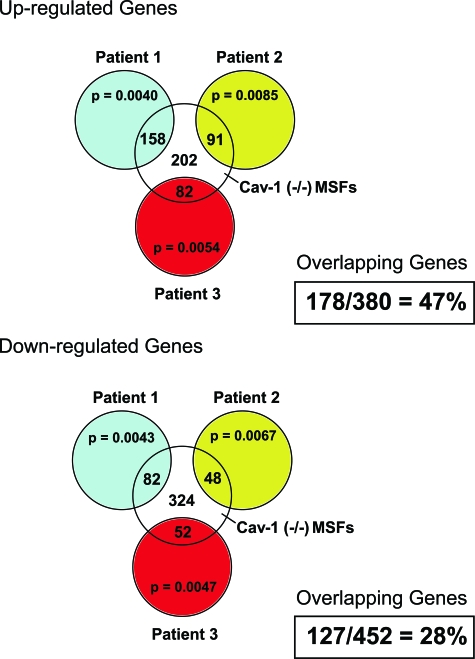
Because of the striking phenotypic similarities between Cav-1−/− MSFs and human CAFs, we also analyzed their transcriptional similarity with CAFs obtained from individual patients.6 This additional analysis was performed because comparison with only the CAF gene signature may underestimate their similarity. Figure 14 shows, based on this more inclusive analysis, that nearly 50% of the genes up-regulated in Cav-1−/− MSFs are also up-regulated in CAFs; similarly, nearly 30% of the genes down-regulated in Cav-1−/− MSFs are also down-regulated in CAFs (Table 5). It is rare to see such concordance between human patient samples and a mouse animal model. For example, comparison of patients 1, 2, and 3 with each other previously yielded a gene signature of 118 up-regulated genes and 66 down-regulated genes.6
Figure 14.
Transcriptional comparison of Cav-1−/− MSFs with CAFs isolated from individual patients. We previously isolated 11 sets of CAFs, with corresponding normal mammary fibroblasts, from the same patients.6 Three of these sets were randomly selected for genome-wide transcriptional profiling.6 These primary cells (CAFs versus normal mammary fibroblasts) were then compared pair-wise to identify genes up-regulated or down-regulated in CAFs from breast cancer patients 1, 2, and 3. These three gene sets were then individually compared with the gene changes observed in wild-type versus Cav-1−/− MSFs. Note that intersection of the up-regulated CAF genes from patients 1, 2, and 3, with up-regulated Cav-1−/− MSF genes, revealed significant overlap. Patients 1, 2, and 3 shared 158, 91, and 82 up-regulated genes with Cav-1−/− MSFs, respectively. These three gene lists were then compiled to yield a master list of 178 unique genes that were up-regulated in both CAFs and Cav-1−/− MSFs. Thus, 178 of the 380 genes that were up-regulated in Cav-1−/− were also up-regulated in CAFs, yielding an overlap of 47%. However, 202 up-regulated Cav-1−/− MSF genes did not show overlap with up-regulated CAFs genes. Identical comparisons were also made for the down-regulated gene sets. Patients 1, 2, and 3 shared 82, 48, and 52 down-regulated genes with Cav-1−/− MSFs, respectively. These three gene lists were then compiled to yield a master list of 127 unique genes that were down-regulated in both CAFs and Cav-1−/− MSFs. Thus, 127 of the 452 genes that were down-regulated in Cav-1−/− were also down-regulated in CAFs, yielding an overlap of 28%. However, 324 down-regulated Cav-1−/− MSF genes did not show overlap with down-regulated CAFs genes. Thus, overall Cav-1−/− MSFs were most closely related to CAFs from patient 1.
Table 5.
