Abstract
IκBγ is one member of a family of proteins that can inhibit the nuclear localization of nuclear factor-κB. However, the other specific functions of IκBγ are still poorly understood, and its effects on tumor metastasis have not yet been characterized. We examined the consequences of targeting IκBγ in melanoma cells using a hammerhead ribozyme. We developed stable transformant B16-F10 melanoma cell lines that express a ribozyme that targets mouse IκBγ (IκBγ-144-Rz). Tail-vein injection of B16-F10 cells that stably express IκBγ-144-Rz into mice resulted in a significant reduction of the metastatic potential of these cells. IκBγ-144-Rz-expressing B16 cells were shown to have increased transcriptional activity of nuclear factor-κB. We then showed that IκBγ-144-Rz-expressing cells demonstrated both reduced invasion and increased apoptosis, suggesting the existence of pathways through which IκBγ promotes melanoma metastasis. Using gene expression profiling, we identified a differentially expressed gene set that is regulated by the stable suppression of IκBγ that may participate in mediating its anti-metastatic effects; we also confirmed the altered expression levels of several of these genes by quantitative real time polymerase chain reaction. Plasmid-mediated expression of IκBγ-144-Rz produced a significant inhibition of the metastatic progression of B16-F10 cells to the lung and resulted in significant anti-invasive and pro-apoptotic effects on murine Lewis lung carcinoma cells. Our results suggest a novel role for IκBγ in promoting the metastatic progression of melanoma.
Nuclear factor (NF)-κB comprises a family of transcription factors that have been demonstrated to play an important role in tumor cell proliferation and metastasis.1 The NF-κB family is comprised of five members: RelA/p65, RelB, c-Rel, p105 (precursor of p50), and p100 (precursor of p52). All of the members share a highly conserved 300-amino acid Rel homology domain in their N-terminal region that is responsible for dimerization, DNA binding, and nuclear localization.2 Inactivated forms of NF-κB family members are associated with a cytoplasmic family of inhibitor proteins known as IκB that share a conserved domain of six to eight ankyrin repeats and include IκBα, IκBβ, IκBγ, BCL-3, p100, and p105. An array of signals causes phosphorylation of IκB molecules, leading to its ubiquitination and subsequent degradation via the proteasome pathway. This allows translocation of NF-κB into the nucleus so it can activate transcription of target genes.3
p105 is retained in the cytoplasm by interaction of its C-terminal sequence with the N-terminal Rel homology domain. Studies have shown that a deletion in its C-terminal allows it to enter the nucleus.4,5 The C-terminal portion of p105 contains ankyrin repeats similar to other members of IκB proteins and encodes a 70-kDa protein known as IκBγ. IκBγ has a unique specificity for NF-κB dimers that contain at least one p50 subunit, preferentially p50-p50 homodimers, but also p50-RelA/p65 heterodimers.6
Although IκBγ has been shown to inhibit NF-κB from localizing into the nucleus to interact with its target genes, the consequences of modulation of IκBγ expression are still incompletely understood. Specifically, the potential function of IκBγ in regulating tumor progression and metastasis has not been examined. In this study, we examine the role of targeted suppression of IκBγ in melanoma metastasis. We report here that ribozyme-mediated down-regulation of IκBγ results in significant suppression in the metastatic potential of B16 melanoma. We also show that inhibition of IκBγ expression results in activation of NF-κB, as well as reduced invasive capacity and increased apoptotic index of melanoma as well as lung cancer cells, and identify genes whose expression is altered by stable suppression of IκBγ that may help mediate the observed anti-tumor effects.
Materials and Methods
Cloning
The pcDNA3.1 plasmid, containing ampicillin- and neomycin-resistance genes and the CMV (cytomegalovirus) promoter, was digested with AflII and XbaI, and ligated with DNA (5′-GACTAGGTGTATTCTGATGAGTCCGTGAGGACGAAACAGGAA-3′) encoding a hammerhead ribozyme (Rz) targeting sequences 130 to 150 of murine IκBγ mRNA, with a cleavage site immediately 3′ to the adenine at position 144 (IκBγ-144-Rz). A mutant ribozyme was generated by altering a single base (G to C) in the ribozyme catalytic core. The DNA encoding the anti-IκB-144-Rz and mutant Rz were cloned and isolated as described previously,7,8 and their sequence was verified using a gene-specific primer (5′-TTTCTTGGGTAGTTTGCATTTT-3′).
Generation of Stable Transformants
The transfection reagent N-[1-(2,3-diolcoyloxy) propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP, 100 mmol/L; Avanti Polar Lipids, Alabaster, AL) and plasmid DNA were added sequentially to D5W (4 nmol of lipid and 1 mg of DNA in 0.5 ml of total volume per 1 × 105 cells) and added drop-wise onto cultured B16-F10 murine melanoma cells (American Type Culture Collection, Manassas, VA) and grown in serum-free medium. Seventy-two hours after transfection, cells were replated at 1:2 to 1:16 dilutions, and grown in selective medium containing 800 μg/ml of geneticin (Mediatech, Herndon, VA) and 10% fetal bovine serum (FBS). Individual colonies were selected, expanded, and analyzed for ribozyme and target gene expression via quantitative real-time polymerase chain reaction. Three clones containing the vector only and mutant IκBγ-144-Rz as well as three clones showing the greatest suppression of IκBγ expression on the initial screen were pooled for further analysis.
