Abstract
The tumor suppressor genes EXT1 and EXT2 are involved in the formation of multiple osteochondromas, which can progress to become secondary peripheral chondrosarcomas. The most common chondrosarcoma subtype is primary central chondrosarcoma, which occurs in the medullar cavity of bone. The EXT1/EXT2 protein complex is involved in heparan sulfate proteoglycan (HSPG) biosynthesis, which is important for signal transduction of Indian hedgehog (IHH), WNT, and transforming growth factor (TGF)-β. The role of EXT and its downstream targets in central chondrosarcomas is currently unknown. EXT1 and EXT2 were therefore evaluated in central chondrosarcomas at both the DNA and mRNA levels. Immunohistochemistry was used to assess HSPG (CD44v3 and SDC2), WNT (β-catenin), and TGF-β (PAI-1 and phosphorylated Smad2) signaling, whereas IHH signaling was studied both by quantitative polymerase chain reaction and in vitro. mRNA levels of both EXT1 and EXT2 were normal in central chondrosarcomas; genomic alterations were absent in these regions and in 30 other HSPG-related genes. Although HSPGs were aberrantly located (CD44v3 in the Golgi and SDC2 in cytoplasm and nucleus), this was not caused by mutation. WNT signaling negatively correlated with increasing histological grade, whereas TGF-β positively correlated with increasing histological grade. IHH signaling was active, and inhibition decreased cell viability in one of six cell lines. Our data suggest that, despite normal EXT in central chondrosarcomas, HSPGs and HSPG-dependent signaling are affected in both central and peripheral chondrosarcomas.
The tumor suppressor genes EXT1 and EXT2 are known for their involvement in peripheral cartilaginous tumors. They cause the hereditary syndrome multiple osteochondromas (OMIM no. 133700), previously known as hereditary multiple exostoses.1,2,3 In multiple osteochondromas patients, benign cartilage capped bony outgrowths (osteochondromas) develop at the surface of bones formed by endochondral ossification. Histologically, they bear a strong resemblance to the normal growth plate. One to five percent of osteochondromas progress to (secondary) peripheral chondrosarcoma.
Although secondary peripheral chondrosarcoma is a rare chondrosarcoma subtype (less than 15% in tertiary referral centers), primary conventional central chondrosarcoma arising in the medullar cavity of bone is far more common (>80% of conventional chondrosarcoma).4,5 Enchondromas are benign cartilage tumors occurring in the center of the bone.4 Enchondromas occur mostly as solitary lesions, although they may occur as multiple lesions in the context of nonhereditary enchondromatosis (Ollier disease) (OMIM no. 166000).6
The protein products of EXT1 and EXT2, Exostosin1 and Exostosin2, are type II transmembrane glycoproteins that form a hetero-oligomeric complex, located in the Golgi membrane. This complex is responsible for elongation of heparan sulfate side chains that are linked to the proteoglycan protein cores, consequently forming heparan sulfate proteoglycans (HSPGs).7,8 HSPGs are involved in sequestering growth factors, anchorage to the extracellular matrix, and several growth signaling pathways.9 Various HSPGs have been described, eg, syndecan (SDC), perlecan, and glypican families and CD44 isoforms containing HS-bearing variable exon 3 (v3).9,10 In Drosophila melanogaster, HSPGs were shown to be essential for gradient formation of the morphogens hedgehog, decapentaplegic, and wingless.11 The human homologues for these morphogens are Indian and Sonic hedgehog, transforming growth factor (TGF)-β/BMP, and WNT, respectively.12 Indian hedgehog (IHH) orchestrates chondrocyte proliferation and differentiation in the human growth plate. IHH signals to its receptor patched (PTCH), which subsequently releases its inhibition on intracellular smoothened (SMO), resulting in the translocation of GLI transcription factors to the nucleus. Here, PTHLH is transcribed together with PTCH and GLI, guaranteeing the preservation of this signaling cascade.13 PTHLH signaling inhibits chondrocyte differentiation and consequently controls longitudinal growth.14,15
Although it is evident that inactivation of the EXT genes is the driving force for the development of benign peripheral cartilaginous tumors,1,2,3,16,17 in the far more common central chondrosarcomas the role of EXT and its downstream targets has not been systematically studied so far. We therefore investigated EXT and its downstream targets in central chondrosarcoma. Results are compared with its rare peripheral counterpart for which EXT involvement is quite well characterized.1,2,3 We studied EXT at the DNA and mRNA level and investigated the expression of HSPGs using immunohistochemistry. Activity of IHH, TGF-β, and WNT signaling, which require HSPGs for proper signaling, was also studied in a large series of central chondrosarcomas including six chondrosarcoma cell cultures.
Materials and Methods
Patient Material
Enchondromas and conventional central chondrosarcomas were selected based on accepted clinicopathological18 and radiological19 criteria. Peripheral, juxtacortical, mesenchymal, dedifferentiated, and clear-cell chondrosarcomas were excluded. In total, specimens of 110 patients were used for this study, including both formalin-fixed, paraffin-embedded (n = 95) and fresh frozen (n = 34) tissue. Both paraffin and frozen series included enchondromatosis (Ollier disease)-related tumors. Details are outlined in Table 1. Histological grading was performed according to Evans and colleagues.20 All specimens were handled according to the ethical guidelines as described in the Code for Proper Secondary Use of Human Tissue in The Netherlands of the Dutch Federation of Medical Scientific Societies.
Table 1.
Clinicopathological Data of the 110 Patients
| Enchondroma
|
Chondrosarcoma
|
|||
|---|---|---|---|---|
| FFPE | Fresh frozen | FFPE | Fresh frozen | |
| Total number of tumors | 33 | 7* | 62 | 27 |
| Grade I | 27 | 11 | ||
| Grade II | 25 | 7 | ||
| Grade III | 10 | 9 | ||
| Male:female | 20:13 | 3:4 | 32:30 | 17:10 |
| Enchondromatosis | 6 | 5 | 6 | 7 |
| Median age at diagnosis years (range) | 37.8 (6.1 to 66.4) | 18.0 (12.0 to 37.0) | 50.0 (17.8 to 78.7) | 40.0 (17.8 to 84.0) |
| Median follow-up months (range) | 65 (6 to 170) | 87 (5 to 247) | ||
FFPE, formalin-fixed, paraffin-embedded.
All fresh-frozen enchondromas were located in the phalanx.
