Abstract
Extensive studies are currently being performed to associate disease susceptibility with one form of genetic variation, namely single nucleotide polymorphisms (SNPs). In recent years another type of common genetic variation has been characterised, namely structural variation, including copy number variations (CNVs). To determine the overall contribution of CNVs to complex phenotypes we have performed association analyses of expression levels of 14,925 transcripts with SNPs and CNVs in individuals who are part of the International HapMap project. SNPs and CNVs captured 83.6% and 17.7% of the total detected genetic variation in gene expression, respectively, but the signals from the two types of variation had little overlap. Interrogation of the genome for both types of variants may be an effective way to elucidate the causes of complex phenotypes and disease in humans.
Understanding the genetic basis of phenotypic variation in human populations is currently one of the major goals in human genetics. Gene expression (the transcription of DNA into messenger RNA) has been interrogated in a variety of species and experimental scenarios to investigate the genetic basis of variation in gene regulation (1-8), and to tease apart regulatory networks (9, 10). In some respects, a comprehensive survey of gene expression phenotypes (steady-state levels of messenger RNA) serves as a proxy for the breadth and nature of phenotypic variation in human populations (11). Much of the observed variation in mRNA transcript levels may be compensated at higher stages of regulatory networks, but an understanding of the nature of genetic variants that affect gene expression will provide an essential framework and model for elucidating the causes of other types of phenotypic variation. Single nucleotide polymorphisms (SNPs) have long been known to be associated with phenotypic variation either through direct causal effects or by serving as proxies for other causal variants with which they are highly correlated (i.e. in Linkage Disequilibrium) (1, 2, 12). An understanding of this association has been facilitated by the validation of millions of SNPs by the International HapMap project (13). However, during the last few years, structural variants, such as copy number variants (CNVs) - defined as DNA segments that are 1 kb or larger in size present at variable copy number in comparison with a reference genome (14)- have attracted much attention (2). It has become apparent that they are quite common in the human genome (15-19) and can have dramatic phenotypic consequences as a result of altering gene dosage, disrupting coding sequences or perturbing long-range gene regulation (20, 21). Evidence has been presented that increased copy number can be positively (18, 22) or negatively (23) correlated with gene expression levels (for example, deletion of a transcriptional repressor could serve to elevate gene expression) but the relative contribution of such large genetic variants (i.e. CNVs) and smaller variants (i.e. SNPs) to phenotypic variation has not been evaluated. It is also still unknown whether SNPs can serve as proxies to CNVs (24, 25), and whether the complex nature of some CNVs requires that they be surveyed directly (26). We have used the phase I HapMap SNPs (13) and the recently described CNV data ascertained in the same HapMap populations (26) to correlate with genome-wide gene expression variation in the same individuals.
Gene expression was interrogated in lymphoblastoid cell lines of all 210 unrelated HapMap individuals (13) from 4 populations (CEU: 60 Utah residents with ancestry from northern and western Europe; CHB: 45 Han Chinese in Beijing; JPT: 45 Japanese in Tokyo; YRI: 60 Yoruba in Ibadan, Nigeria) in 4 technical replicates (see Methods). Out of the 47,294 transcripts that were interrogated, the normalized values for 14,925 transcripts (14,072 genes) were included in the analysis (see Methods and www.sanger.ac.uk/genevar). The SNP genotypes from phase I HapMap (www.hapmap.org; release 16c.1) were used in the analysis (see Methods). CNV data were represented by log2 ratios from comparative genomic hybridization (CGH) of each HapMap individual against a common reference individual on an array comprising 26,574 large-insert clones covering 93.7% of the euchromatic portion of the genome ((26) and www.sanger.ac.uk/humgen/cnv/data). Log2 ratios from two sets of clones were analyzed: the whole set of 24,963 autosomal clones (CGH-clones) and the 1322 autosomal clones corresponding to CNVs present in at least two HapMap individuals (CNV clones) (26). We excluded genes on sex chromosomes due to their imbalance in males and females. We performed linear regression (on each of the 4 populations separately) between normalized quantitative gene expression values and SNP genotypes or clone log2 ratios that were near the gene (SNP position or clone midpoint within 1 Mb and 2Mb, respectively, of the probe midpoint position). We used different window sizes for SNPs and clones because clones are large (median size of ∼170 Kb) and structural variants can exert long-range effects (21), so a 2 Mb window is more appropriate. Statistical significance was evaluated through the use of permutations (27), as previously described (1), and a corrected p-value threshold of 0.001 applied (see Methods). Repeated permutation exercises showed that our permutation thresholds were very stable (see Supplementary Table 4). We test a large number of genes so an additional correction is required. This can either be done by adjusting the threshold to a new corrected threshold above which all genes are expected to be significant (e.g. Bonferoni correction) or by setting the threshold to a value that generates a satisfactory false discovery rate (FDR). We have used the second and we have estimated the FDR based on the number of genes tested and required that in all cases at least 80% of the genes called significant are estimated to be trully significant. Given that there are 14,072 genes that lie within 1Mb of SNPs and within 2Mb of the full set of CGH-clones, and ∼7150 genes that lie within 2Mb from the CNV clones (from 7135 to 7191 depending on the population, due to missing data), we expect this analysis to generate false positive association signals for 14 and 7 genes respectively in each population.