Tissue Culture
For in vitro and in vivo experiments as well as microarray studies, B16-F10 cells were cultured in RPMI medium 1640 with 5% FBS and 800 μg/ml of geneticin (37°C, 5% CO2). This medium was also changed to RPMI 1640 with 1% FBS and 800 μg/ml of geneticin 24 hours before the invasion assay. For fluorescence-activated cell sorting (FACS) analysis, the cells were grown to 100% confluency in RPMI 1640 with 5% FBS and 800 μg/ml of geneticin, and then allowed to grow for 4 days in serum-starved conditions. Murine Lewis lung carcinoma cells were grown in RPMI medium 1640 with 5% FBS. This medium was changed to RPMI 1640 with 1% FBS 24 hours before the invasion assay.
Quantitative Real-Time Polymerase Chain Reaction (PCR) Analysis
Total cellular RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA), processed, and reverse-transcribed. For quantitative PCR (TaqMan assay; Applied Biosystems, Foster City, CA), cDNA was synthesized from 200 ng of DNase I-digested total cellular RNA and the assay was performed as previously described.7 The expression levels of the genes of interest were normalized to the mammalian histone control gene before comparisons.
Protein Extraction and Western Analysis
Total protein was extracted from pooled stable clones of empty vector and IκBγ-144-Rz-transfected cells with RIPA buffer containing protease inhibitors. Fifty μg of protein was electrophoresed in 7.5% Tris-HCl denaturing gels (Bio-Rad, Hercules, CA), and blotted onto nitrocellulose membranes (Bio-Rad). The following antibodies were used: anti-human IκBγ rabbit polyclonal (no. 39078: Active Motif, Carlsbad, CA) and anti-GAPDH mouse monoclonal (MAB374; Chemicon International, Temecula, CA).
FACS Analysis
Rates of apoptosis were analyzed by FACS using Acid-S and Apo-BrdU kit (Phoenix Flow Systems, San Diego, CA). The starved cells (described above) were harvested as per the manufacturer-suggested protocol using Accutase and Accumax (Phoenix Flow Systems) cell detachment buffers. For the BrdU incorporation assay, cells were incubated with BrdU for 4 hours. For each sample (performed in triplicate), 50,000 cells were counted by FACS and the results were analyzed for statistical significance using the Kolmogorov-Smirnov test.
Invasion Assays
The Matrigel assay and Boyden chamber analysis for tumor invasion were performed as previously described.9
NF-κB Activation Assay
One hundred thousand cells from pooled empty vector and pooled IκBγ-144-Rz stable clones were seeded in a 12-well plate and grown overnight in RPMI medium 1640 containing 5% FBS and 800 μg/ml of geneticin. Cells were then transiently co-transfected with a plasmid construct expressing the firefly luciferase gene under the control of NF-κB DNA binding sites (LR0051; Panomics, Fremont, CA) and a plasmid containing the Renilla luciferase gene at a 10:1 ratio using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. The cells were harvested 24 hours after transfection, and the luciferase activity was measured in a luminometer (LB96V MicroLumat Plus; EG&G Berthold, Wellesley, MA) using the dual-luciferase reporter assay system (no. E1910; Promega, Madison, WI) and normalized to Renilla luciferase activity. All assays were performed in triplicate.
Gene Expression Profiling Studies and Statistical Analysis
RNA was extracted from various B16 transformant clones in culture using the RNeasy mini kit (Qiagen) per the manufacturer’s protocol. Ten μg of total target RNA, side by side with 10 μg of universal mouse reference RNA (Stratagene, La Jolla, CA), were converted to aminoallyl-modified cDNA by oligo dt-primed polymerization using SuperScript II reverse transcriptase (Invitrogen), coupled to N-hydroxysuccinimidyl esters of Cy3 or Cy5 (Amersham, Piscataway, NJ), and then hybridized to a microarray slide at 65°C for 12 to 16 hours.10 Slides were then washed and immediately scanned with GenePix 4000B (Molecular Devices, Sunnyvale, CA), by using GenePixPro3 software. After linear normalization, log (base 2) transformation, and supervised hierarchical clustering, the resulting cluster data table was imported into the Significance Analysis of Microarray software package. Delta was chosen to limit the output gene list so that fewer than 5% predicted false-positives would be included.
Microarrays
From the 15,658 cDNAs used in this study, ∼8000 clones were from Incyte Corporation (Wilmington, DE), and the other 8000 from the National Institute on Aging (Bethesda, MD) clone set (originally from Riken, Saitama, Japan). Based on Unigene identifier (http://source.stanford.edu), these clones represent 10,953 unique genes.