Transcriptional Comparison of Cav-1−/− MSFs with CAFs Isolated from Individual Patients
| Genes up-regulated in Cav-1−/− MSFs and CAFs, either from patient 1, 2, or 3 (178 genes) |
| ANLN, AP1S3, ASF1B, ASPM, ATAD2, AURKA, AURKB, BIRC5, BLM, BNC1, BRCA1, BRRN1, BUB1, BUB1B, CALD1, CASC5, CASP1, CBX5, CCNA2, CCNB1, CCNB2, CCNE2, CCNF, CDC20, CDC25C, CDC45L, CDC6, CDC7, CDCA1, CDCA2, CDCA3, CDCA5, CDCA7, CDCA8, CDKN3, CENPA, CENPE, CENPF CENPH, CENPI, CENPK, CENPQ, CEP55, CHEK1, CHTF18, CIT, CKAP2L, CKS1B, CKS2, CSRP1, CTH, DEPDC1B, DHFR, DLG7, DSCR1L1, E2F7, ECT2, ESPL1, EXO1, EZH2, FABP5, FANCA, FANCB, FBXO5, FEN1, FIGNL1, FOXM1, FST, GADD45B, GMNN, GRB10, GSG2, H2AFX, HBEGF, HELLS, HMGB1, HMGB2, HMGB3, HMGN2, HMMR, HN1, IDH2, IL11, IQGAP3, ITGA5, KCTD12, KIF11, KIF20A, KIF22, KIF23, KIF2C, KIFC1, KNTC1, KNTC2, KRT19, LIG1, LMNB1, LSM5, LUZP5, MAD2L1, MASTL, MCM10, MCM3, MCM5, MCM6, MCM7, MCM8, MELK, MID1, MKI67, MLF1, MPHOSPH1, MYBL2, NASP, NEK2, NPR3, NUSAP1, OIP5, PBK, PCNA, PDLIM5, PHF20, PLK1, PLK4, POLD1, POLE, PPIL5, PRC1, PRIM1, PRR11, PTK2, PTTG1, RACGAP1, RAD18, RAD51, RAD51AP1, RECQL4, RFC5, RRM1, RSPO3, SDPR, SGOL1, SHCBP1, SLC7A5, SMC2, SMC4, SMTN, SNRPA1, SORBS1, SPAG5, SPBC24, STIL, STMN2, TACC3, TAGLN2, TBC1D1, TGFB2, TGM2, THY1, TK1, TLN1, TNXB, TOP2A, TPM1, TPM2, TPX2, TRIP13, TRPV2, TTK, TYMS, UBE2C, UBE2S, UHRF1, VRK1, WDHD1, WHSC1, ZFP36L2, ZWILCH |
| Genes down-regulated in Cav-1−/− MSFs and CAFs, either from patient 1, 2, or 3 (127 genes) |
| ABAT, ABHD4, AGT, ALDH1A1, ALDH2, AMPD3, ANK3, AOX1, AQP1, ARHGDIB, ASPA, ASPN, C1S, C3, CAMK1D, CASP1, CASP4, CCL11, CCL2, CD55, CDH13, CDK6, CH25H, CLEC14A, CNNM2, CST6, CTSH, CTSK, DCAMKL1, DOCK4, DPT, DSCR1, DSCR1L1, DSP, EDNRB, EFEMP1, ELTD1, ENPP2, FIP1L1, FLRT2, GAS1, GCH1, GCLC, GPNMB, GPR116, GPR137B, GPR64, GRIA3, HGF, HIST1H3D, HTR2B, ICAM1, IFIT3, IGF2BP3, IL1RL1, IQGAP2, ITGBL1, ITPR2, JAM2, KCTD12, KLHL13, KLHL24, LMO7, LRP11, LSS, MAF, MAGI2, MARK1, MASP1, MGAM, MGST1, MID1, MITF, MOXD1, NGEF, NMNAT2, NOTCH3, NQO1, NRP2, OLR1, PAPPA2, PCMTD1, PLAT, PLSCR4, PLTP, PLXDC1, PPAP2B, PPFIBP2, PRELP, PRG4, PRICKLE2, PRL, PROS1, PRSS35, PTGIR, PTN, PYCARD, RAB3GAP2, RECK, RGL1, RGS5, RRAGD, RSPO3, SEMA3C, SEPP1, SESN3, SFRP2, SFRP4, SLC15A3, SLC39A8, SLC7A11, SMPDL3A, SNED1, SOCS2, SOD3, SORBS2, STMN2, SULF2, TAC1, TEK, TNFAIP2, TNXB, TOB1, TPP1, VIT, ZFP36L1, ZFP36L2 |
Inclusion of a transcript from the Cav-1−/− MSF gene profile in any one of the three patients constituted a hit.