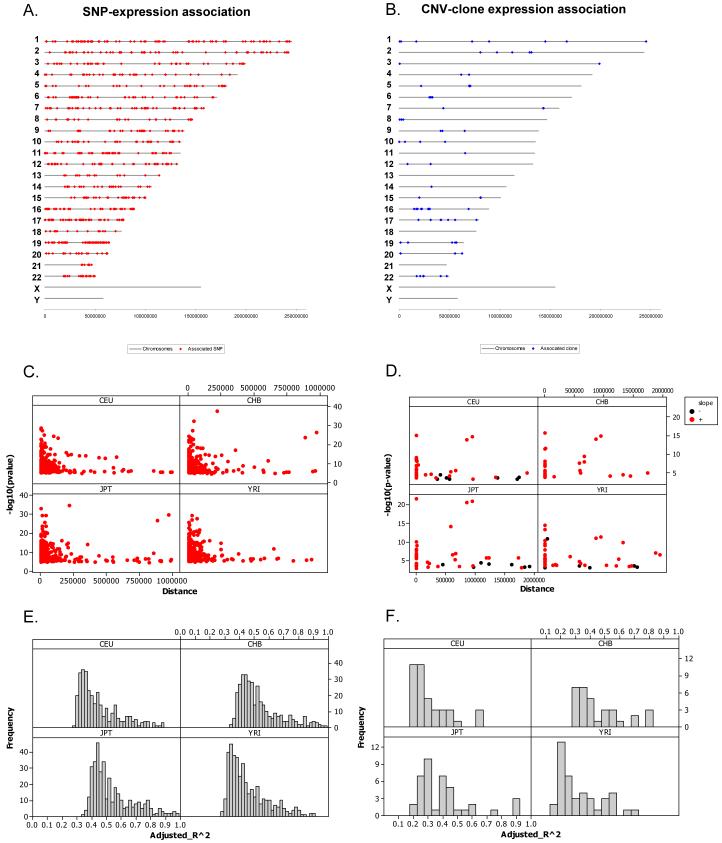
Of the 14,072 genes tested, we detected significant associations with at least one SNP for 323, 348, 370 and 411 genes for CEU, CHB, JPT and YRI, respectively (e.g. Table 1 and Supplementary Table 1). These comprise a total of 888 non-redundant genes of which 331 (37%) were replicated at the same significance level in at least one other population, and of those, 67 (8%) were significant in all 4 populations (Table 2). As expected, we have limited power to detect weak effects due to the small sample sizes: the minimum detected squared regression coefficient (r2) - which reflects the proportion of expression variance accounted for by the linear association with allele counts- was 0.27. However, some very strong effects were detected, that in some cases had an r2 close to 1 (Figure 2). We detected a strong preference for associated SNPs to be close to their respective genes, most of which were within 100 Kb of the interrogated expression probe (Figure 2). In summary, we detected a large number of regions that appear to carry genetic variation affecting gene expression. To evaluate the effect of experimental variation, and hence the robustness of our associations, we compared the list of gene expression associations from our previous study (1) in which we detected 63 expression associations significant at the 0.05 permutation threshold in the CEU population. Of those 63 expression phenotypes, 47 went into the current analysis of which 43 of them (91.5%) were called significant at the same permutation threshold (0.05) in the same population. The previous study was performed with different batches of cells, using RNA extracted in a different laboratory, with RNA levels quantified on a different type of array (custom vs. genome-wide array), so the high degree of experimental and statistical replication strongly suggests that the signals we detected are robust and stable to experimental variation in expression measurements.