Animal Studies
All animal care was in accordance with institutional guidelines and a protocol that was approved by the University of California San Francisco Committee on Animal Research. Groups of 10 45-day-old female C57BL6 mice (Charles River, Wilmington, MA) were inoculated with 3 × 104 stable transformant cells by tail vein injection and the resultant tumors were counted and analyzed using the unpaired, two-sided Student’s t-test as described previously.7 In the therapeutic study, a single intravenous injection of cationic liposome DNA complexes (CLDCs) containing plasmid DNA expressing various constructs was performed 7 days after tumor cell inoculation as previously described.8 For the subcutaneous studies, 1.5 × 106 pooled vector or IκBγ-144-Rz cells were implanted subcutaneously in the posterior flank of C57BL6 mice. Tumor volume and body weight of each animal was measured twice weekly, starting on day 6 of the study. The formula for generating tumor volume is V = (LxW2)/2, where L and W are the tumor length and width.
Results
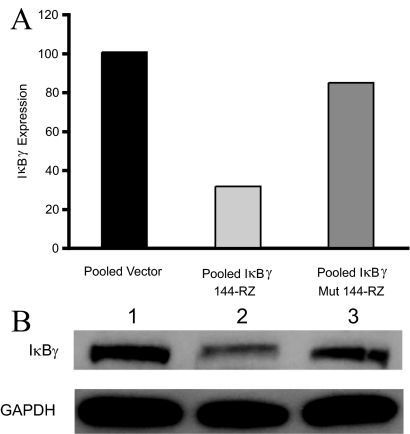
To investigate the role of IκBγ in the metastatic progression of murine melanoma, we developed stable transformant cell lines expressing a hammerhead ribozyme targeting mouse IκBγ at position 144 (IκBγ-144-Rz), driven by the CMV promoter. We also developed stable transformant control cell lines expressing either an empty vector or a disabled, mutant ribozyme (altered at a single base in the IκBγ-144-Rz catalytic core). The suppression of IκBγ expression was initially assessed by quantitative PCR. Three clones that showed inhibition of IκBγ expression were pooled for further analysis, and all subsequent analyses performed with the pooled clones. The level of IκBγ expression in pooled clones containing anti-IκBγ-144-Rz was suppressed by 68% and 62%, respectively, when compared with pooled transformant clones containing vector alone or anti-mutant IκBγ-144-Rz (Figure 1A). Inhibition at the protein level was confirmed by Western blot analysis (Figure 1B).
Figure 1.
A: Expression of IκBγ by quantitative PCR in pooled vector, pooled IκBγ-144-Rz, and pooled mutant ribozyme B16 transformant cells. B: Western analysis of IκBγ and GAPDH expression in pooled vector (lane 1), pooled IκBγ-144-Rz (lane 2), and pooled mutant ribozyme (lane 3) B16 transformant cells.
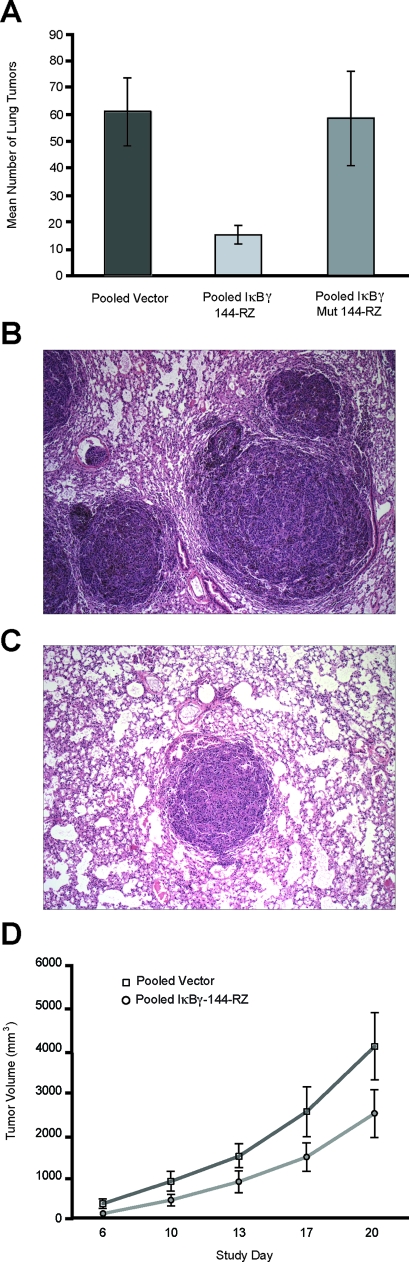
We then examined the in vivo effects of IκBγ suppression on the metastatic potential of B16 melanoma cells. Tail-vein injection of 30,000 pooled transformant cells stably expressing IκBγ-144-Rz into immunocompetent C57BL6 mice resulted in 74% (P < 0.00001) and 75% (P < 0.001) fewer total number of metastatic lung tumors when compared with mice injected with stable transformant cells containing mutant IκBγ-144-Rz or vector alone, respectively (Figure 2A). Histological analysis revealed that the metastatic tumors that developed in the lungs of mice inoculated with anti-IκBγ ribozyme-expressing B16 cells were smaller, with smaller nuclei, and more well circumscribed than tumors formed by their control counterparts (Figure 2, B and C).
Figure 2.
