Abstract
Many different proteins associated with the cell cycle, including cyclins, cyclin-dependent kinases, and proto-oncogenes such as c-MYC (MYC), are increased in degenerating neurons. Consequently, an ectopic activation of the cell cycle machinery in neurons has emerged as a potential pathogenic mechanism of neuronal dysfunction and death in many neurodegenerative diseases, including Alzheimer’s disease. However, the exact role of cell cycle re-entry during disease pathogenesis is unclear, primarily because of the lack of relevant research models to study the effects of cell cycle re-entry on mature neurons in vivo. To address this issue, we developed a new transgenic mouse model in which forebrain neurons (CaMKII-MYC) can be induced to enter the cell cycle using the physiologically relevant proto-oncogene MYC to drive cell cycle re-entry. We show that such cell cycle re-entry results in neuronal cell death, gliosis, and cognitive deficits. These findings provide compelling evidence that dysregulation of cell cycle re-entry results in neurodegeneration in vivo. Our current findings, coupled with those of previous reports, strengthen the hypothesis that neurodegeneration in Alzheimer’s disease, similar to cellular proliferation in cancer, is a disease that results from inappropriate cell cycle control.
Neurons in the normal brain are viewed as being quiescent, not advancing past the G0 period of the cell division cycle. However, in Alzheimer’s disease (AD), multiple lines of evidence suggest that neurons vulnerable to degeneration emerge from this postmitotic state—phenotypically suggestive of cells that are cycling, rather than the normal terminally differentiated nondividing state.1,2,3,4,5 Notably, cell cycle alterations are not limited to AD. For example, accumulation of hyperphosphorylated pRb (ppRb) and altered localization of E2F-1 also occurs in neurons in Parkinson’s disease and amyotrophic lateral sclerosis, suggesting that neurons re-enter the G1 phase of the cell cycle.6,7,8 The successful duplication of DNA, at least in AD,9,10,11 indicates that some neurons successfully complete S phase and precludes the possibility that the re-expression of various cell cycle markers observed is merely an epiphenomena caused by reduced proteosomal activity.12
The causes or consequences of neuronal cell cycle are incompletely understood. Nonetheless, it is well known that the activation of cell cycle processes is part of the mechanism by which loss of trophic support during development leads to neuronal cell death, and there is also evidence for a role for cell cycle regulators in neuronal death evoked by various stressors.13,14 In support of the importance of cell cycle reactivation in mediating cell death, when a powerful oncogene, SV40 T antigen, is expressed specifically in maturing Purkinje cells or in forebrain neurons in transgenic mice, the cells replicate their DNA (ie, initiate cell cycle), but then subsequently degenerate and die.15
Unfortunately, studies performed to date in cell culture do not faithfully reproduce age-related neurodegenerative diseases because the cell culture studies use embryo-derived primary neuronal cultures that do not accurately model adult neurons. Furthermore, in traditional transgenic mouse models,15 the expression of transgenes during embryogenesis, which results in neurodegeneration at an early age, is more akin to a developmental error than an age-dependent process associated with neurodegenerative disease. Although a recently developed inducible SV40 T-antigen transgenic model16 addressed such developmental issues, SV40 T antigen is not a physiologically relevant entity in any neurodegenerative disease. As such, a faithful and physiologically relevant model of neuronal cell cycle activation in mature neurons was lacking. To generate such a faithful model, in this study, we used the tetracycline-controlled transactivator (tTA) system to generate bitransgenic mice (CaMKII-MYC) that can be induced to overexpress human c-MYC (MYC) under the control of the CaMKII promoter that drives high transgene expression in forebrain neurons.17 MYC is well known for its oncogenic activity and overexpression of MYC is commonly associated with tumorigenesis. Indeed, previously we used the Tet system to generate mice that conditionally expressed a human MYC transgene in hematopoietic cells and hepatocytes18,19 and found that the sustained expression of the MYC transgene induced cell cycle progression and culminated in the formation of malignant T-cell lymphomas, acute myeloid leukemias, and hepatomas. Interestingly, and of physiological relevance to AD, phospho-MYC is increased in dystrophic neurites and neurons with neurofibrillary tangles in AD and MYC is induced in degenerating neurons in animal models of trauma and ischemia.20 Therefore, MYC expression in transgenic mice in our model is not only a tool for inducing cell cycle re-entry in neurons, but MYC expression itself is pathophysiologically relevant to AD and other neurodegenerative diseases.
Our findings reported in this study show that MYC induction in CaMKII-MYC transgenic mice induced neuronal-specific cell cycle re-entry, neurodegeneration, and, importantly, significant cognitive deficits. These findings continue to strengthen our novel hypothesis that neurodegeneration in AD, like cellular proliferation in cancer, is a disease of inappropriate cell cycle control.