Table 1.
Numbers of genes with significant associations to SNPs (SNP-probe distance < 1Mb), all CGH- clones (clone-probe distance < 2 Mb), or CNV clones (clone-probe distance < 2 Mb) as assessed by permutations, together with the numbers of overlaps between SNP-associated genes and CGH or CNV clone-associated genes (probe-variant distance < 1 Mb for both SNPs and clones)
| permutation threshold 0.01 | |||||
|---|---|---|---|---|---|
| CNV (2Mb) | SNP | CNV (1Mb) + SNP overlap | |||
| CGH-clones | CNV clones | CGH-clones | CNV clones | ||
| CEU | 362 | 138 | 643 | 14 | 15 |
| CHB | 221 | 110 | 673 | 10 | 9 |
| JPT | 319 | 134 | 752 | 13 | 14 |
| YRI | 481 | 166 | 815 | 14 | 11 |
| Non-redundant | 1246 | 451 | 1886 | 28 | 16 |
| permutation threshold 0.01 | |||||
|---|---|---|---|---|---|
| CNV (2Mb) | SNP | CNV (1Mb) + SNP overlap | |||
| CGH-clones | CNV clones | CGH-clones | CNV clones | ||
| CEU | 85 | 40 | 323 | 9 | 8 |
| CHB | 44 | 32 | 348 | 5 | 6 |
| JPT | 58 | 40 | 370 | 8 | 6 |
| YRI | 96 | 42 | 411 | 7 | 6 |
| Non-redundant | 238 | 99 | 888 | 15 | 12 |
| permutation threshold 0.01 | |||||
|---|---|---|---|---|---|
| CNV (2Mb) | SNP | CNV (1Mb) + SNP overlap | |||
| CGH-clones | CNV clones | CGH-clones | CNV clones | ||
| CEU | 32 | 18 | 198 | 5 | 6 |
| CHB | 14 | 19 | 204 | 4 | 4 |
| JPT | 23 | 20 | 217 | 6 | 5 |
| YRI | 27 | 16 | 251 | 2 | 2 |
| Non-redundant | 69 | 39 | 526 | 8 | 8 |
Table 2.
Sharing of associations between populations
| CGH-clone (2Mb) | CNV clone (2Mb) | SNP (1Mb) | |
|---|---|---|---|
| CEU-CHB-JPT-YRI | 5 | 7 | 67 |
| CEU-CHB-JPT | 2 | 4 | 48 |
| CEU-CHB-YRI | 1 | 0 | 11 |
| CEU-JPT-YRI | 1 | 0 | 12 |
| CHB-JPT-YRI | 3 | 3 | 28 |
| CEU-CHB | 1 | 3 | 18 |
| CEU-JPT | 2 | 0 | 15 |
| CEU-YRI | 6 | 6 | 36 |
| CHB-JPT | 4 | 5 | 51 |
| CHB-YRI | 1 | 3 | 18 |
| JPT-YRI | 2 | 3 | 27 |
| CEU only | 67 | 20 | 116 |
| CHB only | 27 | 7 | 107 |
| JPT only | 39 | 18 | 122 |
| YRI only | 77 | 20 | 212 |
| SUM | 238 | 99 | 888 |
| gene associations in at least 2 populations | 28 | 34 | 331 |
| percentage of total | 0.12 | 0.34 | 0.37 |
| gene associations in single populations | 210 | 65 | 557 |
| percentage of total | 0.88 | 0.66 | 0.63 |
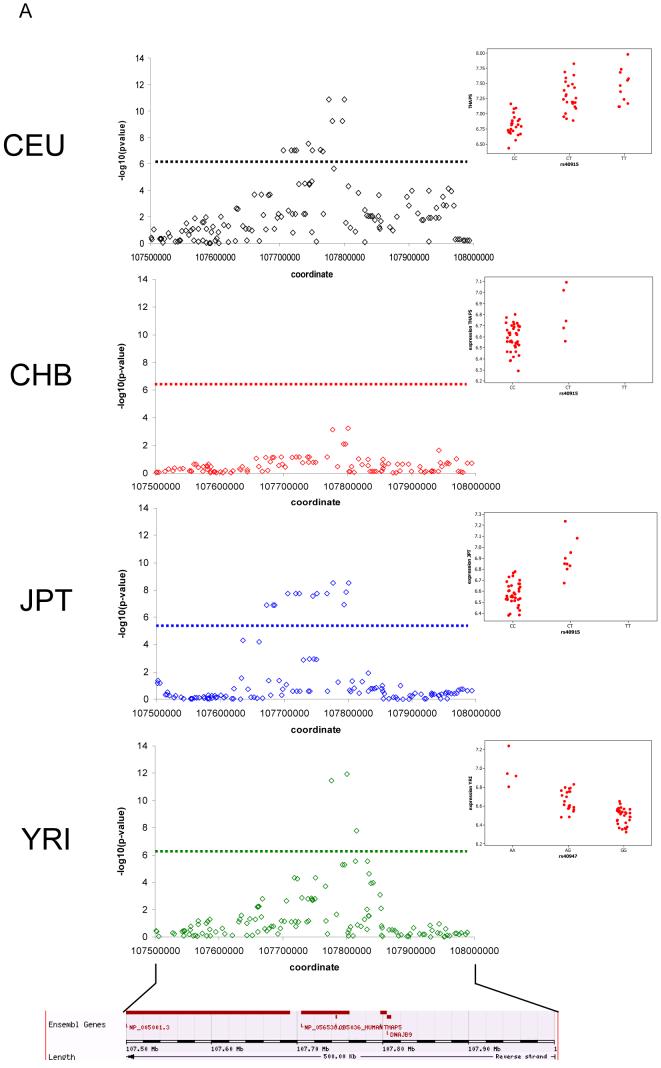
Figure 2.
Genomic location of significant cis-associations for A. SNP-expression associations and B. CNV clone-expression associations. Strength of association as a function of distance between C. SNP and probe and D. CNV and probe. Positive associations between mRNA levels and clone log2 ratios are shown in red, negative associations in black. Distance equal to zero corresponds to the probe residing within the CNV. In each population panel, only the details for the most significant association per significant gene are shown. Distribution of r2 values for the most significant association per significant gene for E. SNP-expression associations and F. clone-expression associations.
