Abstract
Cell-cycle defects are responsible for cancer onset and growth. We studied the expression profile of 60 genes involved in cell cycle in a series of malignant mesotheliomas (MMs), normal pleural tissues, and MM cell cultures using a quantitative polymerase chain reaction-based, low-density array. Nine genes were significantly deregulated in MMs compared with normal controls. Seven genes were overexpressed in MMs, including the following: CDKN2C, cdc6, cyclin H, cyclin B1, CDC2, FoxM1, and Chk1, whereas Ube1L and cyclin D2 were underexpressed. Chk1 is a principal mediator of cell-cycle checkpoints in response to genotoxic stress. We confirmed the overexpression of Chk1 in an independent set of 87 MMs by immunohistochemistry using tissue microarrays. To determine whether Chk1 down-regulation would affect cell-cycle control and cell survival, we transfected either control or Chk1 siRNA into two mesothelioma cell lines and a nontumorigenic (Met5a) cell line. Results showed that Chk1 knockdown increased the apoptotic fraction of MM cells and induced an S phase block in Met5a cells. Furthermore, Chk1 silencing sensitized p53-null MM cells to both an S phase block and apoptosis in the presence of doxorubicin. Our results indicate that cell-cycle gene expression analysis by quantitative polymerase chain reaction can identify potential targets for novel therapies. Chk1 knockdown could provide a novel therapeutic approach to arrest cell-cycle progression in MM cells, thus increasing the rate of cell death.
Malignant pleural mesothelioma (MM) is a fatal disease with increasing incidence worldwide and a poor short-term outcome.1 Similar to other neoplastic diseases, the hallmark of MM is uncontrolled cell growth and proliferation.2 Cell division is tightly regulated by oscillations of cyclin/cyclin-dependent kinase (CDK) complexes. The inhibitory control of these associations is exerted by cyclin-dependent kinase inhibitors (CDKIs).3 Cell-cycle control has important checkpoints such as the G1-S transition, regulated by a hypophosphorylated Rb gene product (pRb). No functional loss of Rb has been described in mesothelioma,4 whereas the INK4a/ARF locus is believed to be involved in mesothelioma oncogenesis.5,6 This locus encodes two proteins, p16INK4a and p14ARF. p16INK4a has a tumor-suppressor effect linked to inhibition of CDKs and induction of G1 phase cell-cycle arrest. Alternatively, p14ARF stabilizes p53 through binding and degradation of MDM2. Mechanisms involved in p16INK4a/ARF locus inactivation as p16 homozygous deletion or promoter-methylation has been reported in 85% and 10% of mesothelioma cell lines and in 22% and 27% of tumor specimens, respectively.5,7,8 Moreover loss of p16 was demonstrated by microarray analysis6,9 or immunohistochemistry10 as an independent predictor of poor survival in patients with MM. Other CDK inhibitors, such as p27 (p27kip1) and p21 (WAF/CIP1) have been recognized as potential prognostic markers in mesothelioma. In particular, there was a positive correlation between higher levels of p27 protein and increased survival times.11,12
Aurora kinases A and B have been related to a more aggressive clinical course of MM and with decreased survival via global gene expression profiling. Only a few studies evaluated the expression of genes involved in the cell-cycle pathway using high-throughput technology.6 Therefore the precise knowledge of alterations in critical pathways in mesothelial cancers could provide effective targets for novel therapies that could improve survival of MM patients.
We aimed to elucidate the expression pattern of 60 genes involved in cell-cycle control using a low-density array platform (microfluidic card, MCF) based on real-time RT-PCR. We studied 45 primary MMs, 3 cell lines, 4 primary cultures and appropriate normal controls. To determine whether the identified gene expression alterations could be used as targets for therapeutic intervention, we performed in vitro silencing of Chk1 in nontumor as well as in tumor mesothelioma cells. Moreover we explored the potential synergistic interactions of Chk1 siRNA with a standard pharmacological treatment (doxorubicin).
Materials and Methods
Patients and Sample Characteristics
Forty-five MM tumor samples were obtained at surgery at four institutions, A.O. San Paolo, Istituto di Ricerca e Cura a Carattere Scientifico Fondazione Ospedale Maggiore, Istituto Clinico Humanitas, and University of Chieti Medical School Hospital. Mesothelioma specimens were obtained from patients who underwent multiple thoracic biopsies for diagnostic purposes. None of them received chemotherapy or radiotherapy before surgery. Informed consent was obtained from all patients under study. Adequacy of samples was evaluated by standard hematoxylin and eosin (H&E) staining on frozen sections. The remaining material was used for diagnostic purposes. Samples containing at least 80% tumor cells and less than 10% inflammatory components were included in the study.
The median age of the patients was 64.7 years (range, 40 to 84 yrs). Ten patients were women (22%) and thirty-five were men (78%). No patient had received chemotherapy or radiotherapy before surgery. According to World Health Organization classification,13 30 MMs were classified as epithelioid (66.7%), 5 as sarcomatoid (11.1%), and 10 cases were biphasic (22.2%). For statistical purposes, sarcomatoid and biphasic MMs were grouped together in the nonepithelioid (non-E) group.
Normal tissue counterparts were obtained by pleural wiping of surgical samples without evidence of pleural disease. Briefly, the mesothelial-lined surfaces of pneumonectomy or lobectomy specimens obtained from surgical procedures were gently wiped with a sterile swab. The swabs were partly smeared and slides stained with Giemsa and cytologically evaluated. Only samples with presence of at least 80% of mesothelial cells and lack of blood and/or of inflammatory components were included in the study. After the morphological evaluation, swabs were submerged in RNA extraction buffer and processed for MCF analysis (see below).
Two mesothelioma cell lines, MSTO-211H and H2452, representative of biphasic and epithelioid subtypes, respectively, and a nontumorigenic mesothelial cell line, Met5a, were obtained from American Type Culture Collection (Manassas, VA) and cultured in a 5% humidified incubator at 37°C following American Type Culture Collection suggested protocol. All cell culture reagents were from Gibco-Invitrogen (Milan, Italy), except for epidermal growth factor, hydrocortisone, and insulin (Sigma-Aldrich, Milan, Italy) required for Met5A culture. Frozen pellets of four previously described primary mesothelioma cell cultures,14 were analyzed as well.