Materials and Methods
Animals
CaMKII-tTA17 and tet-o-MYC mice18 were mated to generate CaMKII-MYC mice. Both CaMKII-tTA and tet-o-MYC mice were established in the FVB strain. A doxycycline-containing diet (200 mg/kg; Bio-Serve, Frenchtown, NJ) was provided before weaning (4 to 6 weeks after birth), and, thereafter, MYC expression was induced by replacing the doxycycline diet with a regular diet for 5 weeks. After behavioral tests, the mice were sacrificed and the brains were processed for immunocytochemistry and biochemical analysis.
Immunocytochemistry
Immunocytochemistry was performed by the ABC method according to the manufacturer’s protocol (Vector Laboratories, Burlingame, CA). Formalin-fixed brains were processed and embedded in paraffin. Six-μm-thick serial sections were cut, mounted onto slides, and rehydrated according to standard protocols. All slides were randomized and blinded with regard to genotype before staining and subsequent analysis. Briefly, slides were immersed in xylene, hydrated through graded ethanol solutions, and endogenous peroxidase activity was eliminated by incubation in 3% hydrogen peroxide for 30 minutes. To reduce nonspecific binding, sections were incubated for 30 minutes in 10% normal goat serum in Tris-buffered saline (50 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.6). After rinsing briefly with 1% normal goat serum in Tris-buffered saline, the sections were incubated overnight at 4°C with one of the following primary antibody; anti-proliferating cell nuclear antigen (PCNA) mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Ki-67 rat monoclonal antibody (DAKO, Carpinteria, CA), anti-cyclin D1 rabbit monoclonal antibodies (Lab Vision, Fremont, CA), and anti-GFAP mouse monoclonal antibody (Chemicon, Billerica, MA). Antibodies were localized using 3–3′-diaminobenzidine as a chromogen (DAKO) after incubation with a secondary antibody.
For double immunocytochemistry, the brain sections were incubated overnight at 4°C with anti-NeuN mouse monoclonal antibody (Millipore, Billerica, MA) or anti-MAP2 mouse monoclonal antibody (Millipore) in addition to anti-PCNA rabbit polyclonal antibody (Abcam, Cambridge, MA), anti-Ki-67 rat monoclonal antibody, or anti-BrdU rat monoclonal antibody (Abcam). Alexa Fluor 488- and 568-coupled secondary antibodies (Invitrogen, Carlsbad, CA) were used for detection. To exclude the possibility of nonspecific reaction, all of the immunocytochemistry experiments contained at least one sample without a primary antibody. Images were acquired through an AxioCam camera on an Axiovert 200M microscope (Zeiss, Thornwood, NY). Images were then analyzed with the Axiovision software (Zeiss). Hematoxylin and eosin (H&E) staining and Nissl stains were also performed for routine histochemical and morphological analyses.
BrdU Incorporation Analysis
DNA synthesis was directly examined by analysis of BrdU incorporation. For this experiment, 50 mg/kg of the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) (Sigma, St. Louis, MO) was intraperitoneally injected into each mouse. A day after injection, mice were anesthetized and perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. Brains were removed, cut in the sagittal plane, embedded in paraffin, sectioned, and mounted on slides. The sections were treated with 2 N hydrochloric acid for 2 hours and then washed two times with 1× Tris-buffered saline (3 minutes each) to neutralize the acid. Incorporated BrdU was detected using an anti-BrdU rat monoclonal antibody.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
The brain region was dissected immediately after collection and used for mRNA isolation using an RNeasy kit (Qiagen, Valencia, CA). RT-PCR was performed using the RETROscript RT-PCR kit (Ambion, Austin, TX) according to the manufacturer’s instructions. Briefly, the purified total RNA (100 ng) was reverse-transcribed with cloned MMLV reverse transcriptase (50 U) by incubating at 44°C for 60 minutes, followed by heating at 92°C for 10 minutes. The resulting single-stranded cDNA was then amplified using a pair of primers for each target gene (25 pmol) and TaqDNA polymerase (2.5 U; Roche, Indianapolis, IN) for the indicated number of cycles of amplification (30 seconds at 94°C for denaturing, 30 seconds at 61°C for primer annealing, and 30 seconds at 72°C for primer extension). The RT-PCR products were then subjected to electrophoresis in a 1.5% agarose gel. The nucleotide sequence used for each primer was as follows: for c-myc: 5′-primer, 5′-TCTGGATCACCTTCTGCTGG-3′; 3′-primer, 5′-CCTCTTGACATTTCTCCTCGG-3′; and for GAPDH: 5′-primer, 5′-ATGTTCCAGTATGACTCCACTCAGG-3′; 3′-primer, 5′-GAAGACACCAGTAGACTCCACGACA-3′. The specificity of PCR was confirmed by measuring the size of PCR product. In addition, the negative control (ie, brain tissues from single transgenic mice) and positive control (ie, SHSY5Y human neuroblastoma cell line) were tested to confirm the specificity of PCR reaction.