Of the 14,072 genes tested, we detected significant associations with at least one of the 24,962 autosomal CGH-clones in 85, 44, 58 and 96 genes in CEU, CHB, JPT and YRI, respectively (238 non-redundant genes), of which 28 (12%) were replicated at the same significance level in at least one other population, and of those, 5 (2%) were significant in all 4 populations (Table 1 and Supplementary Table 2). Not all associated clones were within CNVs defined using the stringent criteria of (26)(119/303 (39%) associated clones were previously defined as CNVs), and it is likely that some of these clones encompass smaller CNVs that are detectable though associations of log2 ratios across a population, but cannot be detected as extreme outliers in their log2 ratios in any one individual (as is required for classification as a CNV in (26)) - see example below. For 36 common (MAF > 0.05) CNVs (encompassing 99 CGH-clones) accurate CNV genotypes were available. We used these genotypes to validate the statistical power of performing association analysis using log2 ratios directly rather than genotypes. There was strong correlation between r-squared values or p-values generated using the log2 ratio signals or the CNV genotypes (Pearson correlation coefficients > 0.9), indicating that log2 ratios can be used directly.
There exists little prior data on CNV-expression associations against which to compare and demonstrate the robustness of our associations. One recent study (18) demonstrated three associations between common deletions and gene expression in a subset of the CEU. Two of these deletions are covered by our CGH data. The reported expression-association caused by the largest of these two deletions is also captured in our analysis (influencing UGT2B17), and we extend this observation to show that this deletion also affects the expression of three other nearby genes (UGT2B7, UGT2B10 and UGT2B11) and that these associations replicate across all four populations. The smaller deletion of only 18 Kb, reported previously (18) as affecting expression of GSTM1, is below the expected resolution of the CGH data. Nonetheless we observe an association that although it does not pass our stringent permutation threshold (0.001), it has significant nominal P-values in all 4 populations (PCEU = 0.0292; PYRI = 0.0018; PJPT = 0.0408; PCHB = 0.0185). This suggests that effects of CNVs far smaller than the CNV calling resolution of the CGH platform can be detected and replicated in multiple populations with our analysis.