Quantitative Real-Time Reverse Transcriptase PCR (q-RT-PCR)
q-RT-PCR was used to study the expression level of EXT1 and EXT2 and the activity of IHH signaling. Of 34 cases fresh frozen tumor tissue was available for RNA isolation, performed as described previously.21 Four growth plate samples, acquired from resections or biopsies for orthopedic clinical conditions not related to cartilaginous tumors, were used as controls. Results were compared with those of 28 peripheral tumors described previously.16 Primers for EXT1, EXT2, PTCH, GLI1, and GLI2 were described previously.22 Four control genes (CYPA, CPSF6, SRPR, and HNRPH1) were selected to normalize the expression data, because of their invariable expression in chondrosarcoma.23 Data of the cell cultures were normalized against HPRT and GAPDH. As a reference for normalization and statistical analysis a calibration curve of a mixture consisting of 15 highly diverse cell lines24 was included. Normalization was performed using Genorm.25
Genomic Analysis
Mutation Analysis of EXT1/2 and SDC2
Two tumors with decreased EXT1 expression (L803 and L1689) were subjected to direct sequencing of the coding sequences of EXT1 and EXT2 and multiplex ligation-dependent probe amplification assay designed for EXT1 and EXT2 as described.26,27 DNA was isolated from fresh-frozen tumor tissue. DNA isolated from another set of seven tumors (four with nuclear and three with cytoplasmic SDC2 expression at immunohistochemistry) and their corresponding normal lymphocytes were screened for mutations by direct sequencing analysis of the five exons of the coding region of the SDC2 gene. Primer sequences are listed in Table 2.
Table 2.
Primer Sequences for SDC2 Mutation Analysis
| Sequence | Tm | Product size | |
|---|---|---|---|
| SDC2-ex1F | 5′-TGTAAAACGACGGCCAGTGTACTCTGCTCCGGATTCGT-3′ | 59 | 266 |
| SDC2-ex1R | 5′-AACAGCTATGACCATGAGGGCTCCTCTCGTAGCTTCA-3′ | 62 | |
| SDC2-ex2F | 5′-TGTAAAACGACGGCCAGTCTCAACATCCTGACTCCCTTG-3′ | 60 | 269 |
| SDC2-ex2R | 5′-AACAGCTATGACCATGAATCCTGCCTGTCTCCTTGAA-3′ | 60 | |
| SDC2-ex3F | 5′-TGTAAAACGACGGCCAGTTCATGATTGCCATGCTCAGT-3′ | 60 | 243 |
| SDC2-ex3R | 5′-AACAGCTATGACCATGAGATAATGCAATGCAATGGAAA-3′ | 59 | |
| SDC2-ex4F | 5′-TGTAAAACGACGGCCAGTTTTCTTCTTTCCAACACATTTCC-3′ | 59 | 270 |
| SDC2-ex4R | 5′-AACAGCTATGACCATGAGTAGGCACCCTCCCACCT-3′ | 60 | |
| SDC2-ex5F | 5′-TGTAAAACGACGGCCAGTTGTCTGCAACCCTTGAATCTC-3′ | 60 | 313 |
| SDC2-ex5R | 5′-AACAGCTATGACCATGATGCAAAAGCTTTATTTTGAAAAGTT-3′ | 60 |
Array Comparative Genomic Hybridization
L803 and L1689 were also analyzed by array comparative genomic hybridization using a high-resolution 8q array containing a tiling bacterial artificial chromosome clone set as described.16 In addition, the seven tumors that were subjected to mutational screening for SDC2 were hybridized to a custom-made HD-Agilent (Agilent Technologies, Amstelveen, The Netherlands) oligonucleotide array containing 4 × 44,000 immobilized 60-mer oligonucleotides. Genomic intervals containing 35 genomic regions of 37 genes selected on their function in cartilage biosynthesis (Supplementary Table S1 at http://ajp.amjpathol.org) were subjected to Agilent’s eArray web tool (Agilent Technologies) to generate oligonucleotide (60 mer) microarray probe sequences. For each gene of interest probes were selected both for exonic and intronic areas with a spacing at 500- to 1000-base intervals, additional probes sequences were selected 5000 bps before and after targeted genes to include potential control elements, such as promoter regions. A total 37,040 oligonucleotide probes were selected in genomic regions, additional probes up to 44,000 were added as internal control selected as a default from the array design options. Slides were printed and produced by Agilent Technologies using standard company protocols available as custom service. Array probes, layout, and access to purchase this microarray from Agilent array can be obtained from the responsible author (K.S.). Test and reference samples were labeled, hybridized, washed, scanned, and analyzed following standard protocols provided by Agilent Technologies.
Immunohistochemistry
For 95 tumors formalin-fixed, paraffin-embedded material was available (Table 1). All were stained for PAI-1 and a subset of 19 tumors for phosphorylated Smad2 to evaluate TGF-β signaling.28,29 In addition all were stained for β-catenin, to evaluate canonical WNT signaling,30 HSPG expression (SDC2 and CD44v3) was evaluated in a subset of 30 tumors. Subsets were representative for the total group of patients. Details of primary antibodies are described in Table 3. PAI1, phosphorylated Smad2, and β-catenin immunohistochemistry were scored semiquantitatively as described previously31 by two observers (Y.M.S., J.V.M.G.) independently. Both were blinded toward clinicopathological data. In brief, scores were given for β-catenin, PAI1, and phosphorylated Smad2 intensity (1 = weak, 2 = moderate, 3 = strong) and for percentage of positive cells (1 = 0 to 24%, 2 = 25 to 49%, 3 = 50 to 74%, and 4 = 75 to 100% positive cells in the total tumor section). To avoid tumors with single positive cells being regarded as positive, cutoff levels for statistical analysis were applied corresponding to general staining pattern (sum of score β-catenin ≥2 and PAI1 ≥3). SDC2 and CD44v3 were evaluated based on presence or absence and localization of staining as described.23 Specificity of the SDC2 antibody for membranous staining was tested using a tissue microarray containing 79 soft tissue tumors of 28 different entities, as was described previously.32
Table 3.
Antibodies Used for IHC, WB, and IF
| Antigen | Manufacturer | Monoclonal/poly-clonal | Positive control | Staining | Antibody concentration | Antigen retrieval* |
|---|---|---|---|---|---|---|
| CD44v3 | Labvision | Monoclonal | Tonsil | Membrane, cytoplasm | 1:400 | Citrate |
| 58K Golgi protein | Abcam Ltd. | Polyclonal | Tonsil | Perinuclear | 1:100 | Citrate |
| Syndecan-2 (10H4) | G. David51 | Monoclonal | Growth plate | Membrane, ECM | 35 μg/ml | None |
| Heterochromatin protein-1γ (2MOD-1G6-AS) | Euromedex | Monoclonal | Any cell | Nuclear | 1:5000 | None |
| β-catenin | BD Transduction | Monoclonal | Skin | Nuclear | 1:800 | Citrate |
| Laboratories | Mamma- | |||||
| PAI-1 | American Diagnostica Inc. | Monoclonal | carcinoma | Cytoplasm | 1:350 | None |
| Phosphorylated Smad 2 | P. ten Dijke29 | Polyclonal | Prostate | Nuclear | 1:2000 | Citrate |
Antigen retrieval was performed using citrate buffer at 98°C for 20 minutes.