A: Total tumor counts in the lungs of mice intravenously injected with pooled vector, pooled IκBγ-144-Rz, or pooled mutant ribozyme B16 transformant cells. Photomicrographs of metastatic B16 tumor cells in the lung transfected with vector only (B) or with vector containing IκBγ-144-Rz (C). D: Tumor volume after subcutaneous injection of pooled vector or pooled IκBγ-144-Rz B16 transformant cells into the flanks of C57BL\6 mice. Original magnifications, ×100.
To investigate the mechanism of the observed decrease in metastatic potential, we examined the effects of IκBγ suppression on the in vitro growth characteristics of B16-F10 melanoma cells. Cell growth curves of the stable transformants containing the IκBγ-144-Rz did not show a significantly decreased growth rate when compared with transformants containing the vector alone (data not shown). However, the ribozyme-expressing clones did display reduced growth compared with control clones in vivo after their subcutaneous injection into C57BL6 mice (Figure 2D).
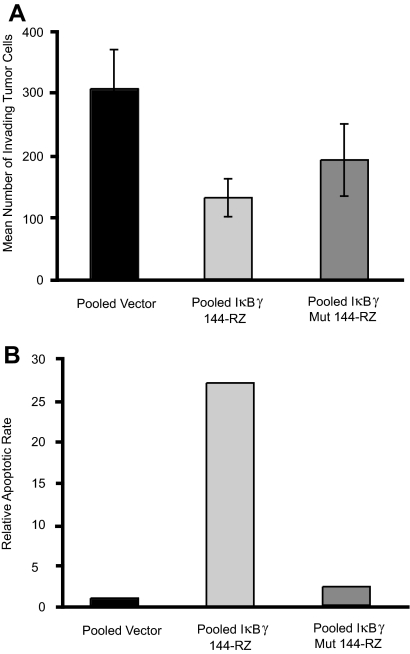
We next examined the effects of targeted suppression of IκBγ on invasion and apoptosis in these B16 transformant cells. To assess invasion, we plated anti-IκBγ ribozyme-expressing or control cells onto Matrigel matrix and counted the invading cells 24 hours later. We found a 55% decrease in the number of invading cells in the IκBγ-144-Rz versus empty vector control clones (P = 0.01) (Figure 3A). There was no statistically significant difference in invasion between the mutant ribozyme clones and the pooled vector clones. Finally, B16 melanoma clones stably expressing IκBγ-144-Rz were examined for apoptotic rates using the terminal dUTP nick-end labeling (TUNEL) assay. Suppression of IκBγ resulted in a 25-fold increase in apoptotic rates versus empty vector control clones in serum-starved conditions (P = 0.001) (Figure 3B). There was no statistically significant difference in apoptotic rates between the mutant ribozyme clones and the pooled vector clones.
Figure 3.
A: Tumor cell invasion into Matrigel by pooled vector, pooled mutant ribozyme, or pooled IκBγ-144-Rz B16 cells. B: Relative apoptotic rate (as determined by percentage of apoptotic cells) in the TUNEL assay in pooled mutant ribozyme or pooled IκBγ-144-Rz B16 transformant cells relative to pooled vector cells.
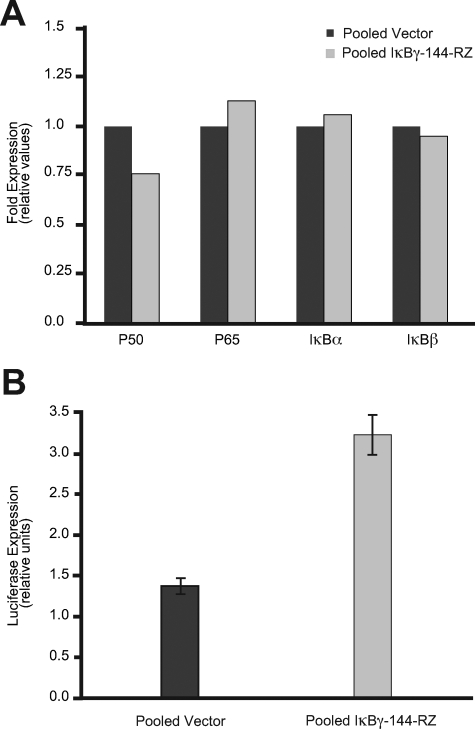
We then assessed the effects of targeting IκBγ on the expression of NF-κB and on the activation of the NF-κB pathway. We analyzed the expression of several members of the NF-κB and IκB families by quantitative PCR. There was no significant change in expression in the p65 subunit of NF-κB, nor that of IκBα and -β (Figure 4A). Expression of p50 was diminished by ∼25%, consistent with targeting p105 mRNA, that also contains the p50 mRNA.6 However, there was no significant down-regulation of p50 protein by Western analysis (data not shown). In addition, we examined the level of NF-κB transcriptional activity. Luciferase assays of a plasmid driven by the NF-κB promoter revealed a 2.3-fold higher level of luciferase activity in the anti-IκBγ-expressing cells when compared with vector control transfectants (Figure 4B).
Figure 4.