Having investigated the potential contribution of CNV to variation in gene expression by using data from all CGH-clones, we interrogated the nature of CNV effects on gene expression in finer detail by performing association tests of 1322 clones within high confidence CNVs (see above) with expression of the 14,072 genes, in order to generate a set of high stringency associations for which the presence of an underlying CNV has already been validated. Significant associations with at least one of the 1322 CNV clones were detected for 40, 32, 40 and 42 genes in CEU, CHB, JPT, YRI, respectively (99 non-redundant genes). Thirty-four of the 99 genes (34%) associated with CNV clones have a significant signal in at least two populations (Table 2), of which 7 (7%) were associated in all populations. Some CNV-clones were associated with more than one gene in the same population, with a notable example being a single CNV-clone associated with expression of 4 genes in all populations (UGT2B genes, see above). CNVs detected by CGH can be classified into five classes: deletion, duplication, deletion and duplication at the same locus, multiallelic, and complex (26); we find all classes of CNV represented among the significant associations. Despite the clear preference for genes to lie close to their associated CNVs (Figure 2), 53% of the expression probes associated with a CGH-clone were located outside the CNVs encompassing that clone (26). This suggests that rather than altering gene dosage, approximately half the CNV effects are caused by disruption of the gene (some parts of the gene, but not the probe, are within in the CNV) or affect regulatory regions and other functional regions that have an impact on gene expression. When we extended our analysis to consider associations between genes and CNVs up to 6Mb apart, we detected a few significant long distance associations beyond 2 Mb (see SOM). These types of long-range effects are becoming more apparent through recent studies looking in detail at specific genomic regions (20, 28). A small minority (5-15%) of the significant CNV-expression associations have a negative correlation between copy number and gene expression, suggesting that not all the detected effects are of the conventional type wherein gene expression levels increase with gene copy number (Supplementary Table 3). Almost all (32/34) of the associations that are shared between populations also exhibit the same direction of correlation in all populations. The two exceptions could result from the CNV being in LD with different regulatory variants in different populations or due to SNP x CNV interactions. However, the strong bias towards positive correlations between copy number and expression levels implies that the vast majority of these associations are attributable to the CNV itself, and not a linked variant.