Terminal dUTP Nick-End Labeling (TUNEL) Analysis
Detection of 3′-OH termini of DNA strand breaks was performed on paraffin sections using an in situ cell death detection kit (Roche) following the recommendations of the manufacturer. Briefly, the tissue sections were treated with proteinase K (20 μg/ml in 10 mmol/L Tris-HCl, pH 7.4) for 30 minutes at 37°C after rehydration. After rinsing slides with PBS, TUNEL reaction mixture containing terminal deoxynucleotidyl transferase and fluorescence-labeled nucleotide was applied for 1 hour at 37°C. Positive signals were observed directly using fluorescence microscopy. The recommended positive and negative controls were applied in adjacent sections.
Behavioral Analysis
All mice (n = 8 in each group) were tested in a modified version of the T-maze essentially as previously described.21 This behavioral task, used to test working memory, is based on spontaneous alternation behavior, which describes the innate tendency of mice to visit arms that were not previously explored. All animals were allowed a 5-minute free run in the maze before testing to familiarize them with the apparatus and to foster alternation behavior. On the testing day, mice were placed in the start arm for 60 seconds before the gate was opened. Once the mouse entered an arm, the door was closed, and the animal was confined in that arm for 30 seconds; thereafter it was returned to the start arm for a new trial. Six consecutive trials were performed and alternation rates across the six trials were expressed as a relative percentage based on the maximal alternation rate of 100%, which occurred when a mouse never entered a repeated arm. Task achievement time (how long it took the animal to enter a goal arm after the gate opened) was also calculated.22
Statistical comparisons were performed by parametric analysis using a one-way analysis of variance and/or Student’s t-test, as appropriate, to determine significant differences across genotypes. The null hypothesis was rejected at P < 0.05.
Results
Bitransgenic animals (CaMKII-MYC) that were maintained on the doxycycline diet to suppress MYC transgene expression (MYC-Off mice) showed minimal expression of MYC (Figure 1). In marked contrast, increased expression, as determined by RT-PCR, of the MYC transgene was significantly increased after the doxycycline-containing diet was replaced by doxycycline-free regular diet (MYC-On mice) and, as expected, was specifically increased in the cerebral cortex and hippocampus (Figure 1).
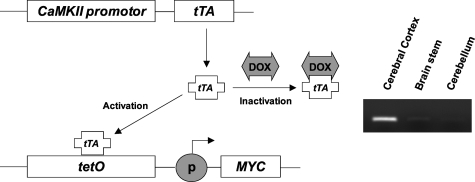
Figure 1.
Inducible expression of MYC in hippocampus by Tet regulatory system. Expression of the tTA peptide is driven by a CaMKII promoter that is active in the forebrain neurons. In its active form, tTA binds to the TetO sequence to drive expression of MYC. Doxycycline (Dox) can inhibit expression of MYC by binding to tTA, rendering it inactive. In RT-PCR analysis of dissected brain tissues, MYC is only detected in cerebral cortex, including hippocampus, after Dox-free regular diet in double-transgenic mice (CaMKII-MYC mice).
To test whether MYC expression induces cell cycle re-entry in neurons, we measured both the expression level of PCNA and Ki-67, well-established markers of cell cycle activity, which increase in nuclei during the cell cycle, and cyclin D1, a key cell cycle regulator. The expression of PCNA, Ki-67, and cyclin D1 is strongly induced in the CA1 region of hippocampus after removal of doxycycline for 5 weeks (Figure 2, A–F; and see Supplemental Figure S1 at http://ajp.amjpathol.org); the most intense staining was observed in the nuclei of pyramidal neurons. In contrast, the hippocampal neurons in the CA1 region in MYC-Off mice showed very low levels of both PCNA and cyclin D1, and undetectable levels of Ki-67. To further confirm the specific expression of these cell cycle markers in neurons, double immunocytochemistry was performed with an anti-NeuN antibody that specifically labels neurons. The immunoreactivity of both cell cycle markers, PCNA and Ki-67, is clearly observed in NeuN-positive neurons (Figure 2). These data strongly indicate a reactivation of the cell cycle in neurons in MYC-On mice.