A: Quantitative PCR analysis of p50, p65, IκBα, and IκBβ expression in pooled vector and pooled IκBγ-144-Rz cells. B: NF-κB activation assay in pooled vector and pooled IκBγ-144-Rz clones. Luciferase activity was assessed by transfecting a NF-κB-Luc reporter vector in pooled vector and pooled IκBγ-144-Rz stable clones. The NF-κB-Luc is a reporter plasmid containing six tandem repeats of the NF-κB consensus DNA-binding sites that measures NF-κB transactivation activity.
Given that B16 harbors activating mutations in β-catenin, we examined whether IκBγ expression was detectable in a panel of human melanoma cells lines, and compared the level of IκBγ and β-catenin. IκBγ was clearly detectable by Western blot analysis, and expressed at various levels in the human melanoma cell lines examined. However, there was no direct correlation between IκBγ and β-catenin expression levels in the cell lines tested (data not shown).
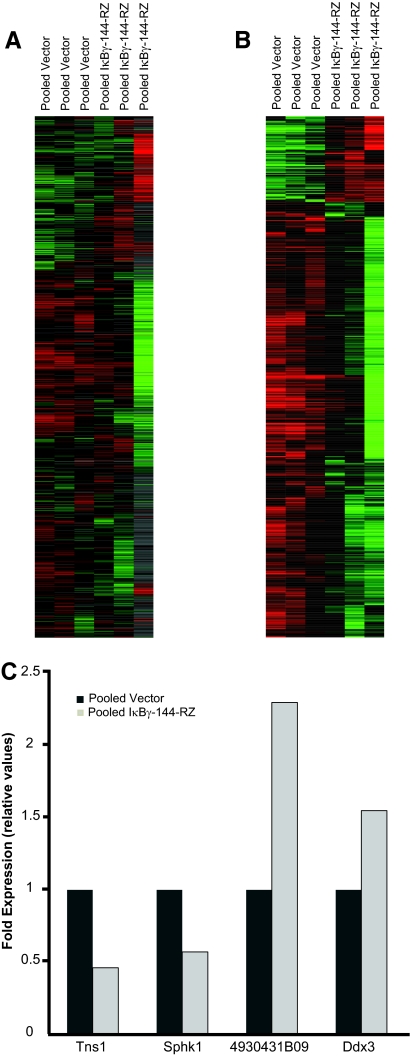
To identify potential downstream genes regulated by inhibition of IκBγ, we performed gene expression profiling and supervised hierarchical analysis of cDNA microarrays of the pooled anti-IκBγ ribozyme-expressing clones compared with pooled vector-only control clones. Significance analysis of microarrays identified 937 genes whose expression was significantly altered by inhibition of IκBγ, of which 783 genes were up-regulated and 154 were down-regulated (Figure 5, A and B). Among the up-regulated genes were several genes encoding tumor suppressor proteins, including metastasis suppressor-1 (MTSS-1), Riken cDNA 4930431B09 (homologous to SNM1 tumor suppressor), Trp53inp1 (transformation related protein 53 inducible nuclear protein 1), DDX3 (DEAD box polypeptide 3), and PIAS-1 (protein inhibitor of activated Stat-1), as well as genes encoding proteins involved in apoptosis, such as EglN3 (Egl nine homolog 3) and Dido-1 (death inducer-obliterator 1). Down-regulated genes included several oncoproteins (MAF, sphingosine kinase-1), and genes involved in cell cycle regulation (cyclin-dependent kinase 2), adhesion, and cell migration (tensin-1). The altered expression of several of these genes was then confirmed by quantitative PCR. Specifically, tensin-1 and sphingosine kinase-1 were confirmed to be down-regulated, whereas DDX3 and Riken cDNA 4930431B09 were up-regulated in anti-IκBγ ribozyme-expressing cells (Figure 5C).
Figure 5.
Gene expression profiling comparing triplicate arrays of RNA isolated from pooled vector (lanes 1 to 3) and pooled IκBγ-144-Rz (lanes 4 to 6) cells. A: The entire gene set in the array. B: The significant gene list derived from SAM analysis. C: Quantitative PCR analysis of Tns1, Sphk1, DDX3, and Riken cDNA 4930431B09 expression as normalized to levels of histone gene expression in pooled vector or pooled IκBγ-144-Rz B16 cells.
