Abstract
The role of repetitive DNA sequences in pericentromeric regions with respect to kinetochore/heterochromatin structure and function is poorly understood. Here, we use a mouse erythroleukemia cell (MEL) system for studying how repetitive DNA assumes or is assembled into different chromatin structures. We show that human gamma-satellite DNA arrays allow a transcriptionally permissive chromatin conformation in an adjacent transgene and efficiently protect it from epigenetic silencing. These arrays contain CTCF and Ikaros binding sites. In MEL cells, this gamma-satellite DNA activity depends on binding of Ikaros proteins involved in differentiation along the hematopoietic pathway. Given our discovery of gamma-satellite DNA in pericentromeric regions of most human chromosomes and a dynamic chromatin state of gamma-satellite arrays in their natural location, we suggest that gamma-satellite DNA represents a unique region of the functional centromere with a possible role in preventing heterochromatin spreading beyond the pericentromeric region.
The centromere is a specialized chromosomal region that plays a critical role in segregating chromosomes during mitotic and meiotic cell division. Centromeric DNA in mammals or other multicellular eukaryotes is characterized by very large arrays of tandemly repeated DNA sequences. Although centromeric DNA repeat sequences are thought to be structurally and/or functionally important, they are poorly conserved between species (Lee et al. 1997).
The centromeres of human chromosomes are characterized by several Mb of alpha-satellite DNA (also known as alphoid DNA), which is composed of a tandem array of a 171-bp repeat unit. Alpha-satellite DNA is the only human centromeric DNA sequence identified to date that forms kinetochores de novo, and thus it is used to form and propagate human artificial chromosomes (HACs) in cultured human and mouse cells (Basu and Willard 2005; Okada et al. 2007). Nonalphoid DNA repeats have also been identified adjacent to alpha-satellite DNA in the pericentromeric regions of human chromosomes. For example, the classical satellites I, II, and III are present in the pericentromeric regions of human chromosomes 3, 4, 9, 13, 14, 15, 21, and 22 (Vissel et al. 1992). A subset of beta-satellite DNA has been detected in the pericentromeric region of human chromosome 9; the pericentromeric regions of the human acrocentric chromosomes include sn5-satellite DNA (Choo 1997). Gamma-satellite DNA has been identified in the pericentromeric regions of human chromosomes 8, X, and Y (Lin et al. 1993; Lee et al. 2000; Schueler et al. 2005). Gamma-satellite DNA is a tandem array of 220-bp GC-rich repeating units, usually forming 10- to 200-kb clusters flanked by alpha-satellite DNA (e.g., at 8q11.1) (Lin et al. 1993).
The centromeres of mouse chromosomes have two types of DNA repeat sequences, the major satellite repeat (∼6-Mb arrays/234 bp per repeat unit) and the minor satellite repeat (∼600-kb arrays/120 bp per repeat unit) (Choo 1997) that are distinct from human centromeric repeats. The major mouse satellite is found in the pericentromeric region, and the minor mouse satellite is found in the centric constriction of the centromere (Choo 1997; Schueler and Sullivan 2006).
Despite the diversity in size and sequence of centromeric and pericentromeric DNA, the overall architecture and composition of centromeric chromatin is similar in different species. One hallmark of all functional centromeres is the presence of the H3 variant, CENP-A (Sullivan and Karpen 2004; Lam et al. 2006; Schueler and Sullivan 2006). CENP-A is associated with the centromere/kinetochore, the large protein/DNA complex that attaches to spindle microtubes during mitosis and that includes highly homogeneous alpha-satellite DNA in human and minor satellite DNA in mouse. CENP-A nucleosomes represent open chromatin domains in the centromere core. In contrast, flanking pericentromeric DNA, consisting of both highly diverged alphoid and nonalphoid DNA repeats in human and major satellite repeats in mouse, is assembled into heterochromatin lacking CENP-A (Schueler and Sullivan 2006). Because all centromeres are closely associated with heterochromatin, it seems that it is likely to have a distinct functional or structural role. For example, pericentromeric DNA may promote an altered chromatin conformation that nucleates or stabilizes the centromere.
Recent data indicate that pericentromeric regions may also regulate gene expression during cellular differentiation or reprogramming. For example, development of mouse hematopoietic cells is regulated by zinc finger proteins encoded by the Ikaros gene (ikzf1) (for review, see Ng et al. 2007) that bind to pericentromeric heterochromatin as well as to regulatory sequences in the upstream promoters of many lymphoid-specific genes. The primary Ikaros transcript is alternatively spliced to generate multiple isoforms of Ikaros. The larger Ikaros isoforms, IK-H, IK-VI, IK-V, IK-IV, and IK-III, contain zinc finger motifs that bind the core DNA sequence 5′-GGGAA-3′ (Georgopoulos 2002). Mouse major-satellite DNA also contains multiple recognition sites for Ikaros (Brown et al. 1997). In actively dividing mouse hematopoietic cells, Ikaros forms nuclear complexes with pericentromeric heterochromatin (Brown et al. 1997). Because Ikaros forms dimers and multimers efficiently, it has been proposed that Ikaros induces heterochromatization or chromatin remodeling of mouse DNA, resulting in repression or activation of target genes (Georgopoulos 2002). Although Ikaros-mediated regulation of gene expression has been proposed in human cells and high affinity Ikaros binding sites have been identified in human gamma-satellite DNA on chromosome 8 (Ronni et al. 2007), it is not yet clear whether Ikaros plays similar roles in regulating gene expression in mouse and human cells.