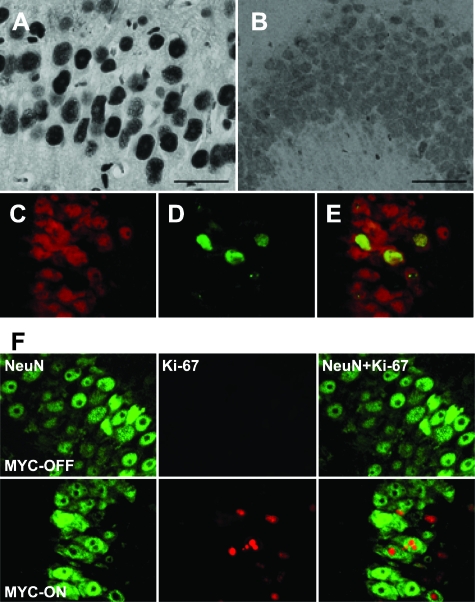
Figure 2.
Cell cycle markers increase in the hippocampal neurons in MYC-On mice. A: The expression of PCNA is dramatically increased in the nuclei of hippocampal neurons (CA1 region) from MYC-On mice. B: In MYC-Off mice, a basal level of PCNA is observed with a diffuse staining pattern in the nucleus and cytoplasm. C–E: Double staining with anti-NeuN antibody (red) clearly demonstrates the expression of PCNA (green) in neurons. F: Similar to PCNA, Ki-67 is strongly increased in the nuclei of pyramidal neurons of the hippocampal CA1 region in MYC-On mice, but not observed in MYC-Off mice. Scale bars = 50 μm.
To extend these data, DNA replication was assessed by analysis of BrdU incorporation. BrdU, which is incorporated during DNA replication, was injected 1 day before sacrificing the mice. After 5 weeks of MYC expression, the number of hippocampal neurons showing nuclear incorporation of BrdU is dramatically increased in the MYC-On mice (Figure 3); the BrdU-positive cells were mostly localized in the pyramidal cell layer, especially the CA1 region, which is confirmed by the co-localization of MAP2, a specific marker for neurons. In contrast, no BrdU-positive cells were found in the pyramidal neurons in MYC-Off mice (Figure 3). That PCNA, Ki-67, cyclin D1, and BrdU immunoreactivity were all observed in the nuclei of the population of hippocampal neurons that express MYC (ie, forebrain neurons), suggests that the cell cycle machinery was switched on and that the neurons progress through S phase. Therefore, the CaMKII-MYC mice appear to be an excellent model for the analysis of the specific events occurring after MYC induction and the subsequent re-entry of postmitotic neurons into the cell cycle.
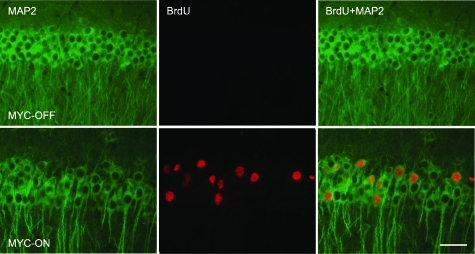
Figure 3.
DNA replication in the hippocampal neurons in MYC-On mice. Incorporation of BrdU during DNA replication is visualized using an anti-BrdU antibody (red). The number of BrdU-positive nuclei is dramatically increased in the hippocampal pyramidal neuron layer (CA1) of MYC-On mice. Double staining with anti-MAP2 antibody (green) clearly demonstrates that the BrdU-positive cells are neurons. In MYC-Off mice, virtually no BrdU-positive nuclei are observed. Scale bar = 50 μm.
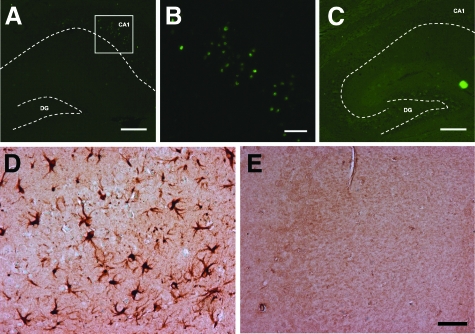
The pathological effects of MYC expression were also examined. Degenerating eosinophilic neurons with a shrunken or pyknotic appearance and condensed chromatin were readily detected in every MYC-On mouse (Figure 4, A and B). Indeed, Nissl stain confirmed massive and selective neuronal loss in the CA1 region in MYC-On mice, but not in MYC-Off mice (Figure 4, C and D). To expand on these findings, we assessed TUNEL staining in MYC-On and MYC-Off mice to measure apoptotic changes. Consistent with the histological data, increased numbers of TUNEL-positive neurons were found in the CA1 region of the hippocampus of MYC-On mice but not in MYC-Off mice (Figure 5, A–C). Furthermore, an antibody to glial fibrillary acidic protein (GFAP) used to detect astrocytes revealed intensely reactive astrocytosis in the hippocampal regions (Figure 5D) in MYC-On mice, but not in other unaffected regions such as the cerebellum (not shown). In contrast to MYC-On mice, reactive astrocytosis was not present in the hippocampus of MYC-Off mice (Figure 5E).