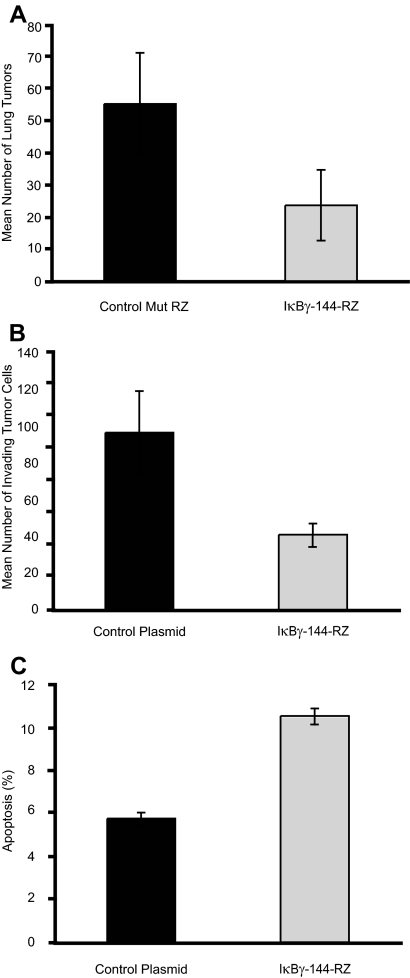
Finally, we assessed the effects of transient, plasmid-based expression of IκBγ-144-Rz. We first examined whether intravenous injection of cationic liposomes complexed to a plasmid (CLDC) encoding IκBγ-144-Rz could alter the metastatic progression of B16-F10 melanoma cells. Systemic administration of CLDC encoding IκBγ-144-Rz after tail vein injection of B16-F10 cells resulted in a significant reduction in the total number of lung metastases (P < 0.03) when compared with tumor-bearing mice treated with CLDC containing a control mutant ribozyme (Figure 6A). Next, we evaluated whether similar phenotypic effects of ribozyme-mediated targeting of IκBγ could be observed in another tumor cell line. In vitro CLDC-based transfection of murine Lewis lung carcinoma cells resulted in significantly reduced tumor cell invasion (P < 0.05) into Matrigel (Figure 6B) and significantly increased apoptosis (P < 0.001, Figure 6C) when compared with tumor cells transfected with a vector only control. Thus, plasmid-based expression of IκBγ-144-Rz in a different tumor type produced similar phenotypic effects in vitro to those observed in the B16 stable transformants.
Figure 6.
Effects of transient, plasmid-mediated expression of IκBγ-144-Rz. A: Analysis of total tumor counts in the lungs of mice treated with CLDC containing IκBγ-144-Rz or disabled control mutant ribozyme. B: Tumor cell invasion into Matrigel of Lewis lung carcinoma cells transfected with vector control or with vector expressing IκBγ-144-Rz. C: Apoptotic rate of Lewis lung carcinoma cells transfected with vector control or with vector expressing IκBγ-144-Rz in the TUNEL assay.
Discussion
In this study, we examined the role of ribozyme-mediated inhibition of IκBγ in melanoma metastasis. Our results showed that stable expression of a hammerhead ribozyme targeting murine IκBγ significantly suppressed the metastatic potential of B16 melanoma cells, when compared with B16 cells expressing either control empty vector or a mutant, disabled anti-IκBγ ribozyme. The anti-IκBγ ribozyme-expressing transformants exhibited a higher level of NF-κB activity, along with significantly reduced invasiveness and a higher apoptotic rate, when compared with control transfectants. The phenotypic effects of the anti-IκBγ ribozyme were also observed on plasmid-mediated expression in Lewis lung carcinoma cells. Gene expression profiling of ribozyme-expressing versus control B16-F10 cells identified the altered expression of a number of genes (confirmed by quantitative PCR) that could help explain the observed in vitro and in vivo phenotypic effects observed after inhibition of IκBγ expression.
Previously, other members of the IκB family have been shown to inhibit the DNA-binding and nuclear translocation activities of the p50 NF-κB subunit,11 whose expression has been described to enhance melanoma cell migration and progression.12 In contrast, the structure of IκBγ protein differs from the other members of this family. It is encoded by the C-terminal portion of the NF-κB p105 gene, a precursor to the p50 NF-κB subunit, suggesting that IκBγ may function differently from other gene family members. However, to date, the function of IκBγ has remained primarily uncharacterized. Prior studies have demonstrated that sequences in the C terminus of p105 contain a transcriptional activation domain.13 IκBγ has been shown to be up-regulated in mast cells14 and brain cells of patients with Alzheimer’s disease,15 supporting its possible role as a transcriptional activator. However, we are unaware of any prior reports examining the role of IκBγ in regulating tumor progression or metastasis.
Our results thus assign novel prometastatic function to IκBγ. Moreover, they suggest that IκBγ promotes the metastatic potential of B16-F10 melanoma cells by virtue of its effects on tumor cell invasion and apoptosis, features that were also observed on ribozyme targeting in Lewis lung carcinoma cells. Interestingly, these anti-tumor effects were shown after activation of NF-κB, which may at first appear surprising. However, these results are consistent with recent studies that describe a tumor suppressor role for NF-κB under specific conditions and in several different model systems.16,17,18,19
For example, functional blockade of NF-κB in the skin of transgenic mice overexpressing IκBα resulted in severe hyperplasia that, in cooperation with activated H-ras, produced invasive neoplasms.20 In addition, mouse embryo fibroblasts from Ikkβ knockout mice exhibited increased rates of proliferation, migration, and invasion.21 Separately, NF-κB has been shown to be involved in mediating p53-induced cell death.22 Our results, demonstrated in both murine B16 melanoma and Lewis lung carcinoma, for the first time, add inhibition of IκBγ to the list of conditions that can unleash the tumor suppressor and pro-apoptotic activity of NF-κB. Because these effects were observed in a cell line with mutant β-catenin, they may not be applicable to melanoma cell lines without β-catenin mutation.