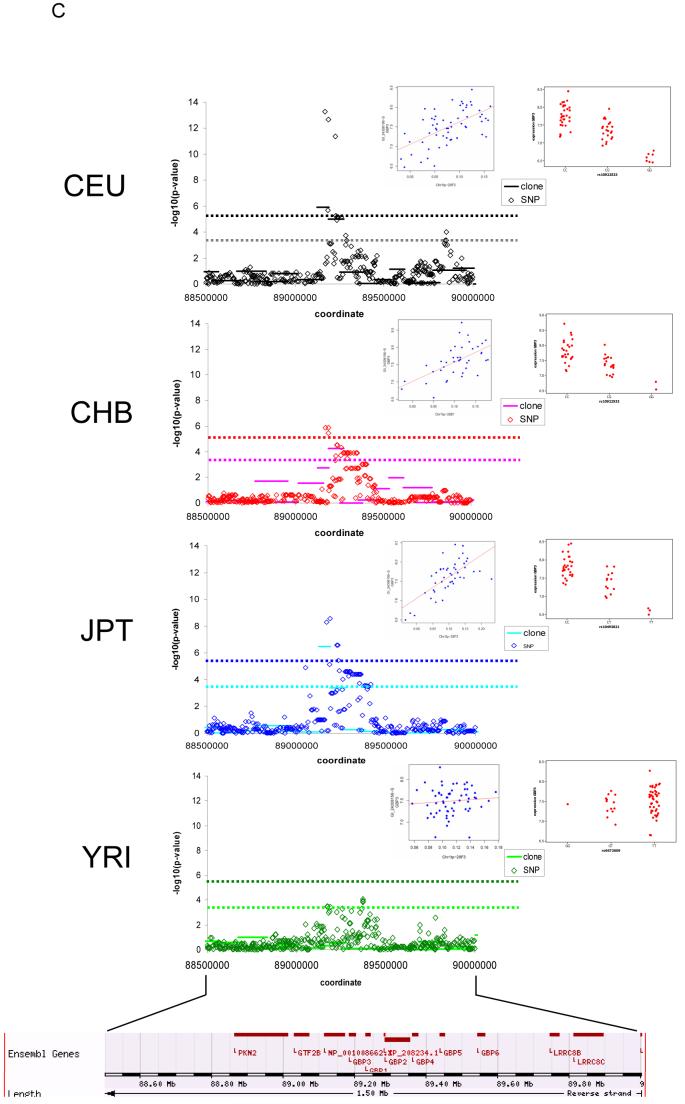
We next determined whether the same associations were also captured by SNPs (e.g. Figure 1C). We only considered those CGH-clones or CNVs within 1 Mb of the probe so that the analysis is comparable to that of the SNPs (total of 188 and 84 genes for CGH-clones and CNVs, respectively). We expect some of the CNVs to be correlated with SNPs via common genealogical history (linkage disequilibrium) and therefore their effect on gene expression would also be captured by SNP associations. Fewer than 20% (in all populations) of the detected CGH-clone associations overlapped with SNP associations (Table 1) even when we included CGH and SNP associations with the same gene but in different populations (28/188 (14%) genes with significant CGH-clone associations also had a SNP association in any population). The same is true of CNV-clone associations: only 15 of 84 genes (18%) with CNV clone associations within 1 Mb also had a SNP association in any population and if we required the association in the same population, only 12 (14%) of genes had a SNP association. On the basis of previous work characterising the patterns of linkage disequilibrium (LD) around CNVs (26), we considered that this low overlap between CNV or CGH-clone associations with SNP associations might be due in part either to a low density of successfully genotyped SNPs around some CNVs or to the suppression of apparent LD by recurrent mutation at some CNVs. Segmental duplications (SDs) are the primary cause of low SNP densities in HapMap Phase I due to the difficulties in developing robust SNP genotyping assays within them (13). We did not observe enrichment of segmentally duplicated sequences within the CGH- and CNV clones that did not share signals with SNPs relative to those CGH- and CNV clones that did share signals with SNPs. However, we observe a 2.5-fold excess of compound CNVs (CNVs with more than one mutation event - based on the classification of the CNVs in (26)) in associations that are not shared with SNPs relative to those that are shared (Fisher’s exact test: P = 0.000064). Thus our analysis suggests that recurrent mutation is a likely factor reducing overlap between CNV and SNP associations.
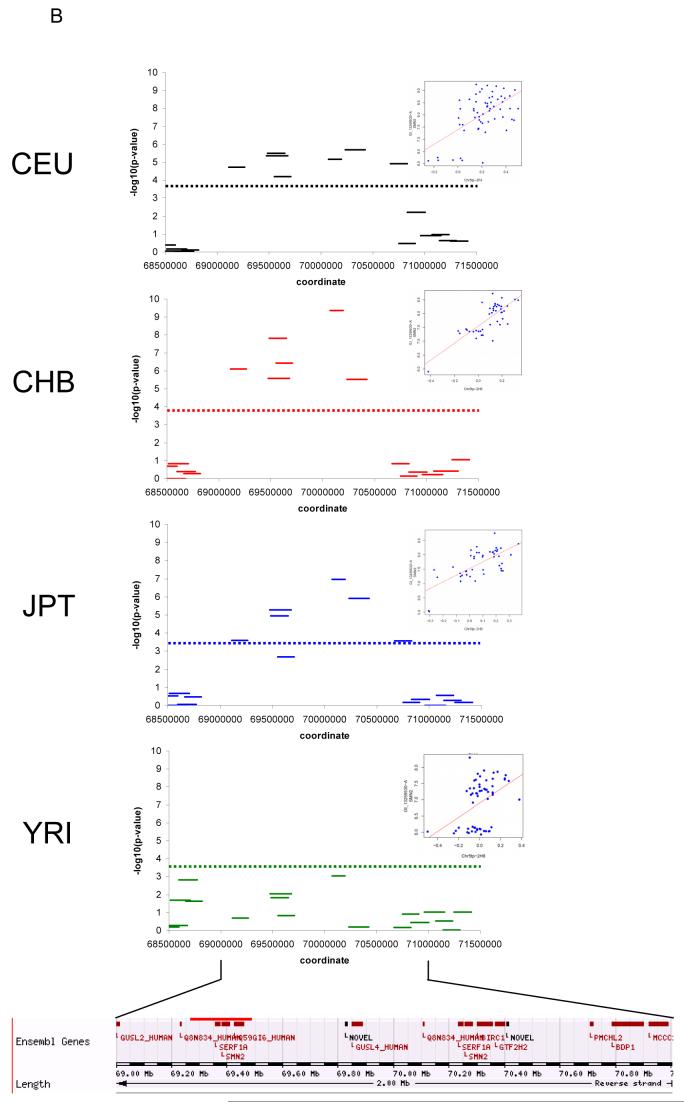
Figure 1.