A separate series of MMs was selected for immunohistochemistry analysis with tissue microarray (TMA) technique (see below). The TMA study population consisted of 87 MM patients. According to World Health Organization classification,13 62 MMs were classified as epithelioid (71%), 21 as biphasic (24%), and 4 cases were sarcomatoid (5%). Pleural biopsies with no neoplastic involvement from patients with pleural effusion were used as controls.
RNA Extraction and cDNA Synthesis
Samples were homogenized by TissueLyser (Qiagen, Valencia, CA) and total RNA was extracted with the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. One μg of total RNA for each sample was reverse-transcribed through a high-capacity cDNA archive kit in a final volume of 100 μl at the following conditions: 25°C for 10 minutes, 37°C for 120 minutes, and 85°C for 5 minutes. All reagents and instrument were from Applied Biosystems (Foster City, CA).
Microfluidic Card Analysis
We configured the microfluidic card to contain primers and probes for 60 cell-cycle-related genes (listed in Supplemental Table S1 at http://ajp.amjpathol.org), and four housekeeping genes (ACTB, TBP, HMBS, and GAPDH). Every sample was evaluated in triplicate and each card was loaded with two samples at a time. One hundred ng of cDNA from each sample were filled per port, according to the manufacturer’s instructions and the cards were run on an ABI Prism 7900HT sequence detection system. Real-time raw data were then transformed in threshold cycles (Ct) by SDS 2.1 software. All genes but one (CUL4A, Hs00180170_m1) could be successfully detected by the instrument (Ct <40) in all of the samples. All reagents, instruments, and software were from Applied Biosystems.
Ct values of all targets were converted into relative quantities (RQ) of mRNA using geNorm software (http://medgen.ugent.be/∼jvdesomp/genorm).15 Briefly, the geometrical average of three user-defined housekeeping genes (ACTB, HMBS, and TBP) was used to calculate the normalization factor for each sample, and the expression value of target genes (RQ) is relative to this number. RQ values were median-normalized and log2-transformed for statistical analysis.
Statistical Analysis
For unsupervised hierarchical clustering log2-transforned RQs of the successfully amplified cell-cycle-related genes were imported in dChip software (http://www.hsph.harvard.edu/∼cli/complab/dchip) Relative abundance of each gene was standardized by software preprocessing steps and hierarchical clustering analysis was then performed using the Euclidean distance as sample distance metric and the average as a linkage method. To investigate differences in cell-cycle gene expression between MMs and normal samples, all genes were analyzed by univariate statistic (Welch t-test). A target was considered differentially expressed between MMs and normal samples if these two criteria were fulfilled: a ratio of mean expression in tumoral and normal samples >2 or <0.5 (fold change, FC >2 or <0.5); and a P value <0.01. For tumor histotype (E-MMs and non-E MMs)-related differences in the expression levels of the 59 cell-cycle genes we used univariate t-statistic setting the threshold for significant difference at P < 0.05.
Tissue Microarray (TMA) Construction
Representative tissue blocks from an independent series of 87 MMs were chosen to construct TMA as previously described.16 From each TMA block a 4-μm-thick section was cut, stained with H&E, and inspected for adequacy before immunohistochemical analysis.
Immunohistochemistry
Four-μm-thick sections cut from TMA blocks were stained with rabbit polyclonal Chk1 primary antibody (1:4000, C9358; Sigma-Aldrich) overnight at 4°C. For antigen retrieval slides were microwaved for 35 minutes in ethylenediaminetetraacetic acid solution. Immunohistochemistry was performed using a DAKO instrument (DAKO, Milan, Italy) and immunostaining was revealed by the DAKO EnVision detection kit with peroxidase/diaminobenzidine as chromogen. All slides were counterstained with hematoxylin.
Immunoreactivity was independently evaluated by two authors (M.F. and S.R.). Chk1 immunostaining was evaluated both at cytoplasmic and at nuclear level. Chk1 immunoreactivity in MMs was compared with normal/reactive mesothelial samples available in four TMA spots. Tumors were classified in the: i) negative group, when no immunoreactivity could be detected; ii) low-expressor group when Chk1 protein presence was comparable to that observed in nonneoplastic spots; iii) overexpressor group, when Chk1 immunoreactivity was higher than normal controls.
siRNA Transfection Experiments
For Chk1 siRNA experiments, MSTO-211H, H2452, and Met5a cells were plated at a density of 2 × 106 cells/per well in six-well plates, and cells were maintained overnight in 2 ml of RPMI 1640 (for MSTO_221H and H2452) or Medium199 (for Met5a) supplemented with 5% fetal bovine serum. One day later, cells were transfected with 100 pmol of either Chk1-directed siRNA (siChk1, siGENOME SMARTpool M-003255) or control nontargeting siRNA (scr, siCONTROL Pool D-001206–13), all from Dharmacon Inc. (Lafayette, CO) using 3 μl of oligofectamine in a final volume of 1 ml of OptiMem medium (both from Invitrogen). Six hours after transfection, OptiMem mixture was replaced with 2 ml of appropriate growth media supplemented with 5% fetal bovine serum either with or without 500 nmol/L doxorubicin (+dox, Sigma-Aldrich). For each experiment, three plates per cell line were prepared simultaneously. After 48 hours, cells from the three matched wells were harvested and pooled for uniformity of subsequent analyses. Cell were trypsinized, washed once in 5 ml of phosphate-buffered saline (PBS), and resuspended in 3 ml of PBS. Then three pellets were made for each cell line for immunoblotting, real-time RT-PCR, and fluorescence-activated cell sorting analyses.