This study analyzes the effect of centromeric and pericentromeric repetitive DNA elements on expression of a chromosomal transgene cassette in cultured mouse erythroleukemia (MEL) cells expressing Ikaros. The results provide insight into possible structural and functional roles of pericentromeric regions in mouse and human chromosomes.
Results
Experimental system for studying antisilencing potential of repetitive DNA elements
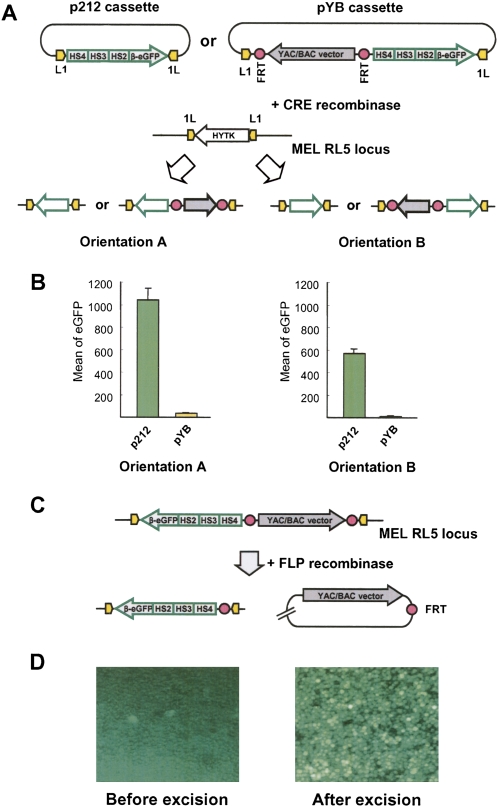
Here, the effects of different repetitive DNA sequences on reporter gene expression were explored using isogenic cell lines that vary in the sequence, but not the location, of the inserted repetitive DNA. These experiments were carried out using the MEL cell line RL5, which carries a counter-selectable marker, HYTK, flanked by inverted lox-P sites. The HYTK-containing cassette is on chromosome 4 (Feng et al. 2005). In our study, the cassette was physically mapped to the Tal1/Pdzk1ip1 (also known as Scl/Map17) locus (Supplemental Fig. S1A). In the presence of Cre recombinase, the loxP-bracketed cassette is exchanged for a reporter gene/repetitive DNA-containing cassette in a high-efficiency in vivo reaction known as recombinase-mediated cassette exchange (RMCE) (Feng et al. 2005). As originally developed, this RMCE system uses the p212 basic targeting cassette that carries the enhanced green fluorescent protein (eGFP) reporter gene under control of the human beta-globin promoter and the human beta-globin locus control region (Feng et al. 2005). When the eGFP transgene (p212 cassette) exchanges into the target locus in RL5 cells, the transgene is expressed at a high level, with higher expression in orientation A than in orientation B (Fig. 1A,B). For this cell line, the level of transgene expression was stable for at least 6 mo without selection, suggesting that the RL5 locus adopts a stable open chromatin structure (data not shown).
Figure 1.
The Cre recombinase-mediated cassette exchange system (RMCE). (A) Cre RMCE was used to exchange the L1-HYTK-1L cassette at the RL5 locus (chromosome 4) in the MEL cells by the p212 or pYB cassettes. Both cassettes have the human beta-globin LCR 234-beta-eGFP and two inverted loxP sites (L1 and 1L). These sites flank the eGFP transgene in p212. In pYB cassette, both eGFP and the YAC/BAC vector backbone sequence are exchanged into the target site. The pYB cassette has two recognition sites for FLP recombinase (FRT). After RMCE, transgenes insert in orientations A or B. (B) Relative eGFP expression is shown in orientation A and orientation B. (C) FLP recombinase-mediated excision of vector DNA sequences. (D) Fluorescence micrograph of MEL cells carrying pYB transgene cassette before or after FLP-mediated excision of the vector backbone sequences.
Our study uses a new cassette, pYB, carrying the eGFP reporter gene (Fig. 1A; Supplemental Figs. S1B, S2). When pYB undergoes Cre-mediated exchange into the target locus in RL5, the eGFP reporter gene and the flanking YAC/BAC vector sequence are coinserted into the mouse chromosome and should result in the reporter gene down-regulation due to epigenetic silencing of the transgene by vector backbone sequences (Chen et al. 2004; Swindle et al. 2004; Suzuki et al. 2006). Indeed, when the pYB-based eGFP transgene exchanges into the target locus, no significant transgene expression was detected (Fig. 1B). Because the YAC/BAC vector sequence in pYB is flanked by FRT sites, it can be excised from the expression cassette by FLP recombinase (Fig. 1C). FLP recombinase-mediated excision of the YAC/BAC vector sequence results in reactivation of transgene expression (Fig. 1D), proving that vector sequence is the cause of transgene silencing. This new system was adapted below to study the effect of mammalian centromeric and pericentromeric repetitive DNA elements on expression of an adjacent transgene in mouse cultured cells.