We calculated the statistical significance for the transcriptome intersection by using hypergeometric probabilities for any two groups of genes. By considering the commonality between human and mouse platforms based on identical transcript identifiers, we generated a P value for the interesting sets. All these comparisons were statistically significant at P < 0.009 (See Figure 14).
Discussion
The process by which normal mammary fibroblasts are converted to tumor-associated fibroblasts remains unknown. Interestingly, we recently observed that human breast cancer-associated fibroblasts show down-regulation of Cav-1 protein expression and exhibit facets of RB functional gene inactivation.6 Thus, we speculated that loss of Cav-1 expression may be a critical initiating event leading toward the cancer-associated fibroblast phenotype. Consistent with this hypothesis, via in vivo transplant studies, we have previously established that the mammary stroma of Cav-1−/− null mice clearly stimulates the growth of both i) normal mammary ductal epithelia and ii) mammary tumor cells.7
Here, we provide the first molecular genetic evidence that loss of Cav-1 expression directly contributes to the cancer-associated fibroblast phenotype. We show that Cav-1−/− MSFs share many properties with human CAFs. Like CAFs, Cav-1−/− MSFs show functional inactivation of RB via hyperphosphorylation. Table 2 shows a list of 55 genes that were commonly up-regulated in both human breast CAFs and Cav-1−/− MSFs. Many of these genes are RB/E2F-regulated genes. In addition, Cav-1−/− MSFs show the over-expression of 96 RB/E2F target genes.
Moreover, we demonstrate that Cav-1−/− MSFs take on the functional characteristics of myofibroblasts, such as: i) contraction/retraction; ii) the up-regulation of muscle-related genes (smooth muscle actin, myosin [heavy and light chains], tropomyosin); and iii) the upregulation of TGF-β ligands, related factors, and responsive genes (TGF-β2/3, procollagen genes, interleukin-11, and CTGF).
CAFs are thought to mediate their effects through paracrine interactions with mammary epithelial-derived tumor cells.4,5 In this regard, we also show that Cav-1−/− MSFs secrete increased amounts of pro-proliferative and pro-angiogenic growth factors, and up-regulate the expression of HGF/scatter factor, a key epithelial morphogen. Functionally, conditioned media prepared from Cav-1−/− MSFs is sufficient to induce an epithelial-mesenchymal transition in 3D cultures of Cav-1+/+ mammary epithelial cells embedded in Matrigel. Thus, Cav-1−/− MSFs fulfill many of the functional criteria already established for the phenotypic behavior of human CAFs. Here, we also show that Cav-1−/− MSFs demonstrate evidence of activated TGF-β signaling and secrete/express increased levels of HGF, vascular endothelial growth factor, and IL-6. Similarly, it has been previously shown that TGF-β-mediated induction of the myofibroblast phenotype induces the enhanced secretion of HGF, vascular endothelial growth factor, and IL-6.25
Tissue fibroblasts show significant plasticity and are able to differentiate into numerous “terminally differentiated” cell types, including adipocytes and muscle cells, among others.26,27,28 Thus, fibroblasts may also be considered as stem-like progenitor cells. With this in mind, we examined the list of 380 transcripts that were up-regulated in Cav-1−/− MSFs and compared them with lists of known ES cell genes and genes controlled by iPS (induced pluripotency)-related transcription factors.11 Interestingly, Cav-1−/− MSFs show the up-regulation of numerous stem/progenitor cell-associated genes (see Tables 2 and 6). Based on this analysis, it is apparent that Rbm39 (a.k.a., CAPER) is the target of Nanog, Sox2, and Myc, three of the iPS transcription factors. Notably, CAPER is highly up-regulated in Cav-1−/− MSFs (∼eightfold) and it is known to function as a co-activator of the AP-1 transcription factor and nuclear receptors (such as estrogen and progesterone).21 Thus, CAPER overexpression is consistent with the idea that Cav-1−/− MSFs may behave more like stem/progenitor cells. In accordance with this idea, we have previously shown that Cav-1−/− mice show an expansion of the epithelial stem/progenitor cell compartment in the skin, mammary gland, and the intestine.7,15,29,30 This may have important implications for understanding the origin(s) of cancer stem cells within the tumor microenvironment. Interestingly, Mishra et al, 2008 have recently suggested that CAFs have many similarities with and may originate from human bone-marrow mesenchymal stem cells.31
Table 6.