Using gene expression profiling and supervised hierarchical analysis of cDNA microarrays, we identified several genes whose expression is altered by stable suppression of IκBγ that may participate in mediating the observed anti-tumor, anti-invasive, and pro-apoptotic effects. Significance analysis of microarrays identified 937 genes whose expression was regulated by inhibition of IκBγ. Several of the up-regulated cDNAs corresponded to tumor suppressor genes. These include MTSS-1, a tumor suppressor gene that has been described to encode an intercellular protein implicated in actin cytoskeleton organization.23 PIAS-1 has been shown to be an activator of p53,24 as well as a negative regulator of NF-κB.25 Other tumor suppressor genes identified included Riken cDNA 4930431B09,26 lethal giant larvae homolog 2,27 DDX3,28 and Trp53inp1.29 Inhibition of IκBγ also resulted in activation of EglN3 and Dido-1 that exhibit pro-apoptotic activity.30,31 In addition, inhibition of IκBγ led to down-regulation of several genes that behave as oncoproteins. Sphingosine kinase-1 is oncogenic and overexpressed in many solid tumors, where it protects the cells from undergoing apoptosis.32 V-Maf is a well-established oncogene that was first identified in an avian transforming virus, and has been shown to be overexpressed in multiple myeloma and T-cell lymphoma.33 Inhibition of IκBγ also caused a decrease in expression of several genes that are involved in cell-cycle control, adhesion, and chemoresistance, including CDK-2, which regulates cell-cycle progression through the G1/S checkpoint and S phase, and is up-regulated in melanoma cells.34
Finally, the gene expression profiling results identified down-regulation of tensin-1 (confirmed by quantitative PCR), which provide a potential downstream effector protein for the effects of ribozyme action on tumor invasion. Tensin-1 is an actin- and phosphotyrosine-binding protein that localizes to focal adhesion molecules and promotes cell migration.35 Recently, tensin-1 was also identified as a down-regulated gene after stable ribozyme-mediated suppression of telomerase RNA in B16 melanoma cells. Targeting telomerase RNA also resulted in reduced invasiveness in the Matrigel assay.36
In summary, our results identify a previously unreported role for IκBγ in promoting the metastatic progression of B16 melanoma. These effects appear to be mediated, at least in part, through its effects on tumor cell invasion and apoptosis. Moreover, our studies suggest that inhibition of IκBγ can unleash the tumor suppressor properties of NF-κB. Finally, these studies have identified several putative downstream effectors of these anti-invasive and pro-apoptotic pathways after inhibition of IκBγ.
Acknowledgments
We thank Rosie Casella for manuscript preparation, the Mouse Pathology Core for processing murine lung tissues, and the Genome Core at the University of California San Francisco Helen Diller Family Comprehensive Cancer Center for assistance with the quantitative PCR studies.
Footnotes
Address reprint requests to Mohammed Kashani-Sabet, M.D., University of California San Francisco Comprehensive Cancer Center, 1600 Divisadero St., 2nd Fl., Box 1706, San Francisco, CA 94115. E-mail: kashanim@derm.ucsf.edu.
Supported by the Herschel and Diana Zackheim Endowment Fund, the American Cancer Society (research scholar grant RSG-03-247-01-MGO to M.K.S.), and the United States Public Health Service (grants CA114337 and CA122947 to M.K.S.).
References
- Amiri KI, Richmond A. Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 2005;24:301–313. doi: 10.1007/s10555-005-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- Blank V, Kourilsky P, Israel A. Cytoplasmic retention, DNA binding and processing of the NF-kappa B p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T, Zabel U, Van Zee K, Muller JM, Fanning E, Baeuerle PA. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- Inoue J, Kerr LD, Kakizuka A, Verma IM. I kappa B gamma, a 70 kd protein identical to the C-terminal half of p110 NF-kappa B: a new member of the I kappa B family. Cell. 1992;68:1109–1120. doi: 10.1016/0092-8674(92)90082-n. [DOI] [PubMed] [Google Scholar]
- Nosrati M, Li S, Bagheri S, Ginzinger D, Blackburn EH, Debs RJ, Kashani-Sabet M. Anti-tumor activity of systemically delivered ribozymes targeting murine telomerase RNA. Clin Cancer Res. 2004;10:4983–4990. doi: 10.1158/1078-0432.CCR-04-0134. [DOI] [PubMed] [Google Scholar]