Pericentromeric repetitive DNA inhibits transgene silencing
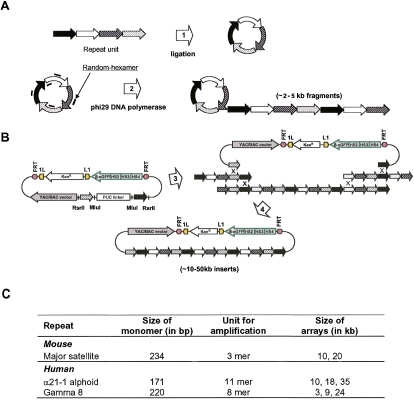
To analyze the effect of repetitive DNA on expression of adjacent genes, large synthetic centromeric and pericentromeric DNA arrays with a defined structure were generated as described by Ebersole and et al. (2005). Briefly, smaller arrays were generated by rolling circle amplification (RCA) and then assembled into longer arrays (up to 35 kb) by homologous recombination during transformation into yeast cells and cloned into pYB to create the reporter cassettes (Fig. 2A,B). Human gamma-satellite DNA arrays (3, 9, or 24 kb) were generated from a tandem repeat of eight 220-bp monomers from chromosome 8 (Lin et al. 1993). Mouse major-satellite DNA arrays (10 or 20 kb) were generated from a trimer of diverged copies of a 234-bp monomer (Choo 1997). Arrays containing 11-mer alpha-satellite 21-I DNA from the centromeric core of chromosome 21 (Okada et al. 2007) were also constructed and incorporated into reporter cassettes (10, 18, or 35 kb) (Fig. 2C). Lastly, as a positive control a cassette was generated that contains two copies of the chicken beta-globin HS4 insulator (cHS4) core. As known, cHS4 is used to protect transgenes from epigenetic gene silencing (Gaszner and Felsenfeld 2006).
Figure 2.
Construction of synthetic DNA arrays for RMCE cassettes. (A) Repeat DNA elements were amplified by rolling circle amplification (RCA). Such amplification generates RCA products of 2–5 kb in size. (B) RCA products were assembled into long arrays in pYB using in vivo recombination in yeast. Repeat-specific hooks in pYB were released by MluI digestion removing the pUC linker. RCA-amplified repeats and linearized vector were cotransformed into VL6-48 yeast cells. End-to-end homologous recombination of RCA products followed by interaction of the recombined fragments with the vector allows recovery of large repeat arrays in yeast. (C) Synthetic arrays used for analysis.
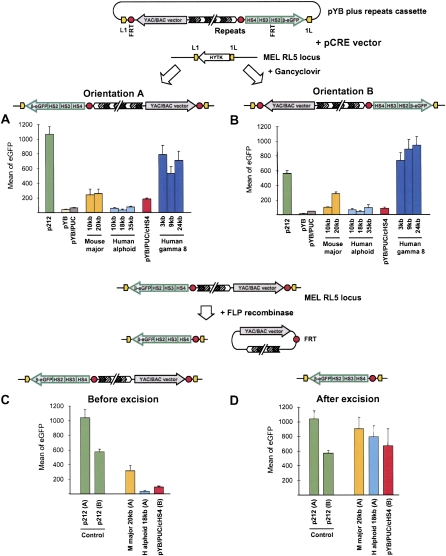
The above cassettes were exchanged into the target locus in RL5 cells. The percentage of correctly targeted ganciclovir-resistant clones varied between 50% and 80%, indicating that targeting efficiency is not particularly sensitive to the size or DNA sequence of the cassette in the size range attempted.
The level of transgene expression was estimated for each repeat type by measuring the mean green fluorescence in the cell population expressing GFP (Fig. 3). Each measurement was the average of values for six or more independent subclones (at least three clones in each orientation). The results showed that mean eGFP expression varied up to 62-fold for different cassettes (Fig. 3A,B), but mean eGFP expression was very similar for different clones with the same cassette in the same orientation. eGFP expression was lowest for cassettes carrying human alphoid DNA, indicating that transgene silencing persists in the presence of human alphoid DNA. For cassettes carrying mouse major-satellite DNA, eGFP expression was approximately sixfold (orientation A) or 14-fold (orientation B) higher than for the control pYB cassette, indicating a modest antisilencing effect. A similar stimulation of eGFP expression was also observed for cassettes carrying chicken beta-globin HS4 insulator DNA. For cassettes carrying gamma-satellite arrays, a much stronger effect was observed, and eGFP expression was 18-fold (orientation A) or 62-fold (orientation B) higher than in control cells. The effect was equally strong for cassettes with 3-, 9-, or 24-kb arrays of gamma-satellite DNA (Fig. 3A,B; Supplemental Table S1). eGFP expression was also analyzed after excision of YAC/BAC vector and repetitive DNA sequences. The results show that excision of the vector and the mouse major, human alphoid, or chicken insulator HS4 DNA arrays stimulated eGFP expression to or nearly to the level of the control cassette p212 (Fig. 3C,D). These results suggest that cassettes carrying human gamma-satellite DNA are not subject to vector DNA-induced silencing of eGFP, while cassettes carrying mouse major-satellite or cHS4 insulator sequences are relatively less-well protected from vector DNA-induced transgene silencing.