Western Blot Analysis
For immunoblotting, cells were solubilized in 150 μl of lysis buffer containing 20 mmol/L Tris, pH 7.2, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 150 mmol/L NaCl, 1 mmol/L ethylenediaminetetraacetic acid, plus 5% protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Protein in all samples were quantified by Bio-Rad (Hercules, CA) assay and for each specimen 50 μg were separated on 12% sodium dodecyl sulfate-polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and analyzed for Chk1 (1 μg/μl, C9358) expression. β-Actin (1:10,000, clone AC-15, A5441) was used as loading control. Both primary antibodies were from Sigma-Aldrich and were used with detection by chemiluminescence (Plus Reagents; GE Health Care, Piscataway, NJ).
Real-Time RT-PCR
For silencing experiments 200 ng of total extracted RNA from MSTO-211H, H2452, and from Met-5a cells were retro-transcribed as described above. Five μl of cDNA were then amplified along with specific primers and probes for CHK1 and β-actin (Hs00176236_m1 and Hs99999903_m1, respectively, TaqMan assays-on-demand). Chk1 expression relative to β-actin was then calculated using the 2^(−ΔΔCt) formula (Applied Biosystems, User Bulletin No. 2, 1997; Updated 2001). For all cell lines, the scramble-transfected cells were used as calibrator (1X) samples, and Chk1 levels in siChk1-cells are expressed as n-fold the corresponding 1X sample. All reagents, instruments, and protocols were from Applied Biosystems.
Cell-Cycle Analysis of siRNA-Incubated MM Cells
Cells transfected with either Chk1 siRNA (siChk1) or control siRNA (scr) alone or in the presence (+dox) of 500 nmol/L doxorubicin, were resuspended in 0.6 ml of 1% fetal bovine serum/PBS and 1.4 ml of cold 100% ethanol was added. Finally, the cells were washed twice with PBS, stained with PI supplemented with RNase (BD Biosciences, Franklin Lakes, NJ), and incubated in the dark at room temperature for 15 minutes. The samples were analyzed for DNA content profile by flow cytometry (Cell Quest, BD Biosciences) and 10,000 events were recorded for each sample. The percentages of cells in sub-G1, G1, S, and G2-M phases were obtained through Cell Quest software.
Results
Gene Expression Analysis
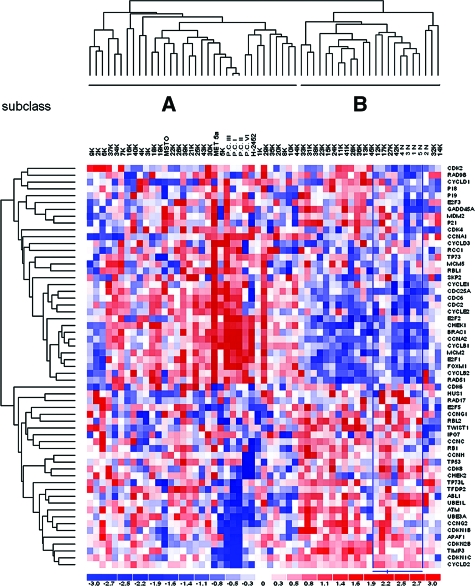
The expression level of 60 cell-cycle-regulated genes was determined in a series of MMs, normal pleura specimens, and MM cell lines. All transcripts but one in the whole dataset could be successfully amplified (Supplemental Table S1 at http://ajp.amjpathol.org). To disclose intrinsic differences among samples, we initially performed unsupervised analysis. Hierarchical clustering of quantitative polymerase chain reaction-array data delineated two main subclasses of samples (A and B sets, Figure 1). The majority of neoplastic samples (27 MMs, 60%) clustered together along with primary and commercial cell lines under the A branch of the dendrogram. The B branch included all five normal pleura specimens and 18 MMs with a cell-cycle gene expression signature similar to normal tissues. Comparison of mesotheliomas belonging to the A or B branches did not reveal differences in tumor histotype or clinicopathological variables (data not shown). After supervised analysis, the expression of nine genes was significantly different between normal (N) and MM (K) specimens (Welch t-test, P < 0.01; Table 1). Seven genes were overexpressed whereas two were underexpressed in MMs. Expression levels of this set of genes in primary cultures (PCs) and the cell lines Met5a (nontumorigenic mesothelial cells), MSTO-211H (biphasic mesothelioma cell line), and H2452 (epithelioid mesothelioma cell line) are also shown in Table 1 for comparison.
Figure 1.
Expression profile of cell-cycle-related genes. Unsupervised hierarchical clustering of 45 MMs (K), 5 normal pleura (N), 3 commercial cell lines (MET5A, H2452, and MSTO-211H), and four primary cultures (PC) is displayed. In the A branch of the dendrogram the majority of tumor samples and all cell cultures were gathered; the B branch included all normal pleura and the remaining MM tissues.
Table 1.
Differentially Expressed Cell Cycle-Related Genes between Normal and MM Specimens
| Gene name | Cell cycle phase | FC K/N | P value K/N | Average N | Average K | Average PC | Met5A RQ | MSTO-211H RQ | H2452 RQ |
|---|---|---|---|---|---|---|---|---|---|
| UBE1L | Ubiquitin-proteasome pathway | 0.4 | 1.4E-05 | 2.93 | 1.14 | 0.23 | 2.32 | 0.57 | 0.40 |
| CCND2 | G1 | 0.5 | 3.0E-06 | 2.38 | 1.21 | 0.02 | 0.00 | 0.63 | 0.00 |
| CHK1 | G2/M | 3.9 | 4.7E-06 | 0.30 | 1.17 | 2.66 | 3.30 | 1.18 | 2.57 |
| CCNH | S | 2.1 | 0.00025 | 0.72 | 1.48 | 1.24 | 0.61 | 2.52 | 0.95 |
| CCNB1 | G2/M | 4.7 | 0.00026 | 0.29 | 1.35 | 6.74 | 3.91 | 0.76 | 5.67 |
| p18-CDKN2C | G1 | 3.0 | 0.00098 | 0.61 | 1.85 | 1.21 | 2.79 | 0.10 | 1.37 |
| CDC2 | G2/M | 5.7 | 0.0021 | 0.25 | 1.43 | 2.93 | 11.64 | 0.54 | 4.54 |
| FOXM1 | G2/M | 3.6 | 0.0044 | 0.34 | 1.22 | 5.34 | 16.17 | 1.37 | 7.78 |
| CDC6 | S | 4.2 | 0.0057 | 0.26 | 1.09 | 3.99 | 5.96 | 1.15 | 3.18 |
RQs in primary and commercial cell lines are indicated as well.