Immunoblotting
To confirm the nuclear localization of SDC2 as shown by immunohistochemistry two fresh central chondrosarcoma samples were digested overnight in dissociation medium containing 0.1% collagenase (Sigma, Zwijndrecht, The Netherlands) and 0.1% dispase (Life Technologies, Breda, The Netherlands), 100 IU/ml. Cell fractionation was performed using two different lysis buffers. To extract cytoplasmic proteins a buffer containing 10 mmol/L HEPES pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EGTA, 0.1 mmol/L EDTA, 1 mmol/L dithiothreitol, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml leupeptin, 1 μg/ml pepstatin A, and 2 μg/ml aprotinin was used, followed by the addition of 10% Nonidet P-40. Subsequently, the extraction of nuclear proteins was performed by vigorously rocking the sample for 15 minutes in the presence of a buffer containing 20 mmol/L HEPES, pH 7.9, 400 mmol/L NaCl, 1 mmol/L EGTA, 1 mmol/L EDTA, 1 mmol/L dithiothreitol, 1 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml leupeptin, 1 μg/ml pepstatin A, and 2 μg/ml aprotinin. Protein concentrations were measured using a DC Protein Assay (Bio-Rad, Hercules, CA). Ten μg of each sample were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were electrophoretically transferred onto polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, MA). Membranes were pre-incubated with 15% skinned milk in Tris-buffered saline/Tween. After incubation with first and secondary antibodies (Table 3), the membranes were developed with enhanced chemiluminescence Western blotting detection reagent (Amersham Biosciences, Buckinghamshire, UK) and visualized by exposure to X-ray films (Hyperfilm ECL, Amersham Biosciences).
Confocal Microscopy and Immunofluorescence
To confirm localization of CD44v3 in the Golgi apparatus, two central chondrosarcomas (with perinuclear CD44v3 expression) were selected for fluorescent double staining using CD44v3 and the 58K Golgi protein antibodies as described previously.22
Hedgehog Activity in Vitro
Chondrosarcoma cell lines CH2879,33 C3842,34 OUMS27,35 and SW1353 (American Type Culture Collection, Manassas, VA), central chondrosarcoma primary cultures L784 and L869, and cyclopamine-responsive pancreatic carcinoma cell line PANC1 (American Type Culture Collection)36 were used to analyze HH signaling. Chondrosarcoma cell lines were cultured in RPMI 1640 and PANC1 in Dulbecco’s modified Eagle’s medium (both Gibco, Invitrogen Life-Technologies, Scotland, UK) supplemented with 10% heat-inactivated fetal calf serum (Gibco, Invitrogen Life-Technologies) at 37°C in a humidified incubator with 95% air and 5% CO2. Cartilaginous phenotype was confirmed by RT-PCR, showing mRNA expression of collagens I, 2B, 3, and 10; Aggrecan; and SOX9.37
For RNA analysis, 2.5 × 105 cells were seeded in a six-well plate. After 24 hours the medium was replaced with serum-starved medium (0.05% fetal calf serum) containing either 10 μmol/L cyclopamine (Toronto Research Chemicals, North York, Canada) or dimethyl sulfoxide 0.1%. Cells were harvested after 24 hours and RNA was isolated using Trizol and microspin column (Qiagen, Hilden, Germany). Cell viability was assessed by WST-1 colorimetric assay (Roche Diagnostics GmbH, Penzberg, Germany), which measures mitochondrial activity. Cells were seeded into 96-well flat-bottom plates (1.5 × 103 cells/well for SW1353 and PANC1 and 5.0 × 103 cells for CH2879, C3842, OUMS27, L784, and L869). After 24 hours dimethyl sulfoxide, cyclopamine at 5 μmol/L and 10 μmol/L, and tomatidine (Toronto Research Chemicals), an inactive but structurally related compound,38 at 10 μmol/L, were added in the presence of 5% fetal calf serum, each condition in quadruplicate. After 3 days of treatment, the metabolic activity of the cells was measured on a Victor3 Multilabel Counter 1420-042 (Perkin Elmer, Waltham, MA) at 450 nm.
Statistical Analysis
Correlations between histological grade and immunohistochemical results were calculated by χ2 test. Differences in cell viability in vitro were calculated by Student’s t-test. P values <0.05 were considered significant.
Results
Normal EXT1 and EXT2 in Central Chondrosarcoma
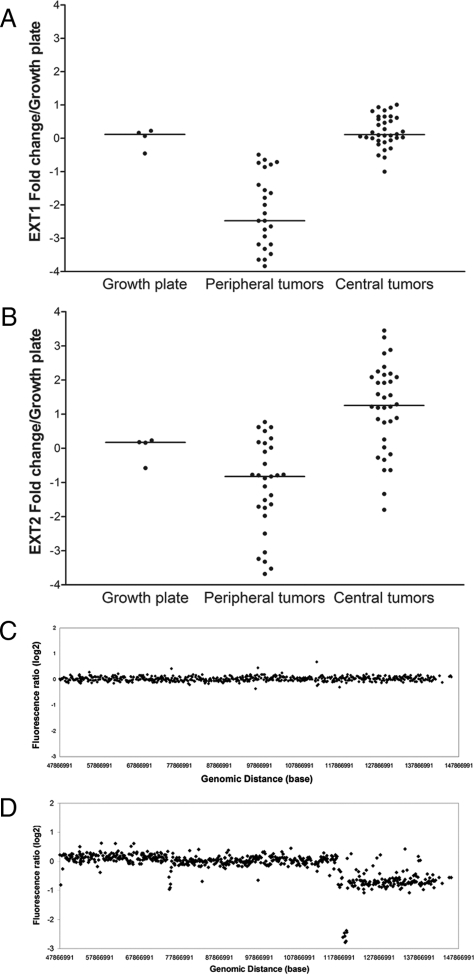
The expression of EXT1 and EXT2 mRNA in central cartilaginous tumors is not decreased as compared with growth plate samples, which is in contrast to the low EXT1 expression levels previously found in peripheral tumors (Figure 1, A and B).22 No correlation of EXT1 or EXT2 expression with histological grade was found (not shown). Direct mutation screening and MLPA for both EXT1 and EXT2 did not reveal any genetic aberrations (not shown). Comparative genomic hybridization on an 8q bacterial artificial chromosome tiling array of the two central chondrosarcomas with the lowest expression of EXT1 did not reveal any alterations on 8q24, the locus for EXT1. Results for L803 are shown in Figure 1, C and D, which are representative for L1689 (not shown). No other gains or losses in 8q were detected. Moreover, no losses were found in the EXT1 and EXT2 genes in the seven chondrosarcomas hybridized at the oligonucleotide Agilent array.
Figure 1.