Figure 3.
eGFP expression in RL5 cells carrying different transgene cassettes. (A,B) Cre recombinase-mediated cassette exchange (RMCE) was used to exchange the L1-HYTK-1L cassette at the RL5 locus (chromosome 4) in the MEL cells by pYB cassette containing different repeats. After RMCE, transgenes insert in orientations A or B. eGFP expression was measured in RL5 cells carrying the indicated cassettes in the indicated orientation. Each bar, measurements of three to six independent clones. (C,D) FLP recombinase-mediated excision of DNA sequences. eGFP expression before and after FLP-mediated excision of the vector backbone and repeat sequences. Bars, eGFP expression in an individual clone. p212 cassette is a control.
The effect of gamma-satellite DNA on transgene expression was also confirmed in RL4 cells, in which the expression cassette is on mouse chromosome 15. Previous studies of transgene expression in RL4 cells indicated that transgenes are subject to gene silencing within approximately 2 mo (Feng et al. 2005). In contrast, when cassettes carrying gamma-satellite DNA were exchanged into RL4 cells, stable transgene expression continued for more than 3 mo (Supplemental Fig. S3).
Antisilencing effect of gamma-satellite arrays is not due to promoter or enhancer or enhancer-blocking activities
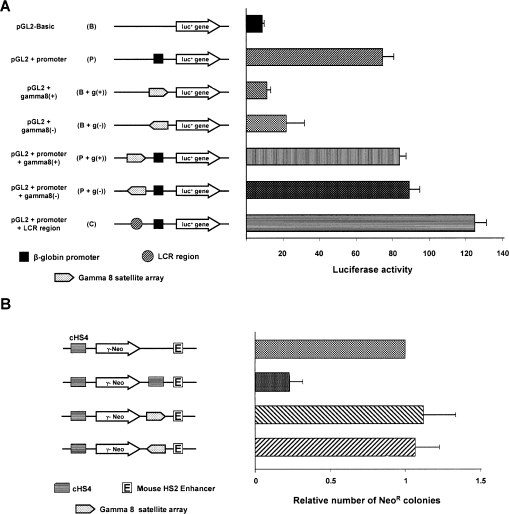
The antisilencing effect cannot be explained by up-regulation of the transgene transcription in the gamma-satellite DNA array constructs. Luciferase reporter assays with a set of pGL2 plasmids (see Methods) did not reveal promoter activity within gamma-satellite DNA in RL5 or NIH3T3 cells (Fig. 4A; Supplemental Fig. S4). A lack of promoter activity was also suggested by Northern analysis. No transcripts initiated from gamma-satellite DNA were detected in RL5 cells targeted with the 9-kb gamma-satellite repeat cassette (data not shown). Also, inversion of gamma-satellite DNA array in the transgene cassette has no effect on transgene expression. The result of the transient assay (Fig. 4A; Supplemental Fig. S4) also does not support enhancer activity for gamma-satellite DNA. Because gamma-satellite monomers contain putative binding sites for the CTCF insulator protein (see below), additional experiments were carried out to elucidate if gamma-satellite DNA functions as an enhancer-blocking insulator (Ohlsson et al. 2001; Gaszner and Felsenfeld 2006). Using an assay developed by Chung et al. (1997), we failed to detect an enhancer-blocking activity of gamma-satellite DNA (Fig. 4B). At the same time, the inclusion of two copies of gamma-satellite DNA into the pCMV/Bsd vector increased the yield of transfectants (Supplemental Fig. S5), suggesting that gamma-satellite DNA may help transgenes to escape from position variegation effects resembling a feature of barrier insulators (Chung et al. 1993).
Figure 4.
Analysis of gamma-satellite DNA. (A) Promoter activity of gamma-satellite 8 DNA. The ∼1.9-kb gamma-satellite 8 DNA fragment was linked to a firefly luciferase reporter gene in a pGL2-basic, or beta-globin promoter vector, or beta-globin promoter plus locus control region (LCR) and was transfected into mouse MEL cells with the Renilla luciferase gene in phRL-CMV as an internal standard. The schematic structures of the vectors are shown in the left panel, and the luciferase activity is shown in the right panel. Values are means ± SD of three individual experiments in triplicate. (B) Enhancer blocking assay. Enhancer blocking assays were performed as previously described (Chung et al. 1993, 1997). The ∼1.9-kb human gamma 8 repeats were inserted into the SacI site between the mouse HS2 enhancer and human gamma-globin promoter-neomycin gene. Before transfection, each construct was linearized by AatII endonuclease. The average colony number obtained is relative to constructs without cHS4 between the enhancer and gamma-neomycin gene.
Gamma-satellite array modulates transgene chromatin structure
Chromatin structure was assessed in the cassettes in RL5 cells by measuring enrichment of lysine 4-dimethylated and trimethylated histone H3 (H3K4me2 and H3K4me3) and lysine 9-trimethylated histone H3 (H3K9me3) in the eGFP transgene. Chromatin immunoprecipitation (ChIP) assays were performed for clones carrying pYB cassettes with 9-kb gamma-satellite or 10-kb alphoid DNA arrays in orientation B (Fig. 5A,B). Previous studies demonstrated that decondensed and transcriptionally active chromatin has a higher level of H3K4me2 and H3K4me3 than condensed, transcriptionally inactive chromatin. In contrast, condensed and transcriptionally inactive chromatin is enriched with H3K9me3 (Peters et al. 2003).