- Kashani-Sabet M, Liu Y, Fong S, Desprez PY, Liu S, Tu G, Nosrati M, Handumrongkul C, Liggett D, Thor AD, Debs RJ. Identification of gene function and functional pathways by systemic plasmid-based ribozyme targeting in adult mice. Proc Natl Acad Sci USA. 2002;99:3878–3883. doi: 10.1073/pnas.002025599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez PY, Lin Co, Thomasset N, Sympson CJ, Bissell MJ, Campisi J. A novel pathway for mammary epithelial cell invasion induced by the helix-loop-helix protein Id-1. Mol Cell Biol. 1998;18:4577–4588. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, Allen RE, Singer MI, Leong SPL, Ljung B, Sagebiel RW, Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JR, Watson E, Buckley S, Hay RT. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1993;21:4516–4523. doi: 10.1093/nar/21.19.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Dai DL, Martinka M, Li G. Prognostic significance of nuclear factor-kappaB p105/p50 in human melanoma and its role in cell migration. Cancer Res. 2006;66:8382–8388. doi: 10.1158/0008-5472.CAN-05-4402. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Gilmore TD. The C terminus of the NF-kappa B p50 precursor and an I kappa B isoform contain transcription activation domains. Nucleic Acids Res. 1992;20:2453–2458. doi: 10.1093/nar/20.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase MG, Klawitter A, Baretton GB. IkappaBgamma is expressed in mast cells. Virchows Arch. 2004;445:515–520. doi: 10.1007/s00428-004-1099-9. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. NF-kappaB precursor, p105, and NF-kappaB inhibitor, IkappaBgamma, are both elevated in Alzheimer disease brain. Neurosci Lett. 2005;373:115–118. doi: 10.1016/j.neulet.2004.09.074. [DOI] [PubMed] [Google Scholar]
- Perkins ND. NF-κB: tumor promoter or suppressor? Trends Cell Biol. 2004;14:64–69. doi: 10.1016/j.tcb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Gilmore TD. Good cop, bad cop: the different faces of NF-κB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- Chen F, Castranova V. Nuclear factor-kappaB, an unappreciated tumor suppressor. Cancer Res. 2007;67:11093–11098. doi: 10.1158/0008-5472.CAN-07-1576. [DOI] [PubMed] [Google Scholar]
- Chen F, Beezhold K, Castranova V. Tumor promoting or tumor suppressing of NF-kappa B, a matter of cell context dependency. Int Rev Immunol. 2008;27:183–204. doi: 10.1080/08830180802130327. [DOI] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, Marinkovich MP, Tao S, Lin Q, Kubo Y, Khavari PA. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- Chen F, Lu Y, Castranova V, Li Z, Karin M. Loss of Ikkbeta promotes migration and proliferation of mouse embryo fibroblast cells. J Biol Chem. 2006;281:37142–37149. doi: 10.1074/jbc.M603631200. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhou K, Zeng X, Lin J, Zhan X. Tyrosine phosphorylation of missing in metastasis protein is implicated in platelet-derived growth factor-mediated cell shape changes. J Biol Chem. 2007;282:7624–7631. doi: 10.1074/jbc.M608448200. [DOI] [PubMed] [Google Scholar]
- Megidish T, Xu JH, Xu CW. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J Biol Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K. Negative regulation of NF-kappaB signaling by PIAS1. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahkter S, Richie CT, Zhang N, Behringer RR, Zhu C, Legerski RJ. Snm1-deficient mice exhibit accelerated tumorigenesis and susceptibility to infection. Mol Cell Biol. 2005;25:10071–10078. doi: 10.1128/MCB.25.22.10071-10078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KL, Justice RW, Bryant PJ. Drosophila in cancer research: the first fifty tumor suppressor genes. J Cell Sci Suppl. 1994;18:19–33. doi: 10.1242/jcs.1994.supplement_18.4. [DOI] [PubMed] [Google Scholar]
- Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Seux M, Nowak J, Bontemps C, Carrier A, Dagorn JC, Pébusque MJ, Iovanna JL, Dusetti NJ. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene. 2005;24:8093–8104. doi: 10.1038/sj.onc.1208951. [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yan H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG, Jr, Schlisio S. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Rojas AM, Sanchez-Pulido L, Fütterer A, van Wely KH, Martinez -AC, Valencia A. Death inducer obliterator protein 1 in the context of DNA regulation. Sequence analyses of distant homologues point to a novel functional role. FEBS J. 2005;272:3505–3511. doi: 10.1111/j.1742-4658.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- Cuvillier O. Sphingosine kinase-1—a potential therapeutic target in cancer. Anticancer Drugs. 2007;18:105–110. doi: 10.1097/CAD.0b013e328011334d. [DOI] [PubMed] [Google Scholar]
- Morito N, Yoh K, Fujioka Y, Nakano T, Shimohata H, Hashimoto Y, Yamada A, Maeda A, Matsuno F, Hata H, Suzuki A, Imagawa S, Mitsuya H, Esumi H, Koyama A, Yamamoto M, Mori N, Takahashi S. Overexpression of c-Maf contributes to T-cell lymphoma in both mice and human. Cancer Res. 2006;66:812–819. doi: 10.1158/0008-5472.CAN-05-2154. [DOI] [PubMed] [Google Scholar]
- Kuźbicki L, Aadowicz E, Chwirot BW. Cyclin-dependent kinase 2 expression in human melanomas and benign melanocytic skin lesions. Melanoma Res. 2006;16:435–444. doi: 10.1097/01.cmr.0000232290.61042.ee. [DOI] [PubMed] [Google Scholar]
- Chen H, Lo SH. Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem J. 2003;370:1039–1045. doi: 10.1042/BJ20021308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri S, Nosrati M, Li S, Fong S, Torabian S, Rangel J, Moore DH, Federman S, Laposa RR, Baehner FL, Sagebiel RW, Cleaver JE, Haqq C, Debs RJ, Blackburn EH, Kashani-Sabet M. Genes and pathways downstream of telomerase in melanoma metastasis. Proc Natl Acad Sci USA. 2006;103:11306–11311. doi: 10.1073/pnas.0510085103. [DOI] [PMC free article] [PubMed] [Google Scholar]