A: EXT1 mRNA expression levels of central chondrosarcoma are comparable with growth plate levels whereas peripheral tumors (published previously16) show a threefold decrease. B: EXT2 mRNA levels are slightly higher in central and slightly lower in peripheral chondrosarcoma, compared with growth plate. Expression levels of the tumors are log2 transformed and expressed as relative to the growth plate levels. C: 8q tiling array comparative genomic hybridization pattern of central chondrosarcoma L803 is shown. Genomic alterations are absent, despite low EXT1 expression by qPCR. L1689 showed similar results as L803 (data not shown). For comparison, an osteochondroma with homozygous loss of multiple clones covering the EXT1 gene and hemizygous loss of a larger part of 8q is shown in D (figure composed of data published previously16).
Aberrant Location of HSPGs SDC2 and CD44v3 in Central Chondrosarcoma
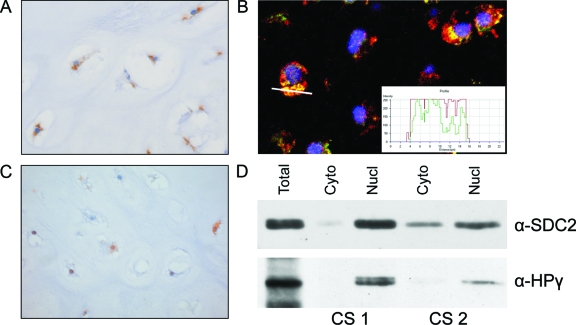
Cytoplasmic staining of CD44v3 was found in 26 of 30 (87%) central tumors (7 of 8 EC and 14 of 22 CS). The staining was dot-like suggesting localization in the Golgi apparatus (Figure 2A), which was confirmed using immunofluorescent confocal microscopy (Figure 2B). Central chondrosarcomas showed cytoplasmic staining of SDC2 in 28 of 29 (97%). Moreover, additional nuclear staining, shown in Figure 2C, was found in half of the tumors (4 of 7 EC and 11 of 23 CS). The soft tissue tumor tissue microarray did not show nuclear staining in any of the specimens (not shown). Control sections of tonsil showing membranous expression pattern of CD44v3 in the epithelial layer and normal growth plate showed SDC2 staining at the cell membrane and in the extracellular matrix surrounding hypertrophic chondrocytes (not shown). Nuclear localization of SDC2 as found by immunohistochemistry was verified in the nuclear component of the cell fractionation by immunoblotting (Figure 2D).
Figure 2.
A: Central chondrosarcoma showing cytoplasmic dot-like accumulation of CD44v3 at immunohistochemistry, suggestive for Golgi retention. B: This was confirmed by IF using a Golgi-specific marker, 58K protein, in green, and CD44v3 in red, resulting in a yellow color when co-localization occurs. The white line indicates the position where the staining profile (inset) has been taken. C: Central chondrosarcoma showing nuclear staining of SDC2, which was found in 50% of chondrosarcomas in addition to cytoplasmic staining. D: Nuclear localization of SDC2 was verified by immunoblotting in two chondrosarcomas. In the isolated nuclear fraction heterochromatin protein 1γ was present, verifying the procedure for separating the nuclear from the cytoplasmic staining. Original magnifications, ×40.
No Aberrations in Other Genes Important for HSPG Formation
Because we found normal EXT in central chondrosarcomas, we searched for aberrations in other regulators of HSPG formation. Mutation screening of the five coding exons of the SDC2 gene did not reveal any mutations in the seven tumors (not shown). Using the oligonucleotide tiling array covering 30 EXT, EXT-like, and other genes involved in HSPG biosynthesis we did not find any specific genomic losses or gains in these seven tumors.
Decreased WNT Signaling and Increased TGF-β Signaling in High-Grade Chondrosarcomas
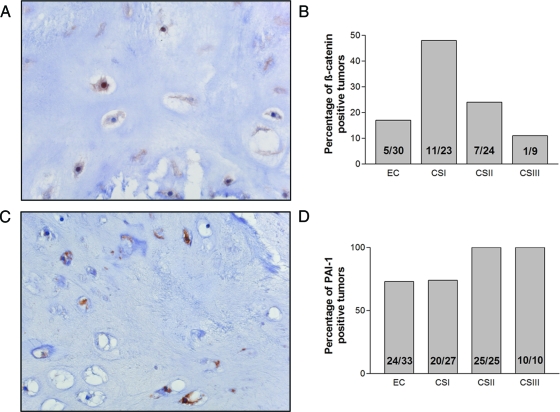
Nuclear staining for β-catenin (Figure 3A) was found in 17% (5 of 30) of enchondromas, in 47% (11 of 23) of grade I, and in 29% (7 of 24) of grade II and in 11% (1 of 9) grade III chondrosarcoma (Figure 3B). The activity of canonical WNT signaling was increased in grade I chondrosarcomas compared with enchondromas. However on further increase in histological grade the activity decreased again (Pearson χ2, P = 0.038). Expression of PAI-1, implicating active TGF-β signaling (Figure 3C), was associated with increased histological grade (Pearson χ2, P = 0.002). All high-grade tumors were positive (35 of 35), whereas the enchondromas and low-grade chondrosarcoma were positive in 73% (24 of 33) and 74% (20 of 27), respectively (Figure 3D). Moreover, nuclear localization of phosphorylated Smad2 was demonstrated in all of 6 enchondromas and 13 chondrosarcomas, suggesting active TGF-β signaling (not shown).
Figure 3.
A and B: The activity of canonical WNT signaling was increased in grade I chondrosarcomas compared with enchondromas. However on further increase in histological grade, the activity decreased again (Pearson χ2, P = 0.038). C and D: PAI-1 expression, indicating active TGF-β signaling, was found in the majority of central chondrosarcomas. PAI-1 expression was correlated to histological grade (χ2 test, P = 0.002). Original magnifications, ×40.
HH Signaling in Central Chondrosarcoma in Vivo and in Vitro
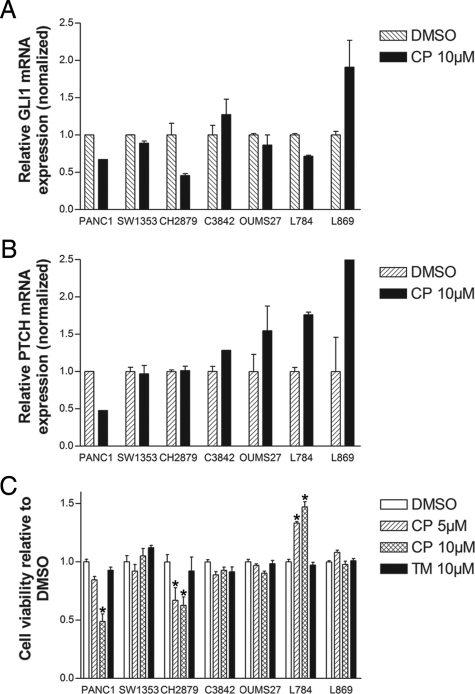
By qPCR active HH signaling was shown in central chondrosarcoma, irrespective of histological grade, because PTCH, GLI1, and GLI2 were expressed at similar levels as in the growth plates (Figure 4, A–C). Cyclopamine treatment resulted in a profound decrease of GLI1 mRNA expression combined with decreased cell viability in only one cell culture (CH2879), whereas all other cultures did not respond by decreased GLI1 levels or cell viability (Figure 5A). In L869, an increase of GLI1 was observed whereas L784 showed increased cell viability. In all chondrosarcoma cell cultures PTCH levels were unchanged or even increased after cyclopamine treatment (Figure 5B). The viability of the tomatidine controls showed that the decrease in viability in PANC1 and CH2879 is not a toxic side effect of the cyclopamine compound (Figure 5C).