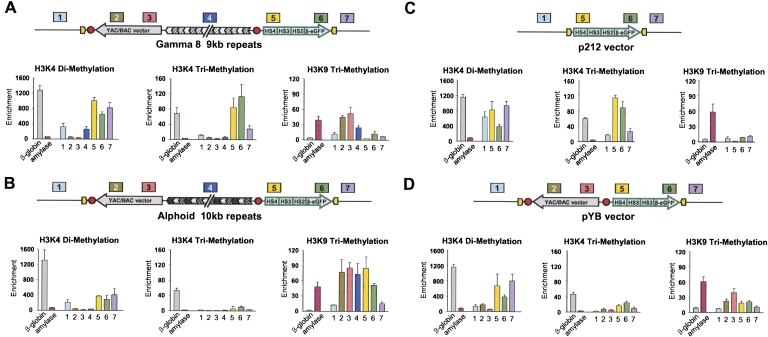
Figure 5.
ChIP analysis of H3K4me2, H3K4me2, and H3K4me3 chromatin in transgene cassettes in RL5 cells. (A) RL5 cells with a transgene cassette carrying a 9-kb gamma-satellite DNA. (B) RL5 cells with a transgene cassette carrying a 10-kb alphoid DNA. Position of the primers for cassettes with gamma-satellite (A) and alpha-satellite (B) DNAs are as follows: (1) centromeric side of RL5 locus; (2) YAC vector backbone; (3) BAC vector backbone; (4) gamma- or alpha-satellite arrays; (5) 5′ end of the locus control region; (6) eGFP coding region; and (7) telomeric side of RL5 locus. Sequences of primers for two control loci, the murine beta-major globin locus and the murine amylase locus, are presented in Supplemental Table S10. (C) RL5 cells with p212 cassette. Positions of the primers are as follows: (1) centromeric side of RL5 locus; (5) 5′ end of the locus control region; (6) eGFP coding region; and (7) telomeric side of RL5 locus. (D) RL5 cells with pYB cassette. Positions of the primers are as follows: (1) centromeric side of RL5 locus; (2) YAC vector backbone; (3) BAC vector backbone; (5) 5′ end of the locus control region; (6) eGFP coding region; and (7) telomeric side of RL5 locus.
ChIP assays showed that in both cassettes H3K9me3, a marker for silent chromatin, was associated with the vector DNA sequence (Fig. 5A,B). This is in agreement with the proposed seeding of heterochromatin by the vector DNA. Enrichment of H3K4me3 in the eGFP transgene correlated with the presence of gamma-satellite DNA but not with the presence of alphoid DNA (Fig. 5A,B). Alphoid DNA was enriched with anti-H3K9me3 but not enriched with H3K4me2 or H3K4me3, suggesting that the inserted alphoid DNA formed heterochromatin. The level of H3K4me2 euchromatic modification was higher at gamma-satellite DNA. At the same time, the level of H3K9me3 was significantly lower compared with that at alphoid DNA, suggesting a more open chromatin structure of the gamma-satellite array. The presence of gamma-satellite DNA did not change the predominant histone modifications on the vector sequence, which remained heterochromatic. Notably, we observed a progressive decrease of H3K9me3 from YAC/BAC vector to transgene (Fig. 5A; probes 2–5) that is in accordance with an active spreading of heterochromatin from the vector sequence toward the transgene with the progressive trapping of heterochromatin within the gamma-satellite array. Control experiments with RL5 cells containing the p212 or pYB targeting cassettes were also performed. As predicted from the transgene expression data, the eGFP sequence in p212 but not in pYB is enriched with H3K4me3 modifications specific for transcriptionally active chromatin (Fig. 5C,D).
Thus, these results suggest that gamma-satellite arrays allow a transcriptionally permissive chromatin conformation in adjacent transgene sequences by arresting the spreading of transcriptionally inactive chromatin from the vector backbone sequence.
Binding of Ikaros-H is required for antisilencing activity of gamma-satellite DNA
Monomers forming gamma-satellite DNA array on the human chromosome 8 are highly diverged (they have average pairwise sequence identities of ∼85%). Previously, it has been shown that some of the monomers contain functional sites for binding of Ikaros proteins that are involved in differentiation along the hematopoietic pathway (Ronni et al. 2007). A computer analysis of a 1.8-kb gamma-satellite DNA fragment used for RCA amplification (carrying eight 220-bp diverged monomers) also predicted the presence of several recognition sites for Ikaros (Supplemental Fig. S6). RT–PCR and Western blot analyses showed that the three largest isoforms of Ikaros, IK-H, IK-VI, and IK-V, are predominately expressed in erythroleukemia MEL cells (Fig. 6A,B). Therefore, we carried out experiments to clarify if Ikaros plays a role in antisilencing activity of gamma-satellite DNA.