ES Cell Related Genes Up-Regulated in Cav-1−/− MSFs
| ES cell expressed genes (41 genes) |
| Aurkb, Birc5, Bub1, Bub1b, Cbx5, Ccna2, Ccnb1, Cdc20, Cdc6, Cdca5, Chek1, Dhfr, Dlg7, Dna2l, Ect2, Ercc6l, Fabp5, Fen1, Gmnn, Hells, Hmgb3, Hmmr, Kif2c, Mcm10, Mcm3, Mcm5, Mcm6, Mcm7, Mybl2, Nasp, Npm3, Oip5, Prim1, Prpf40a, Pttg1, Rad51ap1, Rps24, Slc38a1, Slc7a3, Terf1, Wdhd1 |
| Nanog targets (33 genes) |
| Atad2, Aurka, Bub1b, Cald1, Cbx5, Cdc45l, Cdc7, Cks2, Cyr61, Dlg7, Fanca, H2afx, Hells, Hn1, Kntc1, Lsm5, Nusap1, Oip5, Pbx1, Pcna, Prr11,Rbm39*, Rif1, Rpl17, Set, Slc7a5, Sorbs1, Terf1, Tmem87a, Top2a, Tpx2, Ube2s, Wdhd1 |
| Oct4 targets (11 genes) |
| Bub1b, Hmgb2, Set, Top2a, Cdc7, Sorbs1, Snx5, Atad2, Hn1, Rif1, Prr11 |
| Sox 2 targets (26 genes) |
| Atad2, Bub1b, Cbx5, Cdc45l, Cdc7, Crim1, Cth, Cyr61, H2afx, Hells, Hn1, Kif11, Kif2c, Kntc1, Mphosph1, Nusap1, Oip5, Prr11,Rbm39*, Rif1, Set, Sfpq, Slc7a5, Snrpd3, Top2a, Ube2c |
| Myc targets (36 genes) |
| Aldh18a1, Cbx5, Ccnb1, Cdc25b, Cdc6, Cks2, Exo1, Fabp5, Fbxo5, Foxm1, H2afx, Hmgn2, Hmmr, Kif11, Kif20a, Mad2l1, Mcm3, Mcm5, Mcm7, Mphosph1, Nxt1, Pabpc1, Pcna, Plk1, Rad51ap1,Rbm39*, Rpl18, Smad7, Snrpd3, Snx5, Spag5, Tagln2, Tgfb3, Tpm2, Trip13, U2af1 |
| ES Cell Common Transcriptional Regulators (8 genes) |
| Ezh2, Foxm1, Hmgb2, Hmgb3, Mybl2, Trip13, Wdhd1, Whsc1 |
Note: Underlined genes were up-regulated >5-fold.
Rbm39 = CAPER (Co-Activator of AP-1 and ER).