Figure 4.
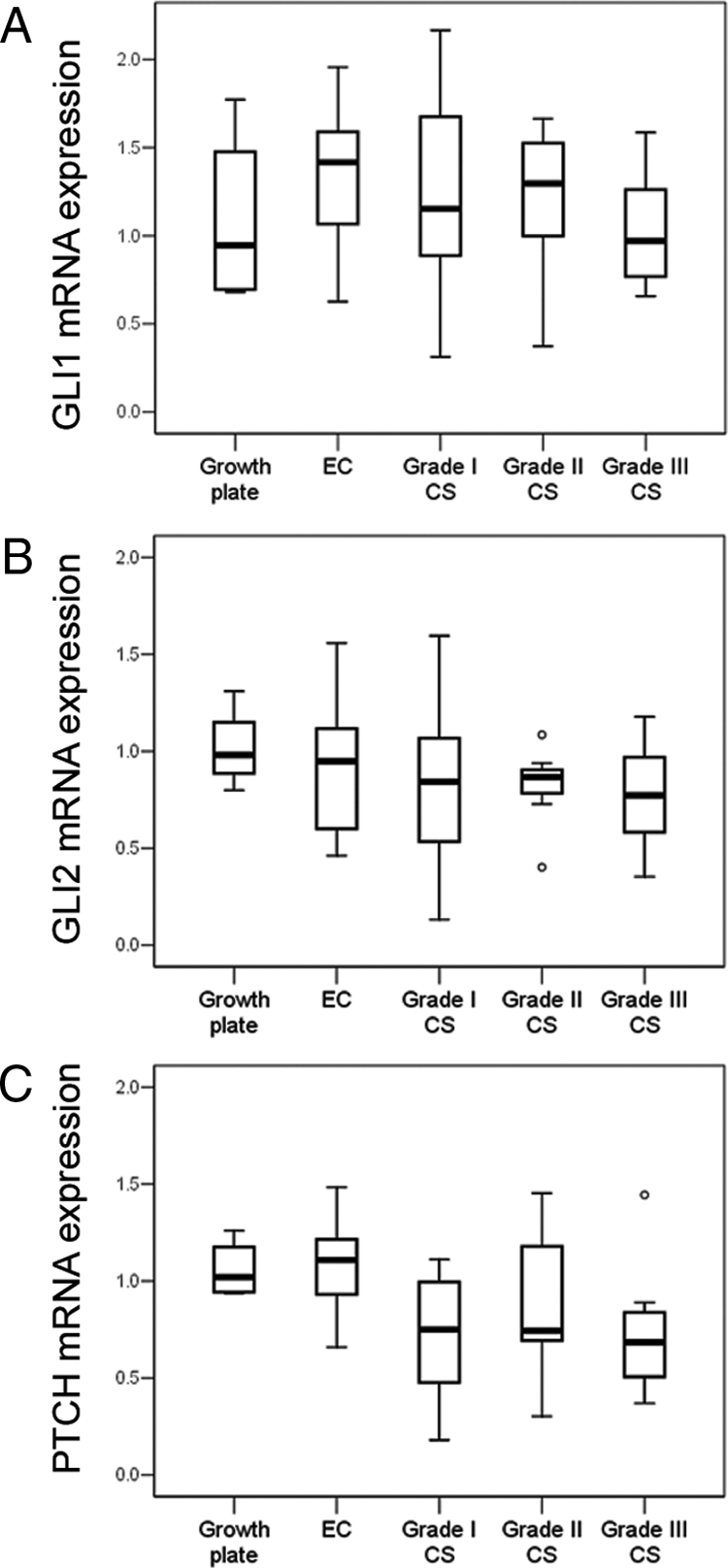
mRNA levels of GLI1 (A), GLI2 (B), and PTCH (C) were measured as a readout of IHH signaling. Levels of all three genes in chondrosarcomas were in the same range of growth plate samples and large variations within the groups were observed. No relation to histological grade was found. Expression levels are log10 transformed.
Figure 5.
A: In vitro the GLI1 mRNA expression decreases to almost half of the level of the dimethyl sulfoxide (DMSO) control in PANC1 and in CH2879 on 24-hour treatment with 10 μmol/L of cyclopamine (CP), although this was not found in the other five chondrosarcoma cell cultures. B: None of the chondrosarcoma cell cultures showed a negative effect on PTCH mRNA expression on 24-hour 10 μmol/L CP treatment. Both experiments were performed in duplicate. C: Both PANC1 and CH2879 demonstrated a decrease in cell viability measured by WST-1 assay, on 72 hours of 5 μmol/L and 10 μmol/L CP treatment. An increase in viability was observed in L784, whereas the other four chondrosarcoma cell cultures did not show an effect of cyclopamine treatment. The black bars represent the inactive compound tomatidine (TM), which demonstrate the selective effect of CP. Data represent three individual experiments performed in quadruplicate. Student’s t-test for paired data, *P < 0.05.
Discussion
We present the first systematic evaluation of EXT and its downstream targets in central chondrosarcoma. We confirm that peripheral and central chondrosarcomas are clear distinct genetical entities also with respect to their EXT expression. Whereas inactivation of EXT is the driving force for the development of benign peripheral cartilage tumors,22 we demonstrate in central cartilage tumors the EXT genes to be normal both at the DNA as well as at the mRNA expression level. The absence of larger deletions in the EXT1 region is in concordance with the previously reported absence of loss of heterozygosity31 and karyotypic aberrations39 at 8q24 in central chondrosarcomas.
The EXT proteins are involved in the biosynthesis of heparan sulfate. Despite normal expression of the EXT genes in central tumors, heparan sulfate proteoglycans (CD44v3 and SDC2) unexpectedly accumulated in the cytoplasm. This is similar to what we previously described for peripheral tumors carrying EXT mutations,22 whereas in other tumors membranous expression of CD44v3 and SDC2 is described.40,41 In addition, 50% of central chondrosarcomas also demonstrated nuclear SDC2 expression, which was not observed in peripheral chondrosarcomas, nor in 79 soft tissue sarcomas. In addition to the general function of SDC2 in modulating extracellular ligands, intracellular actions are implicated for SDC2. In osteosarcoma SDC2 has been shown to induce apoptosis.42,43 We excluded mutations in the nuclear localization signal, located at the first exon of the SDC2 gene, however other mechanisms such as aberrant phosphorylation of the nuclear localization signal or mutations in a kinase responsible for the phosphorylation of the nuclear localization signal might explain the aberrant localization of SDC2 in central chondrosarcoma.