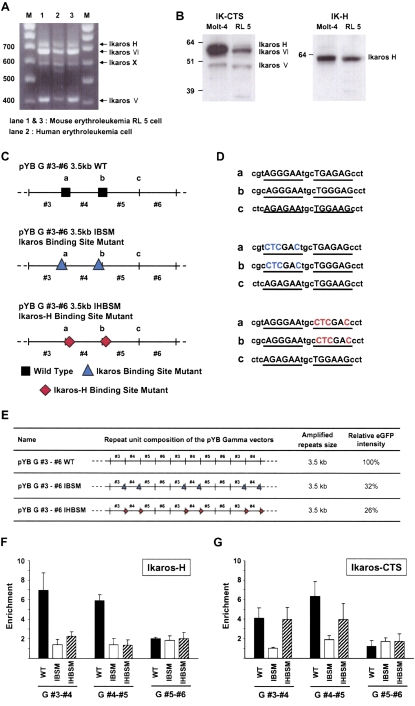
Figure 6.
Mutational analysis of Ikaros binding sites in human gamma-satellite DNA. (A) Analysis of Ikaros transcripts in RL5 cells by RT-PCR using a set of primers specific for alternatively spliced variants. Sequences of primers for RT–PCR are described in Supplemental Table S10. (B) Western analysis of MEL cells extract using IK-CTS antibodies that recognize most of Ikaros isoform and IK-H antibodies that specifically recognize the largest isoform. Note that in MEL cells IK-H is approximately fivefold less abundant than IK-VI isoform. (C) Schemes of three variants of a 4-mer-based gamma-satellite array used for the analysis of antisilencing activity at the RL5 locus. Two conservative bipartite Ikaros binding motifs, “a” and “b,” are present in monomers 3 and 4. In a monomer 5, the motifs are highly diverged and cannot bind Ikaros. Two mutant forms of the array were generated, IHBSM and IBSM, resulting in failure of binding of IK-H isoform and all isoforms of Ikaros, correspondingly. (D) Sequences of original and mutated Ikaros binding sites in 4-mer-based arrays. (E) Effect of IHBSM and IBSM mutations on antisilencing activity of gamma-satellite DNA at the locus RL5. (F,G) ChIP analysis of wild type and IHBSM and IBSM gamma-satellite arrays using IK-CTS and IK-H antibodies. Sequences of primers that specifically amplify two nearest monomers in the arrays are presented in Supplemental Table S10.
The following experiments test whether subfragments of the 8-mer gamma-satellite repeat bind Ikaros, and whether Ikaros plays a role in the antisilencing activity of gamma-satellite DNA. First, the gamma-satellite 8-mer was subcloned as four unique overlapping 4-mers, each of which was amplified into an ∼3.5-kb array and inserted to pYB and the cassette targeted to RL5 cells. eGFP transgene expression was measured in these cell lines, and the results indicated that all four gamma-satellite subclones exhibited antisilencing activity similar to the original gamma-satellite 8-mer (Supplemental Table S1). Note that the 4-mer in each array contains two Ikaros bipartite core motifs 5′-GGGAA-3′ separated by three base pairs (Supplemental Fig. S6). These results are consistent with the recent study by Ronni et al. (2007), showing that a similar bipartite motif binds the largest Ikaros isoform IK-H with higher affinity than IK-VI or IK-V.
One of the 4-mer-based arrays, 3–6, containing monomers 3, 4, 5, and 6, was used for further mutational analysis. We mutagenized bipartite motifs in monomers 3 and 4 within a 4-mer (Fig. 6C). Two types of mutations were introduced into each bipartite motif: one, inactivating the IK-H binding but not effecting binding of other isoforms (mutation IHBSM); another, inactivating the binding of all Ikaros isoforms (mutation IBSM). The mutated 4-mers were amplified into ∼3.5-kb arrays, inserted into the ectopic RL5 site, and then the eGFP transgene expression was measured (Fig. 6E). As can be seen, the presence of intact bipartite motifs is essential for antisilencing activity of gamma-satellite DNA. Inactivation of these motifs recognized by IK-H isoform (IHBSM mutation) had the same dramatic effect on antisilencing activity as the mutation IBSM abolishing binding of all isoforms of Ikaros. ChIP experiments were performed to confirm that IHBSM and IBSM mutations specifically affect the binding of Ikaros to gamma-satellite arrays in vivo. For these experiments, chromatin was immunoprecipitated with antibodies specific for the largest Ikaros isoform, IK-H, or with antibodies IK-CTS that recognize most of the isoforms (see Methods). A promoter region of the Kcnn4 (also known as IKCa1) gene was used as a control for specificity of immunoprecipitation. The results showed that monomers 3 and 4 in the wild type of gamma-satellite array were enriched four- to fivefold in the Ikaros-bound fraction, but the monomers 5 and 6 (lacking Ikaros-binding sites) were not (Fig. 6F,G). As predicted, mutation IHBSM specifically inhibits the binding of the largest Ikaros isoform, IK-H, but not other isoforms, while mutation IBSM results in a failure of binding of all isoforms of Ikaros (Fig. 6F,G). Therefore mutational analysis followed by ChIP experiments clearly showed that antisilencing activity of gamma-satellite DNA in mouse RL5 cells depends on binding of the largest Ikaros isoform, IK-H.