Examples of SNP-expression and clone-expression associations in the four HapMap populations. A. SNP-expression association for THAP5; chr7. Significant associations between SNPs and expression are observed in CEU, JPT, and YRI, but not in CHB. B. clone-expression association for SMN2; chr5. Significant associations between clones and expression are observed in CEU, CHB, and JPT, but not in YRI. C. SNP-expression and clone-expression association for GBP3; chr1. Both SNPs and clones are significantly associated with expression of GBP3 in CEU, CHB, and JPT, but not in YRI. In each plot, dotted lines show the 0.001 permutation significance threshold. For clone-expression associations, all clones in the window are shown, however the significance threshold was determined by permuting data only from those clones in CNVs where the CNV was present in at least two HapMap individuals. All coordinates shown are from Build 35 of the human genome. Inset panels show the relationship between mRNA levels and SNP genotypes or clone log2 ratios, for the most significant clone or SNP in that population, which may differ across populations.
CNV associations that were also detected with SNPs were clearly biased towards large effect sizes (Supplementary Tables 1 and 3). Of the 12 genes with both SNP and CNV associations in the same population, 8 shared the association in 2 or more populations (giving a redundant total across the four populations of 26 shared CNV and SNP associations). The ratio of 8/12 (67%) population shared associations is larger than that observed in all CNV association (34/99 = 34%) potentially suggesting that associations with higher frequency, older CNVs are more likely to be captured by SNPs. For the 26 associations (representing 12 genes; see above) captured both by CNVs and SNPs in the same population, we observed that SNPs and CNVs were themselves highly correlated for 23/26 SNP-CNV pairs (Pearson correlation p-value <0.001) suggesting that for these cases the CNV and SNP captured the same effect, and that only a small fraction of the associations captured both by SNPs and CNVs occurs by chance. In summary, 87/99 (87%) of genes with a significant CNV association are not associated with SNPs.
The large-scale (typically >100kb) copy number variation analysed here appears to be associated with approximately 10-25% as many gene expression phenotypes as captured by ∼700,000 SNPs, and the majority of these effects cannot be explained by altered dosage of the entire gene but by gene disruption and impact on the regulatory landscape of the region where these CNVs occur. When we restrict the analysis to within 1Mb of the probe of the expressed gene, we detected 1061 genes associated with CGH-clones or SNPs, 17.7% of which are associated with CGH-clones, 83.6% with SNPs, and 1.3% with both. Of the 972 genes associated with CNV clones or SNPs, 8.75% are associated with CNV clones, 92.5% with SNPs, and 1.25% with both. While the Phase I HapMap SNPs likely capture a large fraction of the SNP effects in the genome (13), only a small minority of the CNVs in the genome were considered here: CNVs <100 Kb in length are far more numerous than CNVs >100 Kb length (19). As a consequence, 8.75-17.7% is a minimal estimate of the proportion of heritable gene expression variation that is explained by copy number variation.
Our study has attempted to evaluate the relative impact of CNVs and SNPs on phenotypic variation in human populations. Within the limitations of our samples, tissue type, SNP coverage and CNV resolution, each type of genetic variation captures a substantial number of largely mutually exclusive effects on gene expression. We also demonstrate that both CNV and SNP associations are replicated across populations. Replication of association signals is the sine qua non of association studies, and the fact that we observe this even between diverse populations and with small sample sizes highlights the relevance and robustness of the associations we detect. Gene expression is the basis for many crucial functions in the cell, so the relative contribution of these two types of variants is an indication of the nature of the mutational and natural selection processes that contribute to phenotypic diversity and divergence. It is therefore essential that we interrogate both SNPs and CNVs (of all types) to perform a comprehensive exploration of genetic effects on phenotypic variation and disease. It is possible that if a larger number of SNPs were analyzed, or a higher resolution of CNVs was available, we would observe more overlap between the effects attributed to CNVs and SNPs. However, the difficulty of designing robust SNP genotyping assays in structurally dynamic regions of the genome (26) suggests that even with more comprehensive interrogation of SNPs and CNVs, the overlap may not be high enough for one type of variation to be sufficient for exploring the genetic causes of disease. We have also demonstrated that it is not necessary to perform such studies with CNV calls or CNV genotypes but it is possible to use filtered CGH log2 ratios or any other type of high-quality quantitative signal that reflects underlying copy number variation. It has also become apparent that there are many more structural variants that contribute to phenotypic variation than our stringent calling criteria reveal and higher resolution methods are necessary to elucidate their structure and function. Last but not least is the fact that we have only considered simple models of association in small samples so it is very likely that if we apply more complex and realistic models (e.g. epistatic interactions) and/or larger population samples, a larger number of effects would be revealed. The results presented here reinforce the idea that the complexity of functionally-relevant genetic variation ranges from single nucleotides to megabases, and the full range of the effects of all of these variants will be best captured and interpreted by complete knowledge of the sequence of many human genomes. Until this is possible we need to survey all known types of genetic variation to maximise our understanding of human evolution, diversity and disease.