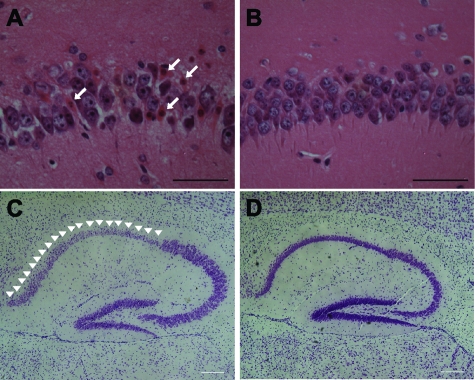
Figure 4.
Selective neuronal cell loss in the CA1 region of the hippocampus in MYC-On mice. A and C: H&E staining and Nissl staining with cresyl violet show a significant level of neurodegeneration in the CA1 region in MYC-On mice. The characteristics of cell death, including condensation of chromatin, pyknotic appearance, and eosinophilic cytoplasm (arrows) are significant in the CA1 region in MYC-On mice (A) whereas no such change is observed in MYC-Off mice (B). Nissl staining clearly demonstrates the selective neurodegeneration in the CA1 region in MYC-On mice (arrowheads, C) but not in MYC-Off mice (D). Scale bars = 100 μm.
Figure 5.
Neurodegeneration and astrocytosis in the hippocampus in MYC-On mice. TUNEL-positive nuclei are specifically localized in the hippocampal CA1 region in MYC-On mice (A, B) but not in MYC-Off mice (C). B: High magnification of the framed portion in A. Immunocytochemistry with an anti-GFAP antibody demonstrates the extensive astrocytosis in the hippocampus in MYC-On mice (D) compared with MYC-Off mice (E). DG, dentate gyrus. Dotted lines outline the hippocampal subfields. Scale bars: 200 μm (A, C); 50 μm (B, D, E).
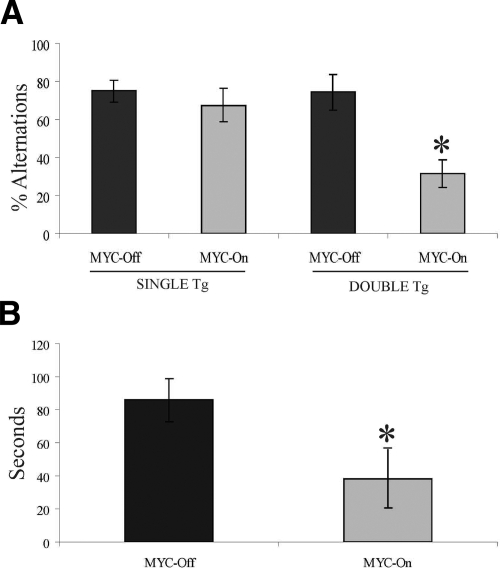
To determine whether the expression of MYC in cortical/hippocampal neurons resulted in cognitive deficits, we tested the spatial working memory of CaMKII-MYC mice using a modified version of the T-maze. The MYC-On mice were significantly impaired compared with MYC-Off mice (P < 0.002). In support of the specificity of our transgene, there was no difference in behavior between single transgenic mice (ie, MYC or CaMKII single-transgenic mice) or MYC-Off double-transgenic mice (Figure 6A). Interestingly, the length of time all animals took to enter a desired arm after each consecutive trial was measured and the MYC-On animals showed much shorter latencies than MYC-OFF mice (Figure 6B). These data suggest that MYC-On mice did not learn the paradigm and/or lack the cognitive flexibility needed to remember the previously visited arm, incorporate that, and consequently switch to the alternate, nonvisited arm. Additionally, these findings may also indicate a lack of inhibitory processes, although further testing should be performed to establish this possibility. Of importance, we also used a Rotarod to measure motor function across groups and found no differences between MYC-On and MYC-Off mice. The data suggests there is no significant difference in motor function between MYC-On and MYC-Off mice.
Figure 6.
Working memory function is decreased in MYC-On mice (n = 8). Mice were placed in the start arm for 60 seconds before the gate was opened. Once the subject entered an arm the door was closed and the animal was confined for 30 seconds; thereafter it was returned to the start arm for a new trial. A: MYC-On mice had significantly more trouble remembering the arm that they had previously visited and re-entering it (*P < 0.002). B: The time elapse was measured from when the door in the start box was opened until the animal entered the chosen arm. MYC-On mice took significantly less time to enter the chosen arm indicating lack of cognitive flexibility (*P < 0.05). Mean percentage of alternations over six trials.