To verify our quantitative ChIP data obtained with mouse RL5 cells, we re-examined if Ikaros proteins indeed bind to gamma-satellite DNA in human cells expressing Ikaros proteins. For this purpose, we chose nonlymphoid leukemia cell line, HEL, in which expression of Ikaros should be present. Western blot analysis confirmed that larger isoforms of Ikaros, IK-H and IK-VI, are indeed abundantly expressed in these cells (data not shown). ChIP experiments clearly demonstrated that Ikaros proteins bind to gamma-satellite DNA on chromosome 8 in human HEL cells (Supplemental Fig. S7). Therefore, our results confirm that in human hematopoietic cells Ikaros binding sites in pericentromeric gamma-satellite arrays are protein-bound.
Gamma-satellite DNA in its natural location is incorporated into chromatin exhibiting a dynamic state
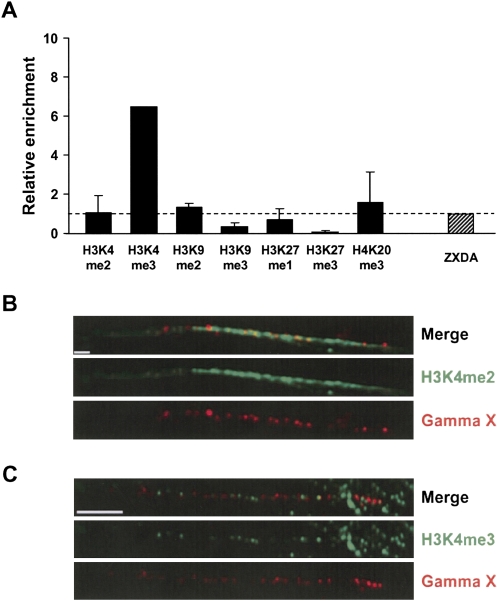
Here, the chromatin organization of gamma-satellite DNA in its natural location was analyzed. Pericentromeric regions of the chromosome X are the best characterized. Chromosome X gamma-satellite DNA is not interspersed with other types of repeats, but rather is organized as a single array of ∼30 kb on the short-arm side of the alphoid- satellite domain (Schueler et al. 2005). First, we analyzed the distribution of H3K4me2, H3K4me3, H3K9me2, H3K9me3, H3K27me1, H3K27me3, and H4K20me3 at gamma-satellite DNA in human primary dermal fibroblasts by ChIP and semiquantitative PCR (see Methods). Gamma-satellite DNA was enriched for H3K4me3 that defines euchromatin but not for histone modifications specific for compact chromatin (Fig. 7A). Using chromatin fibers to optically map the pericentromeric regions, we observed that chromatin containing H3K4 methylation is concentrated at the gamma-satellite array and even extends past the array, suggesting that nearby pericentromeric sequences are also incorporated into open chromatin (Fig. 7B,C).
Figure 7.
Analysis of gamma-satellite DNA in human cells. (A) ChIP analysis of gamma-satellite DNA arrays on chromosome X in human dermal fibroblasts. Enrichment was normalized to enrichment at ZXDA, the most proximal human gene on Xp located in euchromatin. ChIP was performed at least three times and PCRs were done in duplicate for antibodies in each ChIP. (B,C) Gamma satellite is incorporated into chromatin-containing euchromatic histone modifications. Immunofluorescence-FISH on chromatin fibers was performed to spatially define the location of gamma-satellite and histone markers of euchromatin on human chromosomes in human dermal fibroblats. H3K4me2 antibody staining (green) and H3K4me3 antibody staining colocalize with a DNA probe specific for the human X gamma-satellite array (red). While chromatin containing H3K4 methylation is concentrated at the gamma-satellite array, it also extends past the array, suggesting that nearby pericentromeric sequences are also incorporated into open chromatin. Scale bar, 5 microns (B) and 15 microns (C).
Analysis of the gamma-satellite X arrays in fibrosarcoma HT1080 cells revealed heterochromatic modifications (Supplemental Fig. S8), indicating that the chromatin state of the array depends on the genetic background. Cell type-dependent histone modifications were also observed for gamma-satellite DNA arrays in the human chromosome 8 (Supplemental Fig. S8). Interestingly, gamma-satellite DNA was enriched for histone modifications specific for both transcriptionally permissive and compact chromatin. Because of a repetitive structure of the arrays, such a pattern of enrichment indicates the alternation of different domains within one array, as has been observed before for alphoid tandem repeats (Sullivan and Karpen. 2004). Therefore gamma-satellite DNA seems to have a dynamic chromatin structure characteristic of facultative heterochromatin.