RQ, relative quantity; PC, primary MM cell cultures; FC, fold change.
P values indicate the statistical significance at univariate analysis (t-statistic).
MM Histotype Analysis
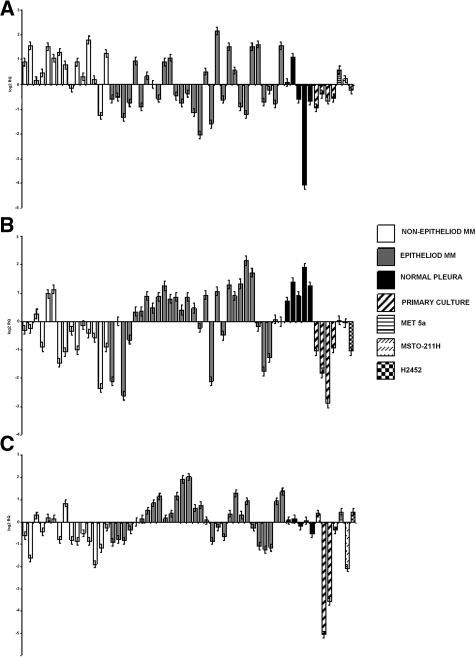
Comparison of epithelioid (E-MMs) to nonepithelioid (non-E MMs) mesotheliomas showed that the mRNA expression level of three genes was significantly associated with tumor histotype (univariate analysis, t-statistic, P value <0.05) (Table 2). Two genes were overexpressed in E-MMs, TFDP2 and ABL1, whereas TWIST1 was overexpressed in the non-E group. The mRNA levels of these genes in all samples are shown in Figure 2, A–C.
Table 2.
Differentially Expressed Genes in Epithelioid (E) and Nonepithelioid (Non-E) MMs
| Gene name | P value | FC E/ Non-E |
|---|---|---|
| TWIST1 | 0.007569093 | 0.6757 |
| TFDP2 | 0.003943293 | 1.8535 |
| ABL1 | 0.038220056 | 1.7324 |
The complete list of cell cycle-related genes was investigated by univariate analysis (t-test).
Figure 2.
MM histotype-related gene expression analysis. The expression levels of the three genes differentially expressed in epithelioid relative to nonepithelioid MMs (P < 0.05) is represented in the whole samples dataset. Twist 1 (A) was overexpressed in nonepithelioid MMs whereas TFD2 and Abl1 (B and C, respectively) were elevated in the epithelioid histotype. Bars represent the relative quantity (RQ) of the gene in each sample (RQ ± SD).
Chk1 Expression Analysis
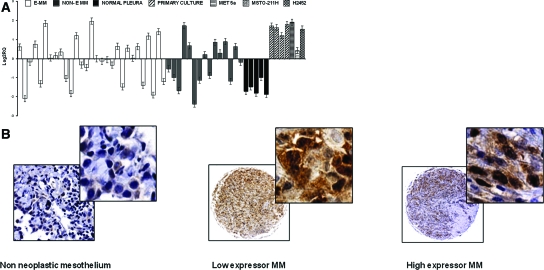
Chk1 was one of the seven genes significantly overexpressed in MMs relative to normal samples (FC = 3, 9; P value <0.001). Chk1 distribution in the two mesothelioma histotypes was heterogeneous although not significantly different, whereas normal pleural tissues consistently displayed low levels of Chk1 (Figure 3A). Twenty-four of forty-five MMs (53%) displayed Chk1 mRNA levels above the median value.
Figure 3.
Chk1 overexpression in MM tissues compared with nonneoplastic mesothelial samples. A: Chk1 mRNA expression profile in tissues and cell lines. Relative quantity (RQ) and SD are represented for each sample. B: Chk1 immunoreactivity is shown for representing cores of nonneoplastic mesothelium, and low- and high-expressor MMs. Expression of Chk1 protein is detectable in both cytoplasmic and nuclear compartments. Original magnifications: ×250 (B, left); ×40 (B, middle and right); ×400 (B, insets).
To confirm mRNA data, we evaluated Chk1 protein expression in an independent series of MMs. Chk1 protein presence in MM cores was detected at both the nuclear and cytoplasmatic level. Nonneoplastic pleural mesothelial cells displayed weak cytoplasmic Chk1 immunoreactivity (Figure 3B). Fourteen MMs (16%; 11 epithelioid, 1 sarcomatous, and 2 biphasic) were negative for Chk1 immunostaining. Comparable immunoreactivity to nonneoplastic cores was detected in 32 MMs (37%; 22 epithelioid and 10 biphasic). Forty-one MMs (47%; 32 epithelioid, 3 sarcomatous, and 6 biphasic) displayed Chk1 overexpression. Representative cores of Chk1 staining are illustrated in Figure 3B. Chk1 overexpression in almost half of the patients was therefore confirmed by two different techniques in two independent sets of MMs (53% and 47% of MMs by MCF and TMA analyses, respectively).
Chk1 Silencing
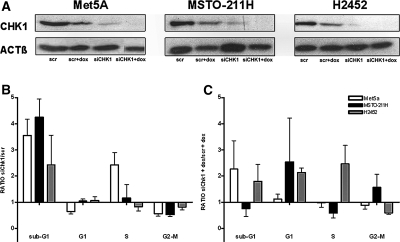
To determine whether down-regulation of Chk1 expression would affect cell-cycle phases, MM commercial cell lines and the nonneoplastic cells Met5a were transfected with either control (scr) or Chk1 (siChk1) siRNAs alone and in combination with doxorubicin. Both Chk1 protein and mRNA levels decreased after 48 hours of siRNA incubation (Figure 4A and Supplemental Figure S1 at http://ajp.amjpathol.org, respectively).