In addition, it is tempting to speculate that, for example, other glycosyltransferases than EXT or the sulfotransferases or epimerases that function to complete the formation of heparan sulfate proteoglycans are affected in central chondrosarcoma. By using a custom-designed oligonucleotide tiling array of 30 EXT, EXT-like, and other genes involved in HSPG biosynthesis we excluded that neither heterozygous nor homozygous losses, nor gains, are found in these genes. Nonetheless, we cannot exclude that these HSPG biosynthesis-related genes might be subjected to point mutations, or other copy number neutral alterations, ie, inversion or methylation.
In Drosophila, HSPGs are essential for gradient formation of the morphogens hedgehog, decapentaplegic, and wingless.11 In humans, the IHH pathway, through PTHLH signaling, is vital for chondrocyte proliferation and differentiation in the growth plate. We show hedgehog signaling to be active in central chondrosarcoma. We previously demonstrated PTHLH signaling, which is downstream of IHH, to be active in central chondrosarcomas as well.44,45 In osteochondromas, IHH signaling is active, whereas activity decreases in peripheral chondrosarcoma with increasing histological grade.23 Tiet and colleagues46 reported active IHH signaling in chondrosarcoma, but did not distinguish between peripheral and central tumors. In a previous pilot series we reported lower levels of IHH signaling in central tumors as compared with the growth plate.45 In the present larger series IHH signaling levels are comparable to growth plate samples. Moreover, in one of the cell lines, CH2879, an important role for hedgehog signaling in cell proliferation could be shown because inhibiting HH signaling using cyclopamine decreased cell viability. IHH signaling has been shown to be activated in many cancers such as medulloblastoma, basal cell carcinoma, small-cell lung cancer, breast cancer, and pancreatic cancer.47 Therefore, targeting IHH signaling seems to be promising in cancer therapy.48 Previously, Tiet and colleagues46 also showed that blocking HH signaling using either cyclopamine or triparanol, an inhibitor of 7-dehydrocholesterol reductase, reduced proliferation and tumor volume in 10 chondrosarcoma xenografts. However, we observed an effect of IHH blockade using cyclopamine only in one of six chondrosarcoma cell cultures. Surprisingly, PTCH was found to be increased in those cultures that were resistant to cyclopamine, which might be induced by other pathways. These data suggest that despite aberrant cellular HSPG distribution, IHH signaling is active in central chondrosarcoma and is vital for cellular proliferation and therefore a putative therapeutic target in a small subset of them. Because HSPGs are thought to be important for the diffusion of HH to its receptor on target cells, it might be that in central chondrosarcoma these diffusion problems are overcome by cell autonomous (autocrine) HH signaling, as was previously also suggested for osteochondromas.49
The activity of other signaling pathways dependent on HSPG was similar to the activity in peripheral chondrosarcoma.23 Our results suggest that active canonical WNT signaling might be important for the transition from enchondroma toward low-grade central chondrosarcoma, however it is not crucial for progression toward higher grade (Figure 6). In contrast, TGF-β signaling was shown to increase with increasing histological grade by both the presence of PAI-1 and the finding of nuclear localization of phosphorylated Smad2. This suggests that TGF-β has a role in either rearrangement of extracellular matrix and/or vessel formation, which are the main characteristics of high-grade chondrosarcoma.50
Figure 6.
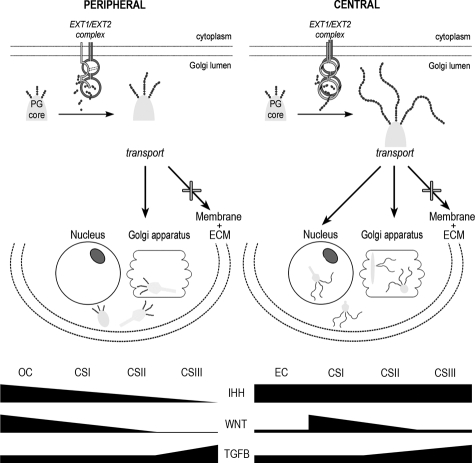
Overview of the EXT/HSPG pathway involved in both peripheral and central chondrosarcomas. Despite the absence of EXT alterations in central chondrosarcomas there is cytoplasmic retention of HSPGs, which is similar to peripheral chondrosarcomas and nuclear SDC2 localization that was specific for central chondrosarcomas. In addition, whereas IHH signaling decreased with grade in peripheral chondrosarcomas, it is still active in high-grade central chondrosarcomas. Active canonical WNT signaling might be important for the transition from benign to malignant central cartilaginous tumor type, however it is not crucial for tumor progression, whereas TGF-β is. Our results therefore suggest that disturbed HSPG function is involved in the histogenesis of both central and peripheral chondrosarcoma, although different steps in HSPG biosynthesis seem affected. (Figure based on Hameetman et al.22).
In conclusion, we present a systematic investigation of EXT and its downstream targets in central chondrosarcoma. We clearly show that the EXT genes are normal in central chondrosarcoma, with nevertheless aberrant localization of HSPGs. Despite this, IHH signaling is active in central chondrosarcoma and is important for proliferation and therefore a potential therapeutic target in a small subset of central chondrosarcomas. The aberrant intracellular accumulation of HSPG in central chondrosarcoma which is similar to peripheral chondrosarcoma and the nuclear SDC2 localization that is exclusively seen in central chondrosarcoma is difficult to explain. Nevertheless, our data, summarized in Figure 6, suggest that a disturbed HSPG functioning is involved in the histogenesis of both central and peripheral chondrosarcoma, although different steps in HSPG biosynthesis seem affected.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Peter ten Dijke, Leiden University Medical Center, for providing the phosphorylated Smad2 antibody; Dr. Thomas Kalinski, Otto-von-Guericke-University, Magdeburg, Germany, for kindly providing us with the C3842 cell line; Masayoshi Namba, Okayama University Medical School, Shikata, Japan, for the OUMS27 cell line; Dr. Alfons de Hooge for his help with the cell fractionation; Ayse Yavas and Ronald Duim for their technical assistance with the mutation analysis for EXT and SDC2, respectively; Jeroen Knijnenburg, Marja van den Burg, and Daniëlle de Jong for expert assistance with the array experiments; and Frans Prins for his help with confocal immunofluorescent microscopy.
Footnotes
Address reprint requests to Judith V.M.G. Bovée, M.D., Ph.D., Department of Pathology, Leiden University Medical Center, Albinusdreef 2, 2333 ZA Leiden, The Netherlands. E-mail: j.v.m.g.bovee@lumc.nl.