In accordance with the hypothesis that fibroblasts can behave as multipotent progenitor cells, NIH 3T3 fibroblasts treated with “conditioned media” from ES cells undergo endothelial cell transdifferentiation.32 Conversely, endothelial cells can transdifferentiate into myofibroblasts under the appropriate conditions.33,34 Importantly, recent mouse genetic studies have clearly documented that this endothelial-mesenchymal transition frequently occurs in vivo, under pathological conditions including cardiac fibrosis and tumorigenesis.35,36 As such, our observation that Cav-1−/− MSFs undergo spontaneous endothelial transdifferentiation is consistent with these findings. Similarly, Cav-1−/− mammary fat pads show dramatically increased vascularization, as compared with wild-type mice. These findings provide additional support for the idea that Cav-1−/− MSFs can promote mammary stromal angiogenesis and/or undergo endothelial cell transdifferentiation in vivo. Thus, these results have broad implications for understanding the role of cancer-associated fibroblasts in promoting tumor angiogenesis in vivo, via their potential to secrete angiogenic growth factors and to directly undergo endothelial cell transdifferentiation.
Pericytes (a.k.a., adventitial cells) are mesenchymal-like vascular mural cells that are associated with connective tissue and are wrapped around the walls of small blood vessels, venules, and capillaries. Interestingly, these undifferentiated pericytes show significant progenitor-like plasticity and can differentiate into several distinct cell types, including fibroblasts, smooth muscle cells, and even macrophages. In addition, pericytes are contractile, express smooth muscle actin, and they function as critical regulators of vascular morphogenesis, angiogenesis, and fibrosis. In this regard, pericytes show striking phenotypic similarities with myofibroblasts and/or cancer-associated fibroblasts. However, pericytes are difficult to define and their origins are still not well understood. Given the abundance of CD31(+) microvasculature in Cav-1−/− mammary fat pads, it is quite possible that Cav-1−/− MSFs may originate from pericytes. In further support of this idea, pericytes, tumor-associated myofibroblasts, mesenchymal stem cells, and endothelial progenitor cells all express a common tumor-endothelial marker (Tem1), namely endosialin.37,38,39,40,41,42 Thus, this interesting possibility undoubtedly deserves further study.
Supplementary Material
Footnotes
Address reprint requests to Drs. Federica Sotgia and Michael P. Lisanti, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson University, 233 South 10th Street, Philadelphia, PA, 19107; E-mail: federica.sotgia@jefferson.edu or michael.lisanti@kimmelcancercenter.org.
Supported by grants from the Elsa U. Pardee Foundation and the W.W. Smith Charitable Trust, and a Research Scholar Grant from the American Cancer Society to F.S. I.M. was supported by a Post-doctoral Fellowship from the Susan G. Komen Breast Cancer Foundation. P.G.F. was supported by a grant from the W.W. Smith Charitable Trust, and a Career Catalyst Award from the Susan G. Komen Breast Cancer Foundation. M.P.L. was supported by grants from the NIH/NCI (R01-CA-80250; R01-CA-098779; R01-CA-120876), the American Association for Cancer Research, and the Department of Defense-Breast Cancer Research Program (Synergistic Idea Award). This project is funded, in part, under a grant with the Pennsylvania Department of Health (to F.S. and M.P.L.). R.G.P. was supported by grants from the NIH/NCI (R01-CA-70896, R01-CA-75503, R01-CA-86072, and R01-CA-107382) and the Dr. Ralph and Marian C. Falk Medical Research Trust. The Kimmel Cancer Center was supported by the NIH/NCI Cancer Center Core grant P30-CA-56036 (to R.G.P.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
A guest editor acted as editor-in-chief for this manuscript. No person at Thomas Jefferson University or Albert Einstein College of Medicine was involved in the peer review process or final disposition for this article.