Figure 4.
Chk1 siRNA sensitizes MM cells to apoptosis and to doxorubicin toxicity. Cells were transfected with either control siRNA (scr) or Chk1 siRNA (siChk1) alone or in combination with 500 nmol/L doxorubicin (+dox) and harvested after 48 hours for immunoblotting and FACS analysis. A: Met5A, MSTO-211H, and H2452 cells lysates were immunoblotted for Chk1 detection. Actin was probed as loading control. B: Chk1 knockdown increases cell death rate in MM cells. Met5A, MSTO-211H, and H2452 cells transfected with either siChk1 or scr were stained with PI and analyzed for DNA content profile by FACS analysis. The percentages of cells in sub-G1, G1, S, and G2-M phases were obtained through Cell-Quest software. The percentage of cells treated with siChk1 relative to scr-incubated cells (ratio) is shown for each cell line. C: Chk1 siRNA sensitizes p53-null cell to doxorubicin toxicity. Met5A, MSTO-211H, and H2452 cells were transfected as described above and incubated with 500 nmol/L doxorubicin. The percentage of cells in each phase was determined and values of siChk1-treated cells are expressed as ratios relative to corresponding controls (see text and Supplemental Figure S2 for details at http://ajp.amjpathol.org). Results represent mean values from three independent experiments and error bars indicate SD.
In the absence of a DNA-damaging agent, down-regulation of Chk1 protein perturbed cell-cycle phases compared with control siRNA. Fluorescence-activated cell sorting analysis showed an increase of apoptotic cells in all cell lines (Figure 4B and Supplemental Figure S2 at http://ajp.amjpathol.org). Furthermore we observed an S phase block and decreased G1 and G2/M phases in Met5a. Conversely, in MSTO-211H and H2452 cells, G1, S, and G2/M phases were unaffected (Figure 4B).
Incubation with doxorubicin alone modified cell-cycle phases in all cell lines. In particular, all cells displayed an increase in sub-G1 (apoptosis) phase (Supplemental Figure S2 at http://ajp.amjpathol.org). When the combination of silencing and doxorubicin was performed, there was an increase of apoptosis in Met5a and in H2452 cell lines, and a G1 block in MSTO-211H cells. H2452 was the only cell line to display an S phase block (Figure 4C and Supplemental Figure S2 at http://ajp.amjpathol.org).
Discussion
The present study, based on the microfluidic card technology, identified specific cell-cycle gene expression alterations in a series of MM tissues and cells. In particular, the expression levels of nine genes were significantly different in MM samples relative to normal pleura. These alterations included a significant downexpression of Ube1L, a putative tumor suppressor gene frequently deleted in lung cancer,17 as well as increased expression of Chk1, cyclins H and B1, FoxM1, Cdc6, and cyclin inhibitor p18/CDKN2C. Of notice among the seven overexpressed genes four (Chk1, p34-CDC2, cyclin B1, and FOXM1) are involved in G2/M phase regulation of the cell cycle. One of these genes, Chk1, was then chosen for siRNA-mediated knockdown in MM cell lines. Chk1 is a serine/threonine kinase involved in DNA damage-induced checkpoint and in cell-cycle progression through both S and G2/M phases.
We focused our study on Chk1 based on its importance as central regulator of cell-cycle progression and cellular response to DNA-damaging chemotherapy.18,19 Although both Chk1 and FOXM1 were overexpressed in all three cell lines and down-regulated in all nonneoplastic pleural tissues, Chk1 showed a more significant P value. Finally, its selective chemical inhibitor (as UCN01) is now under clinical trial. To determine whether MM pharmacological approach could benefit from Chk1 inhibition, we first confirmed Chk1 overexpression at protein level in an independent set of MMs and then we knocked-down Chk1 in vitro in combination or not with doxorubicin treatment. Interestingly, the same proportion of Chk1-overexpressing MMs could be observed at both gene and protein expression investigations.
In response to silencing alone or in association with doxorubicin treatment, our data showed different cell-cycle profiles in the three cell lines. Inhibition of Chk1 in the absence of the DNA-damaging agent increased cell death in all cell lines indicating that Chk1 played also a role in survival of MM cells. This finding is in agreement with a previous report documenting the induction of apoptosis in H1299 cells by Chk1 anti-sense20 although other studies observed no increase of apoptotic cells after siRNA treatment.21
Chk1 knockdown induced an S phase block in Met5a cells, and a weak arrest in S phase in MSTO-211H cells only. This effect may be explained by the different genetic background of these three cell lines. In fact, in the nontumorigenic cells Met5a there is a preservation of both wild-type p16/CDKN2A and p53 proteins (Sanger Institute, Somatic Mutation in Cancer Database). Conversely, both H2452 and MSTO-211H harbor a mutation for CDKN2A. Moreover, H2452 cells bear a truncating mutation in p53 gene, whereas MSTO-211H cells do not.22 Because CDKN2A and p53 are both involved in cell-cycle regulation and G1/S transition, different genetic abnormalities can therefore induce different cell-cycle pattern after Chk1 silencing.
MM cell line sensitivity toward the DNA-damaging agent doxorubicin was also ascertained. Doxorubicin alone increased cell death in all cell lines, as measured by sub-G1 cells, although to a lesser extent in the p53-mutated H2452. In this cell line there was a sustained fraction of cells in mitosis (G2/M), confirming the previous findings about the inefficiency of DNA-damaging agents to induce cell division arrest and apoptosis in tumor cells lacking p53.23 When the combination of Chk1-siRNA and doxorubicin was added in H2452 cells, G2/M phase decreased and higher apoptotic and S phase-blocked cells were present. In the p53-proficient cell line MSTO-211H the combination doxorubicin/siChk1 did not increase either cell death rate (sub-G1 phase) or replicating cells (S phase), suggesting that in presence of intact p53 pathway Chk1 silencing does not improve anti-tumor drug effectiveness. Taken together, these observations indicate that incubation of MSTO-211H and H2452 cells with Chk1 siRNA sensitize them to apoptosis induction, and Chk1 silencing improves the response of p53-null H2452 cells to the DNA-damaging drug doxorubicin.