Supported by the Netherlands Organization for Scientific Research (grants 908-02-018 to Y.M.S. and J.V.M.G.B. and 917-76-315 to J.V.M.G.B.), the EuroBoNeT Consortium (grant 018814 to Y.M.S., K.S., A.M.C.J., P.C.W.H., and J.V.M.G.B.), an European Commission granted Network of Excellence for studying the pathology and genetics of bone tumors.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Ahn J, Ludecke H-J, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1). Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- Wuyts W, Van Hul W, Wauters J, Nemtsova M, Reyniers E, Van Hul E, De Boulle K, De Vries BBA, Hendrickx J, Herrygers I, Bossuyt P, Balemans W, Fransen E, Vits L, Coucke P, Nowak NJ, Shows TB, Mallet L, Van den Ouweland AMW, McGaughran J, Halley DJJ, Willems P. Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet. 1996;5:1547–1557. doi: 10.1093/hmg/5.10.1547. [DOI] [PubMed] [Google Scholar]
- Stickens D, Clines G, Burbee D, Ramos P, Thomas S, Hogue D, Hecht JT, Lovett M, Evans GA. The EXT2 multiple exostoses gene defines a family of putative tumour suppressor genes. Nat Genet. 1996;14:25–32. doi: 10.1038/ng0996-25. [DOI] [PubMed] [Google Scholar]
- Bertoni F, Bacchini P, Hogendoorn PCW. Chondrosarcoma. Fletcher CDM, Unni KK, Mertens F, editors. Lyon,: IARC Press,; World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. 2002:pp 247–251. [Google Scholar]
- Mulder JD, Schütte HE, Kroon HM, Taconis WK. Amsterdam: Elsevier,; Radiologic Atlas of Bone Tumors. 1993:pp 139–171. [Google Scholar]
- Ollier M. De la dyschondroplasia. Bull Soc Chir Lyon. 1899;3:22–23. [Google Scholar]
- McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, Tufaro F. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Knudson CB, Knudson W. Cartilage proteoglycans. Semin Cell Dev Biol. 2001;12:69–78. doi: 10.1006/scdb.2000.0243. [DOI] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Transducing hedgehog: the story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amling M, Neff L, Tanaka S, Inoue D, Kuida K, Weir E, Philbrick WM, Broadus AE, Baron R. Bcl-2 lies downstream of parathyroid hormone related peptide in a signalling pathway that regulates chondrocyte maturation during skeletal development. J Cell Biol. 1997;136:205–213. doi: 10.1083/jcb.136.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Eerden BCJ, Karperien M, Gevers EF, Lowik CWGM, Wit JM. Expression of Indian hedgehog. PTHrP and their receptors in the postnatal growth plate of the rat: evidence for a locally acting growth restraining feedback loop after birth. J Bone Miner Res. 2000;15:1045–1055. doi: 10.1359/jbmr.2000.15.6.1045. [DOI] [PubMed] [Google Scholar]
- Hameetman L, Szuhai K, Yavas A, Knijnenburg J, van Duin M, Van Dekken H, Taminiau AHM, Cleton-Jansen AM, Bovée JVMG, Hogendoorn PCW. The role of EXT1 in non hereditary osteochondroma: identification of homozygous deletions. J Natl Cancer Inst. 2007;99:396–406. doi: 10.1093/jnci/djk067. [DOI] [PubMed] [Google Scholar]
- Bovée JVMG, Cleton-Jansen AM, Taminiau AHM, Hogendoorn PCW. Emerging pathways in the development of chondrosarcoma of bone and the implications for targeted treatment. Lancet Oncol. 2005;6:599–607. doi: 10.1016/S1470-2045(05)70282-5. [DOI] [PubMed] [Google Scholar]
- Eefting D, Schrage YM, Geirnaerdt MJ, Le Cessie S, Taminiau AHM, Bovee JVMG, Hogendoorn PCW. Assessment of interobserver variability and histological parameters to improve reliability in classification and grading of central cartilaginous tumors. Am J Surg Pathol. 2009;33:50–57. doi: 10.1097/PAS.0b013e31817eec2b. [DOI] [PubMed] [Google Scholar]
- Geirnaerdt MJA, Hermans J, Bloem JL, Kroon HM, Pope TL, Taminiau AHM, Hogendoorn PCW. Usefulness of radiography in differentiating enchondroma from central grade I chondrosarcoma. Am J Roentgenol. 1997;169:1097–1104. doi: 10.2214/ajr.169.4.9308471. [DOI] [PubMed] [Google Scholar]
- Evans HL, Ayala AG, Romsdahl MM. Prognostic factors in chondrosarcoma of bone: a clinicopathologic analysis with emphasis on histologic grading. Cancer. 1977;40:818–831. doi: 10.1002/1097-0142(197708)40:2<818::aid-cncr2820400234>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Baelde HJ, Cleton-Jansen AM, van Beerendonk H, Namba M, Bovée JVMG, Hogendoorn PCW. High quality RNA isolation from tumours with low cellularity and high extracellular matrix component for cDNA microarrays: application to chondrosarcoma. J Clin Pathol. 2001;54:778–782. doi: 10.1136/jcp.54.10.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameetman L, David G, Yavas A, White SJ, Taminiau AHM, Cleton-Jansen AM, Hogendoorn PCW, Bovee JVMG. Decreased EXT expression and intracellular accumulation of heparan sulphate proteoglycan in osteochondromas and peripheral chondrosarcomas. J Pathol. 2007;211:399–409. doi: 10.1002/path.2127. [DOI] [PubMed] [Google Scholar]
- Hameetman L, Rozeman LB, Lombaerts M, Oosting J, Taminiau AHM, Cleton-Jansen AM, Bovée JVMG, Hogendoorn PCW. Peripheral chondrosarcoma progression is accompanied by decreased Indian hedgehog (IHH) signalling. J Pathol. 2006;209:501–511. doi: 10.1002/path.2008. [DOI] [PubMed] [Google Scholar]
- Rozeman LB, Hameetman L, van Wezel T, Taminiau AHM, Cleton-Jansen AM, Hogendoorn PCW, Bovée JVMG. cDNA expression profiling of central chondrosarcomas: Ollier disease resembles solitary tumors and alteration in genes coding for energy metabolism with increasing grade. J Pathol. 2005;207:61–71. doi: 10.1002/path.1813. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink GR, White SJ, Gabelic S, Hogendoorn PCW, Breuning MH, Bakker E. Mutation screening of EXT1 and EXT2 by direct sequence analysis and MLPA in patients with multiple osteochondromas: splice site mutations and exonic deletions account for more than half of the mutations. Eur J Hum Genet. 2004;13:470–474. doi: 10.1038/sj.ejhg.5201343. [DOI] [PubMed] [Google Scholar]
- White SJ, Vink GR, Kriek M, Wuyts W, Schouten J, Bakker B, Breuning MH, den Dunnen JT. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- Sawdey M, Podor TJ, Loskutoff DJ. Regulation of type 1 plasminogen activator inhibitor gene expression in cultured bovine aortic endothelial cells. Induction by transforming growth factor-beta, lipopolysaccharide, and tumor necrosis factor-alpha. J Biol Chem. 1989;264:10396–10401. [PubMed] [Google Scholar]
- Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin CH, Funa K, ten Dijke P. The L45 loop in type I receptors for TGF-beta family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- Hinck L, Nelson WJ, Papkoff J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovée JVMG, Cleton-Jansen AM, Kuipers-Dijkshoorn N, Van den Broek LJCM, Taminiau AHM, Cornelisse CJ, Hogendoorn PCW. Loss of heterozygosity and DNA ploidy point to a diverging genetic mechanism in the origin of peripheral and central chondrosarcoma. Genes Chromosom Cancer. 1999;26:237–246. [PubMed] [Google Scholar]
- Willems SM, Schrage YM, Baelde JJ, Briaire-de B, Mohseny A, Sciot R, Bovee JVMG, Hogendoorn PCW. Myxoid tumours of soft tissue: the so-called myxoid extracellular matrix is heterogeneous in composition. Histopathology. 2008;52:465–474. doi: 10.1111/j.1365-2559.2008.02967.x. [DOI] [PubMed] [Google Scholar]
- Gil-Benso R, Lopez-Gines C, Lopez-Guerrero JA, Carda C, Callaghan RC, Navarro S, Ferrer J, Pellin A, Llombart-Bosch A. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: comparative histologic and genetic studies with its tumor of origin. Lab Invest. 2003;83:877–887. doi: 10.1097/01.lab.0000073131.34648.ea. [DOI] [PubMed] [Google Scholar]
- Kalinski T, Krueger S, Pelz AF, Wieacker P, Hartig R, Ropke M, Schneider-Stock R, Dombrowski F, Roessner A. Establishment and characterization of the permanent human cell line C3842 derived from a secondary chondrosarcoma in Ollier’s disease. Virchows Arch. 2005;446:287–299. doi: 10.1007/s00428-004-1194-y. [DOI] [PubMed] [Google Scholar]
- Kunisada T, Miyazaki M, Mihara K, Gao C, Kawai A, Inoue H, Namba M. A new human chondrosarcoma cell line (OUMS-27) that maintains chondrocytic differentiation. Int J Cancer. 1998;77:854–859. doi: 10.1002/(sici)1097-0215(19980911)77:6<854::aid-ijc10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Chatel G, Ganeff C, Boussif N, Delacroix L, Briquet A, Nolens G, Winkler R. Hedgehog signaling pathway is inactive in colorectal cancer cell lines. Int J Cancer. 2007;121:2622–2627. doi: 10.1002/ijc.22998. [DOI] [PubMed] [Google Scholar]
- Cleton-Jansen AM, van Beerendonk HM, Baelde HJ, Bovée JVMG, Karperien M, Hogendoorn PCW. Estrogen signaling is active in cartilaginous tumors: implications for antiestrogen therapy as treatment option of metastasized or irresectable chondrosarcoma. Clin Cancer Res. 2005;11:8028–8035. doi: 10.1158/1078-0432.CCR-05-1253. [DOI] [PubMed] [Google Scholar]
- Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- Bovée JVMG, Sciot R, Dal Cin P, Debiec-Rychter M, Van Zelderen-Bhola SL, Cornelisse CJ, Hogendoorn PCW. Chromosome 9 alterations and trisomy 22 in central chondrosarcoma: a cytogenetic and DNA flow cytometric analysis of chondrosarcoma subtypes. Diagn Mol Pathol. 2001;10:228–236. doi: 10.1097/00019606-200112000-00004. [DOI] [PubMed] [Google Scholar]
- Döme B, Somlai B, Ladanyi A, Fazekas K, Zoller M, Timar J. Expression of CD44v3 splice variant is associated with the visceral metastatic phenotype of human melanoma. Virchows Arch. 2001;439:628–635. doi: 10.1007/s004280100451. [DOI] [PubMed] [Google Scholar]
- Roskams T, De Vos R, David G, Van Damme B, Desmet V. Heparan sulphate proteoglycan expression in human primary liver tumours. J Pathol. 1998;185:290–297. doi: 10.1002/(SICI)1096-9896(199807)185:3<290::AID-PATH91>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Modrowski D, Orosco A, Thevenard J, Fromigue O, Marie PJ. Syndecan-2 overexpression induces osteosarcoma cell apoptosis: implication of syndecan-2 cytoplasmic domain and JNK signaling. Bone. 2005;37:180–189. doi: 10.1016/j.bone.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Orosco A, Fromigue O, Bazille C, Entz-Werle N, Levillain P, Marie PJ, Modrowski D. Syndecan-2 affects the basal and chemotherapy-induced apoptosis in osteosarcoma. Cancer Res. 2007;67:3708–3715. doi: 10.1158/0008-5472.CAN-06-4164. [DOI] [PubMed] [Google Scholar]
- Bovée JVMG, Van den Broek LJCM, Cleton-Jansen AM, Hogendoorn PCW. Up-regulation of PTHrP and Bcl-2 expression characterizes the progression of osteochondroma towards peripheral chondrosarcoma and is a late event in central chondrosarcoma. Lab Invest. 2000;80:1925–1933. doi: 10.1038/labinvest.3780202. [DOI] [PubMed] [Google Scholar]
- Rozeman LB, Hameetman L, Cleton-Jansen AM, Taminiau AHM, Hogendoorn PCW, Bovée JVMG. Absence of IHH and retention of PTHrP signalling in enchondromas and central chondrosarcomas. J Pathol. 2005;205:476–482. doi: 10.1002/path.1723. [DOI] [PubMed] [Google Scholar]
- Tiet TD, Hopyan S, Nadesan P, Gokgoz N, Poon R, Lin AC, Yan T, Andrulis IL, Alman BA, Wunder JS. Constitutive hedgehog signaling in chondrosarcoma up-regulates tumor cell proliferation. Am J Pathol. 2006;168:321–330. doi: 10.2353/ajpath.2006.050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Tian H, de Sauvage FJ. The hedgehog signaling pathway in cancer. Clin Cancer Res. 2006;12:5924–5928. doi: 10.1158/1078-0432.CCR-06-1736. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Benoist-Lasselin C, de Margerie E, Gibbs L, Cormier S, Silve C, Nicolas G, Lemerrer M, Mallet JF, Munnich A, Bonaventure J, Zylberberg L, Legeai-Mallet L. Defective chondrocyte proliferation and differentiation in osteochondromas of MHE patients. Bone. 2006;39:17–26. doi: 10.1016/j.bone.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ayala G, Liu C, Nicosia R, Horowitz S, Lackman R. Microvasculature and VEGF expression in cartilaginous tumors. Hum Pathol. 2000;31:341–346. doi: 10.1016/s0046-8177(00)80248-8. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van der Schueren B, Marynen P, Cassiman JJ, Van den Berghe H. Spatial and temporal changes in the expression of fibroglycan (syndecan-2) during mouse embryonic development. Development. 1993;119:841–854. doi: 10.1242/dev.119.3.841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.