Supplementary Material
Acknowledgments
We thank A.Clark, and J. Pritchard for comments on earlier versions of the manuscript, M. Smith for assistance with software development and M. Gibbs, J. Orwick and C. Gerringer for technical support. Funding was provided by the Wellcome Trust to ETD, MEH, PD, CTS and NC, NIH ENDGAME to ETD and ST, CRUK to ST, NT, the Leukemia and Lymphoma Society and the Brigham and Women’s Hospital Department of Pathology to CL and MRC to MD. ST is a Royal Society Wolfson Research Merit Award holder. SWS is supported by grants from Genome Canada/Ontario Genomics Institute and is a Scholar of the Canadian Institutes of Health Research and the Howard Hughes Medical Institute.
References
- 1.Stranger BE, et al. PLoS Genet. 2005 Dec;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung VG, et al. Nature. 2005 Oct 27;437:1365. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doss S, Schadt EE, Drake TA, Lusis AJ. Genome Res. 2005 May;15:681. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brem RB, Kruglyak L. Proc Natl Acad Sci U S A. 2005 Feb 1;102:1572. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storey JD, Akey JM, Kruglyak L. PLoS Biol. 2005 Aug;3:e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oleksiak MF, Roach JL, Crawford DL. Nat Genet. 2005 Jan;37:67. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monks SA, et al. Am J Hum Genet. 2004 Dec;75:1094. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadt EE, et al. Nature. 2003 Mar 20;422:297. [Google Scholar]
- 9.Chesler EJ, et al. Nat Genet. 2005 Mar;37:233. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 10.Bystrykh L, et al. Nat Genet. 2005 Mar;37:225. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 11.Dermitzakis ET, Stranger BE. Mamm Genome. 2006 Jun;17:503. doi: 10.1007/s00335-006-0005-y. [DOI] [PubMed] [Google Scholar]
- 12.Pastinen T, Hudson TJ. Science. 2004 Oct 22;306:647. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 13.IHMC Nature. 2005 Oct 27;437:1299. [Google Scholar]
- 14.Feuk L, Marshall CR, Wintle RF, Scherer SW. Hum Mol Genet. 2006 Apr 15;15(Suppl 1):R57. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- 15.Iafrate AJ, et al. Nat Genet. 2004 Sep;36:949. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 16.Sebat J, et al. Science. 2004 Jul 23;305:525. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 17.Tuzun E, et al. Nat Genet. 2005 May 15;37:727. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 18.McCarroll SA, et al. Nature Genetics. 2006;38:86. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 19.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. Nat Genet. 2006 Jan;38:75. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 20.Lupski JR, Stankiewicz P, editors. Genomic Disorders: the genomic basis of disease. Humana Press; Totawa, New Jersey: 2006. [DOI] [PubMed] [Google Scholar]
- 21.Kleinjan DA, van Heyningen V. Am J Hum Genet. 2005 Jan;76:8. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somerville MJ, et al. N Engl J Med. 2005 Oct 20;353:1694. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JA, et al. Ann Neurol. 2006 Feb;59:398. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- 24.Locke DP, et al. American Journal of Human Genetics. 2006;79:275. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Nat Genet. 2006 Jan;38:82. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 26.Redon R, et al. Nature. 2006 Nov 23;444:444. [Google Scholar]
- 27.Doerge RW, Churchill GA. Genetics. 1996 Jan;142:285. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merla G, et al. Am J Hum Genet. 2006 Aug;79:332. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.