Further supporting Chk1 knockdown as potential therapeutic approach in MM to impair cell-cycle progression and to sensitize tumor cells to chemotherapeutic agents, the present investigation documents a general increase in mRNA levels of other genes involved in the G2/M phase, such as FoxM1, cyclin B1, and its partner p34-Cdc2. FoxM1 is a transcription factor that regulates the expression of G2/M-specific genes, including cyclin B, eventually responsible for entry into mitosis.24 FoxM1 has been previously found overexpressed in cervical squamous carcinoma25 and in human breast cancer,26 thereby underscoring its importance in cell proliferation and transformation.
Cyclin B1 (CCNB1) complexes with p34 (cdc2), its catalytic partner of the mitosis-promoting factor (MPF) kinase complex, to induce the entry into mitosis and G2-M transitions. Although cyclin B1 overexpression plays a crucial role in different tumors,27 its involvement in mesothelioma, together with p34-cdc2, was demonstrated only in vitro,28,29,30 and a decrease in cell proliferation was associated with reduced cyclin B1/cdc2 activity.28 We found overexpression of S phase players such as cyclin H (CCNH) and cdc6 transcripts, whereas a G1-protein such as cyclin D2 was underexpressed as previously reported in mesothelioma.31,32 Indeed a G1-cyclin-dependent kinase inhibitor p18 (CDKN2C) was overexpressed.
Finally, we investigated differences in gene expression related to histotype. We found two genes associated with epithelioid MMs (TFDP2 and ABL1) and one (TWIST 1) with nonepithelioid MMs. TFDP2 interacts with retinoblastoma gene (RB) and induces gene activation. RB interacts with E2F family of transcription factors to arrest cells in G1.33 The involvement of TFDP2 in mesothelioma cancerogenesis has not been demonstrated yet, but previous reports34,35 showed that it is overexpressed in HPV-positive head and neck squamous cell carcinomas, along with up-regulated retinoblastoma-binding protein (p18) and replication factor-C genes. In addition it plays a role in uncontrolled proliferation of ovarian cancer cells.36
ABL1 is a proto-oncogene coding for a cytoplasmic and nuclear tyrosine kinase. Chromosomal rearrangements of ABL1 or viral transduction are responsible for malignant transformation, such as in myeloid leukemia. Abl1 overexpression has not been previously reported in MM tissues but its role has been investigated in oral squamous cell carcinomas in which it is correlated to a worse outcome.37 Moreover chromosomal rearrangements of Abl1 into the chimeric Bcr/Abl oncogene are the target for tyrosine kinase inhibitor compounds such as Imatinib, STI571, or CP57148B. This observation is noteworthy and should be further investigated for therapeutic purposes.
Of interest, TFDP2 and Abl1 are genes involved in cell-cycle regulation at G1-S checkpoint through interaction with pRb pathway.38 Our finding suggests that epithelioid MMs could deregulate pRb pathway to trigger cell-cycle progression. TWIST1 encodes a nuclear protein with a helix-loop-helix domain shared by DNA binding proteins and acts as a transcription factor. Twist1 play a crucial role in regulating the metastatic process39 and it is involved in the epithelial-mesenchymal transition (EMT) through the loss of adhesion mediated by E-cadherin.40,41,42 This role is supported by our result, because we documented Twist1 overexpression in biphasic and sarcomatoid MMs compared with the epithelioid group.
Previous gene expression studies in MM patients performed on an oligonucleotide array-based platform did not reveal a number of significant differences in expression of cell-cycle-related genes between MMs and normal counterparts,43 indeed down-regulation of genes involved in just pRb pathway could be identified.44 In particular, overexpression of some cell-cycle control genes (Aurora kinase A and B) as well as loss of p16/INK4A were recently related to a more aggressive clinical course of MMs.6
Microfluidic card technology has the recognized advantage to be specific and accurate in mRNA quantification.45 The low-density array platform we used is based on real-time RT-PCR technology, which is now considered the gold standard for gene expression quantitative studies. Therefore this technology could be useful to investigate gene expression signatures in MM, with higher accuracy and lower interlaboratory variability than array-based platforms.45
In conclusion, we identified deregulation of cell-cycle-related genes mostly occurring at the G2/M phase. We confirmed up-regulation at protein level of an overexpressed gene (Chk1) in an independent set of MM patients. Moreover, we showed that the siRNA-mediated knockdown of Chk1 could sensitize MM cells to cell-cycle arrest and apoptosis induction. Deregulation of cell-cycle checkpoints is now recognized as a salient feature of the malignant transformation process and checkpoint alterations in tumors provide an opportunity for developing a therapeutic strategy that combines conventional cancer treatment with inhibitors of cell-cycle checkpoints.46,47 This is based on the premise that pharmacological impairment of checkpoint function may selectively sensitize tumors with intrinsic checkpoint defects to drug-induced genotoxic stress.19,48 Moreover, chemical Chk1 inhibitors (as UCN01) are now under clinical testing in combination with standard chemotherapy for solid tumors (NCT00036777),49,50,51 and lymphomas (phase II, NCI-04-C-0173). Consistent with this issue, we showed that inhibition of Chk1 in MM cancer cells potentiate the effects of DNA-damaging agents, in particular in p53-deficient ones. This finding could offer a novel therapeutic approach for patients with mesothelioma.
Supplementary Material
Footnotes
Address reprint requests to Silvano Bosari, M.D., Division of Pathology A.O.S. Paolo, Via A. Di Rudinì 8, 20142 Milano, Italy. E-mail: silvano.bosari@unimi.it.
Supported by Ministero dell ’Istruzione, dell, ’Università e della Ricerca (grant 2004065489 to G.C., S.B., and A.M.).