Discussion
The underlying cause of the neuronal damage that occurs in AD and other neurodegenerative diseases is not fully understood. However, one likely contributor is the cell cycle re-entry found in the vulnerable neurons in postmortem brain of AD patients.23,24,25 Unfortunately, determining the consequence of cell cycle re-entry in postmitotic neurons has been technically challenging and the models used to date have not been pathophysiologically relevant. To explore this phenomenon and to establish such a relevant model system, we developed a transgenic mouse model in which cell cycle re-entry could be induced in mature neurons. Using the MYC oncogene, which is expressed in neurons in AD,20 we were able to induce cell cycle re-entry as determined by the expression of classical cell-cycle marker proteins (ie, PCNA, Ki-67, and cyclin D1), as well as DNA replication as evidenced by BrdU incorporation, specifically in hippocampal pyramidal neurons. Although BrdU data in isolation should be interpreted with caution because it can also reflect other processes, in light of the other cell cycle markers, it is unlikely that the BrdU data represents anything other than cell cycle activity. As such, this is the first physiologically relevant model of cell cycle re-entry in mature neurons and it allowed us to determine the effect of cell cycle re-entry on postmitotic neurons.
The pathological effect of MYC expression was neurodegeneration. In all of the MYC-On mice, eosinophilic neurons that were shrunken and pyknotic with condensed chromatin were readily observed. In addition, there were increased numbers of TUNEL-positive neurons as well as marked astrocytosis. Although the expression of MYC mRNA is found in all forebrain neurons, the morphological correlates were confined to the CA1 region of the hippocampus suggesting either a restricted expression of MYC protein in the CA1 region, similar to other CamKII transgenic lines,26 or that CA1 neurons are more susceptible to neurodegeneration than other forebrain neurons as shown in many studies.27 Of importance, the pathological changes found in MYC-On transgenic animals were correlated with cognitive deficits. In a modified T-maze test, there was clear evidence of a decline in spatial working memory in the MYC-On mice compared with the single transgenic or MYC-Off mice.
Traditional transgenic models that have been used to study the role of oncogenes in tumorigenesis continuously overexpress the transgenes. For example, when a powerful oncogene, the SV40 T antigen, is expressed specifically in maturing Purkinje cells in transgenic mice, the cells replicate their DNA, but subsequently degenerate and die.15 Similarly, expression of the SV40 T antigen driven by the rhodopsin promoter causes photoreceptor degeneration, again associated with cell cycle reactivation and DNA synthesis.28 Although these data are consistent with our conclusion that cell cycle reactivation in neurons leads to neurodegeneration, the effect of the SV40 T antigen and dysregulation of cell cycle re-entry during development cannot be excluded in those studies. Furthermore, although a new inducible SV40 T antigen transgenic model was recently developed16 to avoid those problems, SV40 T antigen is not a physiologically relevant entity in neurodegenerative disease and further study is required to delineate its exact mechanism of action. Importantly, our CaMKII-MYC mice preclude initial and developmentally specific consequences of oncogene expression and hence represent a valuable model for studying the pathogenesis of age-related neurodegenerative diseases. In contrast to other approaches for induction of cell cycle re-entry in neurons such as the SV40 T antigen,16 MYC is a physiologically relevant entity that has been shown to be increased in vulnerable neurons in patients with AD.20 Like our studies, this AD-related expression of MYC is restricted to increases in specific neuronal populations, dystrophic neurites, and neurofibrillary tangles without global increases in expression by immunoblot.20,29 The latter is perhaps not surprising given that any neuron expressing MYC would, based on our study, rapidly degenerate.
Indeed, it is important to bear in mind that pathological studies represent the terminal stages of disease. As such it does not exclude the possibility that MYC might be temporally induced in the early stage of disease. In this regard, it should be noted that only 5 weeks induction of MYC is enough to induce cell cycle re-entry and neurodegeneration in our mice. In support of this, MYC is temporally induced in the early stage during traumatic brain injury30 and in cerebral ischemia.31,32,33
In conclusion, using these CaMKII-MYC animals, we demonstrated that activation of an AD-associated oncogene in forebrain neurons leads to cell cycle re-entry, neurodegeneration, gliosis, and cognitive deficits. The establishment of this model provides a working platform to test genetic and pharmacological approaches to block cell cycle re-entry.34
Supplementary Material
Acknowledgments
We thank Dr. Eric Kandel (Columbia University, New York, NY) for providing CamKII-tTA transgenic mice.
Footnotes
Address reprint requests to Hyoung-gon Lee, Ph.D., or Mark A. Smith, Ph.D., Department of Pathology, Case Western Reserve University, 2103 Cornell Rd., Cleveland, OH 44106. E-mail: hyoung-gon.lee@case.edu and mark.smith@case.edu.