References
- Sappino AP, Skalli O, Jackson B, Schurch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41:707–712. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- Serini G, Gabbiani G. Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res. 1999;250:273–283. doi: 10.1006/excr.1999.4543. [DOI] [PubMed] [Google Scholar]
- Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. I. Paracrine cells important in health and disease. Am J Physiol. 1999;277:C1–C9. doi: 10.1152/ajpcell.1999.277.1.C1. [DOI] [PubMed] [Google Scholar]
- Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol. 1999;277:C183–C201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- Mercier I, Casimiro MC, Wang C, Rosenberg AL, Quong J, Allen KG, Danilo C, Sotgia F, Bonuccelli G, Jasmin JF, Xu H, Bosco E, Aronow B, Witkiewicz A, Pestell RG, Knudsen ES, Lisanti MP. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 down-regulation and RB tumor suppressor functional inactivation: implications for the response to hormonal therapy. Cancer Biol Ther. 2008;7:1212–1225. doi: 10.4161/cbt.7.8.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Sotgia F, Lee H, Hassan G, Di Vizio D, Bonuccelli G, Capozza F, Mercier I, Rui H, Pestell RG, Lisanti MP. Stromal and epithelial caveolin-1 both confer a protective effect against mammary hyperplasia and tumorigenesis: caveolin-1 antagonizes cyclin D1 function in mammary epithelial cells. Am J Pathol. 2006;169:1784–1801. doi: 10.2353/ajpath.2006.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang C, Jiao X, Katiyar S, Casimiro MC, Prendergast GC, Powell MJ, Pestell RG. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem. 2008;283:7007–7015. doi: 10.1074/jbc.M706992200. [DOI] [PubMed] [Google Scholar]
- Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, Pestell RG. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci USA. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Cordero KE, Larios JM, Miller RS, Johnson MD, Chinnaiyan AM, Lippman ME, Rae JM. Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors. Genome Biol. 2006;7:R28. doi: 10.1186/gb-2006-7-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhou M, Wong WH. Integrated analysis of microarray data and gene function information. Omics. 2004;8:106–117. doi: 10.1089/1536231041388320. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Williams TM, Cohen AW, Minetti C, Pestell RG, Lisanti MP. Caveolin-1-deficient mice have an increased mammary stem cell population with up-regulation of Wnt/beta-catenin signaling. Cell Cycle. 2005;4:1808–1816. doi: 10.4161/cc.4.12.2198. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Williams TM, Schubert W, Medina F, Minetti C, Pestell RG, Lisanti MP. Caveolin-1 deficiency (−/−) conveys premalignant alterations in mammary epithelia, with abnormal lumen formation, growth factor independence, and cell invasiveness. Am J Pathol. 2006;168:292–309. doi: 10.2353/ajpath.2006.050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, Lowe SW, Knudsen ES. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Zhang XL, Bitzer M, von Gersdorff G, Bottinger EP, Lisanti MP. Caveolin-1 regulates transforming growth factor (TGF)-beta/SMAD signaling through an interaction with the TGF-beta type I receptor. J Biol Chem. 2001;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem. 2002;277:1229–1234. doi: 10.1074/jbc.M110417200. [DOI] [PubMed] [Google Scholar]
- Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol Cell Biol. 2002;22:3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Zhang HZ, Bennett JM, Smith KT, Sunil N, Haslam SZ. Estrogen mediates mammary epithelial cell proliferation in serum-free culture indirectly via mammary stroma-derived hepatocyte growth factor. Endocrinology. 2002;143:3427–3434. doi: 10.1210/en.2002-220007. [DOI] [PubMed] [Google Scholar]
- Cat B, Stuhlmann D, Steinbrenner H, Alili L, Holtkotter O, Sies H, Brenneisen P. Enhancement of tumor invasion depends on transdifferentiation of skin fibroblasts mediated by reactive oxygen species. J Cell Sci. 2006;119:2727–2738. doi: 10.1242/jcs.03011. [DOI] [PubMed] [Google Scholar]