S.R. and E.F. contributed equally to the study.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Robinson BW, Musk AW, Lake RA. Malignant mesothelioma. Lancet. 2005;366:397–408. doi: 10.1016/S0140-6736(05)67025-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Kratzke RA, Otterson GA, Lincoln CE, Ewing S, Oie H, Geradts J, Kaye FJ. Immunohistochemical analysis of the p16INK4 cyclin-dependent kinase inhibitor in malignant mesothelioma. J Natl Cancer Inst. 1995;87:1870–1875. doi: 10.1093/jnci/87.24.1870. [DOI] [PubMed] [Google Scholar]
- Cheng JQ, Jhanwar SC, Klein WM, Bell DW, Lee WC, Altomare DA, Nobori T, Olopade OI, Buckler AJ, Testa JR. p16 alterations and deletion mapping of 9p21-p22 in malignant mesothelioma. Cancer Res. 1994;54:5547–5551. [PubMed] [Google Scholar]
- López-Ríos F, Chuai S, Flores R, Shimizu S, Ohno T, Wakahara K, Illei PB, Hussain S, Krug L, Zakowski MF, Rusch V, Olshen AB, Ladanyi M. Global gene expression profiling of pleural mesotheliomas: overexpression of Aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer Res. 2006;66:2970–2979. doi: 10.1158/0008-5472.CAN-05-3907. [DOI] [PubMed] [Google Scholar]
- Hirao T, Bueno R, Chen CJ, Gordon GJ, Heilig E, Kelsey KT. Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis. 2002;23:1127–1130. doi: 10.1093/carcin/23.7.1127. [DOI] [PubMed] [Google Scholar]
- Wong L, Zhou J, Anderson D, Kratzke RA. Inactivation of p16INK4a expression in malignant mesothelioma by methylation. Lung Cancer. 2002;38:131–136. doi: 10.1016/s0169-5002(02)00178-2. [DOI] [PubMed] [Google Scholar]
- Ladanyi M. Implications of P16/CDKN2A deletion in pleural mesotheliomas. Lung Cancer. 2005;49(Suppl 1):S95–S98. doi: 10.1016/j.lungcan.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Borczuk AC, Taub RN, Hesdorffer M, Hibshoosh H, Chabot JA, Keohan ML, Alsberry R, Alexis D, Powell CA. P16 loss and mitotic activity predict poor survival in patients with peritoneal malignant mesothelioma. Clin Cancer Res. 2005;11:3303–3308. doi: 10.1158/1078-0432.CCR-04-1884. [DOI] [PubMed] [Google Scholar]
- Beer TW, Shepherd P, Pullinger NC. p27 immunostaining is related to prognosis in malignant mesothelioma. Histopathology. 2001;38:535–541. doi: 10.1046/j.1365-2559.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- Bongiovanni M, Cassoni P, De Giuli P, Viberti L, Cappia S, Ivaldi C, Chiusa L, Bussolati G. p27(kip1) immunoreactivity correlates with long-term survival in pleural malignant mesothelioma. Cancer. 2001;92:1245–1250. doi: 10.1002/1097-0142(20010901)92:5<1245::aid-cncr1444>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Lyon: IARC Press,; World Health Organization Classification of Tumours. Pathology, Genetics, Tumours of the Lung, Pleura, Thymus and Heart. 2004:pp 126–127. [Google Scholar]
- Catania A, Colombo G, Carlin A, Garofalo L, Gatti S, Buffa R, Carboni N, Rosso L, Santambrogio L, Cantalamessa L, Lipton JM. Autocrine inhibitory influences of alpha-melanocyte-stimulating hormone in malignant pleural mesothelioma. J Leukoc Biol. 2004;75:253–259. doi: 10.1189/jlb.0603264. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034.1–RESEARCH0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis M, Pellegrini C, Cannone M, Arizzi C, Coggi G, Bosari S. Quantitative PCR and HER2 testing in breast cancer: a technical and cost-effectiveness analysis. Am J Clin Pathol. 2008;129:563–570. doi: 10.1309/1AKQDQ057PQT9AKX. [DOI] [PubMed] [Google Scholar]
- Pitha-Rowe I, Petty WJ, Feng Q, Koza-Taylor PH, Dimattia DA, Pinder L, Dragnev KH, Memoli N, Memoli V, Turi T, Beebe J, Kitareewan S, Dmitrovsky E. Microarray analyses uncover UBE1L as a candidate target gene for lung cancer chemoprevention. Cancer Res. 2004;64:8109–8115. doi: 10.1158/0008-5472.CAN-03-3938. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Semizarov D, Sowin TJ, Rosenberg SH, Zhang H. Novel indication for cancer therapy: Chk1 inhibition sensitizes tumor cells to antimicotics. Int J Cancer. 2005;115:528–538. doi: 10.1002/ijc.20770. [DOI] [PubMed] [Google Scholar]
- Luo Y, Rockow-Magnone SK, Kroeger PE, Frost L, Chen Z, Han EK, Ng SC, Simmer RL, Giranda VL. Blocking Chk1 expression induces apoptosis and abrogates the G2 checkpoint mechanism. Neoplasia. 2001;3:411–419. doi: 10.1038/sj.neo.7900175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Xiao Z, Chen J, Ng SC, Sowin T, Sham H, Rosenberg S, Fesik S, Zhang H. Human Chk1 expression is dispensable for somatic cell death and critical for sustaining G2 DNA damage checkpoint. Mol Cancer Ther. 2003;2:543–548. [PubMed] [Google Scholar]
- Manfredi JJ, Dong J, Liu WJ, Resnick-Silverman L, Qiao R, Chahinian P, Saric M, Gibbs AR, Phillips JI, Murray J, Axten CW, Nolan RP, Aaronson SA. Evidence against a role for SV40 in human mesothelioma. Cancer Res. 2005;65:2602–2609. doi: 10.1158/0008-5472.CAN-04-2461. [DOI] [PubMed] [Google Scholar]
- Macip S, Kosoy A, Lee SW, O'Connell MJ, Aaronson SA. Oxidative stress induces a prolonged but reversible arrest in p53-null cancer cells, involving a Chk1-dependent G2 checkpoint. Oncogene. 2006;25:6037–6047. doi: 10.1038/sj.onc.1209629. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Chan D, Yu S, Chiu P, Yao K, Liu V, Cheung A, Ngan H. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- Bektas N, Haaf A, Veeck J, Wild PJ, Luscher-Firzlaff J, Hartmann A, Knuchel R, Dahl E. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer. 2008;8:42. doi: 10.1186/1471-2407-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeno S, Noguchi T, Kikuchi R, Uchida Y, Yokoyama S, Muller W. Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma. Cancer. 2002;94:2874–2881. doi: 10.1002/cncr.10542. [DOI] [PubMed] [Google Scholar]