Supported by the National Institutes of Health (grants AG031364, AG030096 and AG028679) and Alzheimer’s Association (NIRG-07-60164).
H.L. and G.C. contributed equally to this study.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
M.A.S. is, or has in the past been, a paid consultant for, owns equity or stock options in, and/or receives grant funding from Neurotez, Neuropharm, Edenland, Panacea Pharmaceuticals, and Voyager Pharmaceuticals. G.P. is a paid consultant for and/or owns equity or stock options in Takeda Pharmaceuticals, Voyager Pharmaceuticals, Panacea Pharmaceuticals, and Neurotez Pharmaceuticals.
References
- Arendt T, Holzer M, Grossmann A, Zedlick D, Bruckner MK. Increased expression and subcellular translocation of the mitogen activated protein kinase kinase and mitogen-activated protein kinase in Alzheimer’s disease. Neuroscience. 1995;68:5–18. doi: 10.1016/0306-4522(95)00146-a. [DOI] [PubMed] [Google Scholar]
- Vincent I, Jicha G, Rosado M, Dickson DW. Aberrant expression of mitotic cdc2/cyclin B1 kinase in degenerating neurons of Alzheimer’s disease brain. J Neurosci. 1997;17:3588–3598. doi: 10.1523/JNEUROSCI.17-10-03588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Smith AD. Expression of cell division markers in the hippocampus in Alzheimer’s disease and other neurodegenerative conditions. Acta Neuropathol. 1997;93:294–300. doi: 10.1007/s004010050617. [DOI] [PubMed] [Google Scholar]
- McShea A, Harris PL, Webster KR, Wahl AF, Smith MA. Abnormal expression of the cell cycle regulators P16 and CDK4 in Alzheimer’s disease. Am J Pathol. 1997;150:1933–1939. [PMC free article] [PubMed] [Google Scholar]
- Zhu X, McShea A, Harris PL, Raina AK, Castellani RJ, Funk JO, Shah S, Atwood C, Bowen R, Bowser R, Morelli L, Perry G, Smith MA. Elevated expression of a regulator of the G2/M phase of the cell cycle, neuronal CIP-1-associated regulator of cyclin B, in Alzheimer’s disease. J Neurosci Res. 2004;75:698–703. doi: 10.1002/jnr.20028. [DOI] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Dorsey R, Chalovich EM, Hammond RR, Achim CL. Expression patterns of retinoblastoma protein in Parkinson disease. J Neuropathol Exp Neurol. 2003;62:68–74. doi: 10.1093/jnen/62.1.68. [DOI] [PubMed] [Google Scholar]
- Thakur A, Siedlak SL, James SL, Bonda DJ, Rao A, Webber KM, Camins A, Pallas M, Casadesus G, Lee HG, Bowser R, Raina AK, Perry G, Smith MA, Zhu X. Retinoblastoma protein phosphorylation at multiple sites is associated with neurofibrillary pathology in Alzheimer disease. Int J Clin Exp Pathol. 2008;1:134–146. [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Bowser R. Alterations in G(1) to S phase cell-cycle regulators during amyotrophic lateral sclerosis. Am J Pathol. 2003;162:823–835. doi: 10.1016/S0002-9440(10)63879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A, Arendt T. Aneuploidy and DNA replication in the normal human brain and Alzheimer’s disease. J Neurosci. 2007;27:6859–6867. doi: 10.1523/JNEUROSCI.0379-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Siedlak SL, Wang Y, Perry G, Castellani RJ, Cohen ML, Smith MA. Neuronal binucleation in Alzheimer disease hippocampus. Neuropathol Appl Neurobiol. 2008;34:457–465. doi: 10.1111/j.1365-2990.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- Szweda PA, Friguet B, Szweda LI. Proteolysis, free radicals, and aging. Free Radic Biol Med. 2002;33:29–36. doi: 10.1016/s0891-5849(02)00837-7. [DOI] [PubMed] [Google Scholar]
- Giovanni A, Wirtz-Brugger F, Keramaris E, Slack R, Park DS. Involvement of cell cycle elements, cyclin-dependent kinases, pRb, and E2F x DP, in B-amyloid-induced neuronal death. J Biol Chem. 1999;274:19011–19016. doi: 10.1074/jbc.274.27.19011. [DOI] [PubMed] [Google Scholar]
- Park DS, Obeidat A, Giovanni A, Greene LA. Cell cycle regulators in neuronal death evoked by excitotoxic stress: implications for neurodegeneration and its treatment. Neurobiol Aging. 2000;21:771–781. doi: 10.1016/s0197-4580(00)00220-7. [DOI] [PubMed] [Google Scholar]