- Rosen OM, Smith CJ, Fung C, Rubin CS. Development of hormone receptors and hormone responsiveness in vitro. Effect of prolonged insulin treatment on hexose uptake in 3T3–L1 adipocytes, J Biol Chem. 1978;253:7579–7583. [PubMed] [Google Scholar]
- Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3–L1 cells, J Biol Chem. 1978;253:7570–7578. [PubMed] [Google Scholar]
- Strakova Z, Livak M, Krezalek M, Ihnatovych I. Multipotent properties of myofibroblast cells derived from human placenta. Cell Tissue Res. 2008;332:479–488. doi: 10.1007/s00441-008-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozza F, Williams TM, Schubert W, McClain S, Bouzahzah B, Sotgia F, Lisanti MP. Absence of caveolin-1 sensitizes mouse skin to carcinogen-induced epidermal hyperplasia and tumor formation. Am J Pathol. 2003;162:2029–2039. doi: 10.1016/S0002-9440(10)64335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hassan GS, Williams TM, Minetti C, Pestell RG, Tanowitz HB, Frank PG, Sotgia F, Lisanti MP. Loss of caveolin-1 causes the hyper-proliferation of intestinal crypt stem cells, with increased sensitivity to whole body gamma-radiation. Cell Cycle. 2005;4:1817–1825. doi: 10.4161/cc.4.12.2199. [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW, Banerjee D. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingh J, Lambers E, Hamada H, Bord E, Thorne T, Goukassian I, Krishnamurthy P, Rosen KM, Ahluwalia D, Zhu Y, Qin G, Losordo DW, Kishore R. Cell-free embryonic stem cell extract-mediated derivation of multipotent stem cells from NIH3T3 fibroblasts for functional and anatomical ischemic tissue repair. Circ Res. 2008;102:e107–e117. doi: 10.1161/CIRCRESAHA.108.176115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M, Gerth J, Stein G, Wolf G. Transdifferentiation of endothelial and renal tubular epithelial cells into myofibroblast-like cells under in vitro conditions: a morphological analysis. Cells Tissues Organs. 2005;180:204–214. doi: 10.1159/000088937. [DOI] [PubMed] [Google Scholar]
- Paranya G, Vineberg S, Dvorin E, Kaushal S, Roth SJ, Rabkin E, Schoen FJ, Bischoff J. Aortic valve endothelial cells undergo transforming growth factor-beta-mediated and non-transforming growth factor-beta-mediated transdifferentiation in vitro. Am J Pathol. 2001;159:1335–1343. doi: 10.1016/s0002-9440(10)62520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- Becker R, Lenter MC, Vollkommer T, Boos AM, Pfaff D, Augustin HG, Christian S. Tumor stroma marker endosialin (Tem1) is a binding partner of metastasis-related protein Mac-2 BP/90K. FASEB J. 2008;22:3059–3067. doi: 10.1096/fj.07-101386. [DOI] [PubMed] [Google Scholar]
- Christian S, Winkler R, Helfrich I, Boos AM, Besemfelder E, Schadendorf D, Augustin HG. Endosialin (Tem1) is a marker of tumor-associated myofibroblasts and tumor vessel-associated mural cells. Am J Pathol. 2008;172:486–494. doi: 10.2353/ajpath.2008.070623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadyen JR, Haworth O, Roberston D, Hardie D, Webster MT, Morris HR, Panico M, Sutton-Smith M, Dell A, van der Geer P, Wienke D, Buckley CD, Isacke CM. Endosialin (TEM1. CD248) is a marker of stromal fibroblasts and is not selectively expressed on tumour endothelium. FEBS Lett. 2005;579:2569–2575. doi: 10.1016/j.febslet.2005.03.071. [DOI] [PubMed] [Google Scholar]
- MacFadyen J, Savage K, Wienke D, Isacke CM. Endosialin is expressed on stromal fibroblasts and CNS pericytes in mouse embryos and is downregulated during development. Gene Expr Patterns. 2007;7:363–369. doi: 10.1016/j.modgep.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S, Kataoka S, Ishida I, Roberts BL, Teicher BA. Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvasc Res. 2008;76:180–188. doi: 10.1016/j.mvr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Bagley RG, Rouleau C, St Martin T, Boutin P, Weber W, Ruzek M, Honma N, Nacht M, Shankara S, Kataoka S, Ishida I, Roberts BL, Teicher BA. Human endothelial precursor cells express tumor endothelial marker 1/endosialin/CD248. Mol Cancer Ther. 2008;7:2536–2546. doi: 10.1158/1535-7163.MCT-08-0050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.