- Vivo C, Levy F, Pilatte Y, Fleury-Feith J, Chretien P, Monnet I, Kheuang L, Jaurand MC. Control of cell cycle progression in human mesothelioma cells treated with gamma interferon. Oncogene. 2001;20:1085–1093. doi: 10.1038/sj.onc.1204199. [DOI] [PubMed] [Google Scholar]
- Kettunen E, Vivo C, Gattacceca F, Knuutila S, Jaurand MC. Gene expression profiles in human mesothelioma cell lines in response to interferon-gamma treatment. Cancer Genet Cytogenet. 2004;152:42–51. doi: 10.1016/j.cancergencyto.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Sun X, Wei L, Liden J, Hui G, Dahlman-Wright K, Hjerpe A, Dobra K. Molecular characterization of tumour heterogeneity and malignant mesothelioma cell differentiation by gene profiling. J Pathol. 2005;207:91–101. doi: 10.1002/path.1810. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Godleski JJ, Roelofs CR, Longacker JL, Bueno R, Sugarbaker DJ, Marsit CJ, Nelson HH, Kelsey KT. Asbestos burden predicts survival in pleural mesothelioma. Environ Health Perspect. 2008;116:723–726. doi: 10.1289/ehp.11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Toyooka S, Shivapurkar N, Shigematsu H, Miyajima K, Takahashi T, Stastny V, Zern AL, Fujisawa T, Pass HI, Carbone M, Gazdar AF. Aberrant methylation profile of human malignant mesotheliomas and its relationship to SV40 infection. Oncogene. 2005;24:1302–1308. doi: 10.1038/sj.onc.1208263. [DOI] [PubMed] [Google Scholar]
- Zheng N, Fraenkel E, Pabo CO, Pavletich NP. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 1999;13:666–674. doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht NF, Burk RD, Adrien L, Dunne A, Kawachi N, Sarta C, Chen Q, Brandwein Gensler M, Prystowsky MB, Childs G, Smith RV, Belbin TJ. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213:283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, Netterville JL, Levy S, Yarbrough WG, Chung CH. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- Reimer D, Sadr S, Wiedemair A, Concin N, Hofstetter G, Marth C, Zeimet AG. Heterogeneous cross-talk of E2F family members is crucially involved in growth modulatory effects of interferon-gamma and EGF. Cancer Biol Ther. 2006;5:771–776. doi: 10.4161/cbt.5.7.2750. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Lin SC, Chen YJ, Chang KM, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45:721–731. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000;19:5643–5650. doi: 10.1038/sj.onc.1203878. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Puisieux A, Valsesia-Wittmann S, Ansieau S. A twist for survival and cancer progression. Br J Cancer. 2006;94:13–17. doi: 10.1038/sj.bjc.6602876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng GZ, Zhang W, Wang LH. Regulation of cancer cell survival, migration, and invasion by Twist: AKT2 comes to interplay. Cancer Res. 2008;68:957–960. doi: 10.1158/0008-5472.CAN-07-5067. [DOI] [PubMed] [Google Scholar]
- Gordon GJ, Rockwell GN, Jensen RV, Rheinwald JG, Glickman JN, Aronson JP, Pottorf BJ, Nitz MD, Richards WG, Sugarbaker DJ, Bueno R. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol. 2005;166:1827–1840. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal S, Wiewrodt R, Malden LD, Amin KM, Matzie K, Friedberg J, Kucharczuk JC, Litzky LA, Johnson SW, Kaiser LR, Albelda SM. Gene expression profiling of malignant mesothelioma. Clin Cancer Res. 2003;9:3080–3097. [PubMed] [Google Scholar]
- Schramm A, Vandesompele J, Schulte JH, Dreesmann S, Kaderali L, Brors B, Eils R, Speleman F, Eggert A. Translating expression profiling into a clinically feasible test to predict neuroblastoma outcome. Clin Cancer Res. 2007;13:1459–1465. doi: 10.1158/1078-0432.CCR-06-2032. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol Cancer Ther. 2006;5:1935–1943. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- Tse AN, Rendahl KG, Sheikh T, Cheema H, Aardalen K, Embry M, Ma S, Moler EJ, Ni ZJ, Lopes de Menezes DE, Hibner B, Gesner TG, Schwartz GK. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–139. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- Edelman MJ, Bauer KS, Jr, Wu S, Smith R, Bisacia S, Dancey J. Phase I and pharmacokinetic study of 7-hydroxystaurosporine and carboplatin in advanced solid tumors. Clin Cancer Res. 2007;13:2667–2674. doi: 10.1158/1078-0432.CCR-06-1832. [DOI] [PubMed] [Google Scholar]
- Jimeno A, Rudek MA, Purcell T, Laheru DA, Messersmith WA, Dancey J, Carducci MA, Baker SD, Hidalgo M, Donehower RC. Phase I and pharmacokinetic study of UCN-01 in combination with irinotecan in patients with solid tumors. Cancer Chemother Pharmacol. 2008;61:423–433. doi: 10.1007/s00280-007-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RP, Lewis LD, Beelen AP, Olszanski AJ, Johnston N, Rhodes CH, Beaulieu B, Ernstoff MS, Eastman A. Modulation of cell cycle progression in human tumors: a pharmacokinetic and tumor molecular pharmacodynamic study of cisplatin plus the Chk1 inhibitor UCN-01 (NSC 638850). Clin Cancer Res. 2006;12:7079–7085. doi: 10.1158/1078-0432.CCR-06-0197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.