- Feddersen RM, Ehlenfeldt R, Yunis WS, Clark HB, Orr HT. Disrupted cerebellar cortical development and progressive degeneration of Purkinje cells in SV40 T antigen transgenic mice. Neuron. 1992;9:955–966. doi: 10.1016/0896-6273(92)90247-b. [DOI] [PubMed] [Google Scholar]
- Park KH, Hallows JL, Chakrabarty P, Davies P, Vincent I. Conditional neuronal simian virus 40 T antigen expression induces Alzheimer-like tau and amyloid pathology in mice. J Neurosci. 2007;27:2969–2978. doi: 10.1523/JNEUROSCI.0186-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Blanco R, Carmona M, Puig B. Phosphorylated c-MYC expression in Alzheimer disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Neuropathol Appl Neurobiol. 2001;27:343–351. doi: 10.1046/j.1365-2990.2001.00348.x. [DOI] [PubMed] [Google Scholar]
- Casadesus G, Webber KM, Atwood CS, Pappolla MA, Perry G, Bowen RL, Smith MA. Luteinizing hormone modulates cognition and amyloid-beta deposition in Alzheimer APP transgenic mice. Biochim Biophys Acta. 2006;1762:447–452. doi: 10.1016/j.bbadis.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Piérard C, Liscia P, Valleau M, Drouet I, Chauveau F, Huart B, Bonneau D, Jouanin JC, Beaumont M, Beracochea D. Modafinil-induced modulation of working memory and plasma corticosterone in chronically-stressed mice. Pharmacol Biochem Behav. 2006;83:1–8. doi: 10.1016/j.pbb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- McShea A, Lee HG, Petersen RB, Casadesus G, Vincent I, Linford NJ, Funk JO, Shapiro RA, Smith MA. Neuronal cell cycle re-entry mediates Alzheimer disease-type changes. Biochim Biophys Acta. 2007;1772:467–472. doi: 10.1016/j.bbadis.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Evans TA, Raina AK, Delacourte A, Aprelikova O, Lee HG, Zhu X, Perry G, Smith MA. BRCA1 may modulate neuronal cell cycle re-entry in Alzheimer disease. Int J Med Sci. 2007;4:140–145. doi: 10.7150/ijms.4.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina AK, Garrett MR, Previll LA, Obrenovich ME, Hartzler AW, Webber KM, Casadesus G, Lee HG, Perry G, Zhu X, Smith MA. Oncogenic parallels in Alzheimer disease. Int J Neuroprotec Neuroregen. 2006;2:80–85. [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Guthrie PB, Kater SB. Intrinsic factors in the selective vulnerability of hippocampal pyramidal neurons. Prog Clin Biol Res. 1989;317:333–351. [PubMed] [Google Scholar]
- al-Ubaidi MR, Hollyfield JG, Overbeek PA, Baehr W. Photoreceptor degeneration induced by the expression of simian virus 40 large tumor antigen in the retina of transgenic mice. Proc Natl Acad Sci USA. 1992;89:1194–1198. doi: 10.1073/pnas.89.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Blanco R. N-myc and c-myc expression in Alzheimer disease, Huntington disease and Parkinson disease. Brain Res Mol Brain Res. 2000;77:270–276. doi: 10.1016/s0169-328x(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Movsesyan V, Ahmed F, Cernak I, Schinelli S, Stoica B, Faden AI. Cell cycle inhibition provides neuroprotection and reduces glial proliferation and scar formation after traumatic brain injury. Proc Natl Acad Sci USA. 2005;102:8333–8338. doi: 10.1073/pnas.0500989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Fujimura M, Noshita N, Chang YY, Chan PH. SOD1 down-regulates NF-kappaB and c-Myc expression in mice after transient focal cerebral ischemia. J Cereb Blood Flow Metab. 2001;21:163–173. doi: 10.1097/00004647-200102000-00008. [DOI] [PubMed] [Google Scholar]
- Nakagomi T, Asai A, Kanemitsu H, Narita K, Kuchino Y, Tamura A, Kirino T. Up-regulation of c-myc gene expression following focal ischemia in the rat brain. Neurol Res. 1996;18:559–563. doi: 10.1080/01616412.1996.11740470. [DOI] [PubMed] [Google Scholar]
- McGahan L, Hakim AM, Robertson GS. Hippocampal Myc and p53 expression following transient global ischemia. Brain Res Mol Brain Res. 1998;56:133–145. doi: 10.1016/s0169-328x(98)00038-2. [DOI] [PubMed] [Google Scholar]
- Woods J, Snape M, Smith MA. The cell cycle hypothesis of Alzheimer’s disease: suggestions for drug development. Biochim Biophys Acta. 2007;1772:503–508. doi: 10.1016/j.bbadis.2006.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.