The ability of gamma-satellite DNA arrays to contribute alternative chromatin structures in nonhematopoietic cells (i.e., where Ikaros is not expressed) suggests that another protein(s) may bind to these repeats and effect their chromatin status. Computer analysis revealed the presence of putative binding sites for the insulator protein CTCF (Kim et al. 2007) in most of gamma-satellite monomers (Supplemental Fig. S9A). Electrophoretic mobility shift assays (EMSA) showed a specific interaction in vitro between CTCF and gamma-satellite monomers from chromosome 8, X, and Y (Supplemental Figs. S10A–C, S9B; Supplemental Table S2) that is not sensitive to DNA methylation (Supplemental Fig. S11). However, based on chromatin immunoprecipitation with N-terminal CTCF antibodies, gamma-satellite DNA was not significantly enriched in the CTCF-bound fraction compared with the MYC promoter (Supplemental Fig. S10D,E). Therefore, additional studies with different CTCF antibodies and other methods of preparing soluble chromatin containing multiple DNA repeats in the input fraction are required to clarify whether repetitive CTCF sites mapped in vitro may also play a role in shaping chromatin structure and functioning of gamma-satellite arrays in vivo. Therefore, additional studies are required to clarify if CTCF protein plays a role in shaping the chromatin structure of gamma-satellite arrays.
Collectively, these results indicate that chromosomal gamma-satellite DNA arrays embedded into presumptive constitutive heterochromatin retain the feature of facultative heterochromatin and may be assembled both into euchromatin and a compact heterochromatin-like form.
Gamma-satellite DNA is conserved in pericentromeric regions of primate genomes
Given conservation between gamma-satellite DNAs in chromosomes 8, X, and Y, we performed a computer search of gamma-satellite repeat sequences in the sequenced human genome. This analysis revealed the presence of gamma-satellite arrays in pericentromeric regions of 16 other human chromosomes (Fig. 8; Supplemental Table S3). (The failure to detect gamma-satellite DNA in four chromosomes may be due to the poor characterization and/or annotation of pericentromeric regions.) Using gamma-satellite DNA divergence, we designed unique probes for several human chromosomes, including chromosomes 12 and 21, for which the presence of gamma-satellite DNA was not previously reported. As seen from Supplemental Figure S12, the probes position the repeats in the pericentromeric regions. Blocks of gamma-satellite DNA repeats were also detected in chimpanzee and rhesus macaque genomes. While the poor quality of assembled pericentromeric sequences did not allow positioning of all the repeats, in the cases where these repeats were mapped, they were localized in pericentromeric regions similar to that in humans. These data are summarized in Supplemental Table S4, A through D.
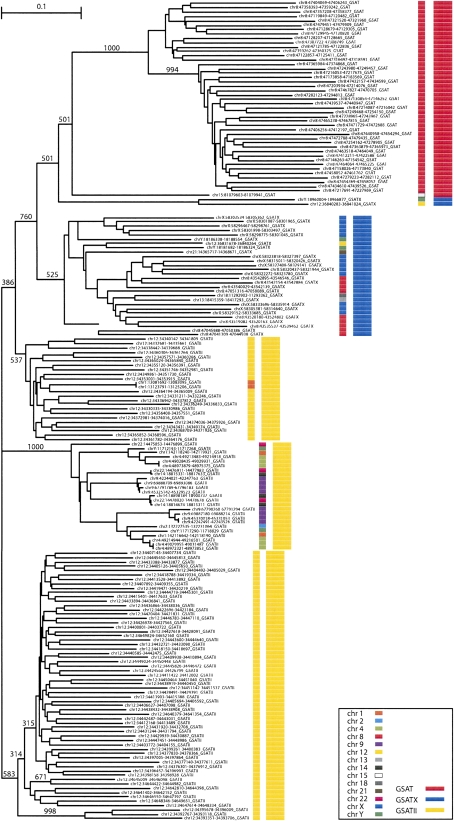
Figure 8.
Phylogenetic tree of human gamma-satellite repeats SAT, GSATII, and GSATX copies were extracted from the UCSC human genome annotation. From each gamma-satellite array, one monomer was selected and aligned in MAFFT (Katoh et al. 2005). Partial GSAT copies were excluded from the analysis. The tree was obtained using the neighbor-joining method implemented in ClustalW (Chenna et al. 2003) and 1000 bootstrap replicates.
All human gamma-satellite arrays consist of three subfamilies of repeats, GSAT, GSATX, and GSATII. Sequence identity between the gamma-satellite subfamilies is ∼60% (Supplemental Fig. S13). Gamma-satellite repeats contain consensus binding sites for Ikaros (Supplemental Figs. S6 and S14). These sites are more frequent in monomers of GSAT subfamily. A significant fraction of GSAT, GSATX, and GSATII monomers contain two-core motifs separated with 3–5 base pairs. This configuration of consensus sequences exhibits a high affinity for the IK-H isoform.
Based on these results and those presented elsewhere (Schueler et al. 2005), the organization of gamma-satellite DNA seems to be conserved in primates.
Discussion
The goal of this study was to explore the effect of repetitive centromeric and pericentromeric DNA elements on vector DNA-induced silencing of a transgene and on transgene chromatin structure in MEL cells. This analysis exploited a recombination-based cloning system in RL5 cells, with which DNA cassettes with a selectable marker and reporter gene could be inserted and/or exchanged into a target site in mouse chromosome 4 (or 15) (Feng et al. 2005). Long arrays of centromeric and pericentromeric repetitive sequences with a defined structure were also generated for these studies (Ebersole et al. 2005). The results demonstrated that human and mouse pericentromeric DNA can prevent vector DNA-induced epigenetic silencing of transgene expression (i.e., antisilencing). The strongest antisilencing activity was associated with gamma-satellite DNA from human chromosome 8. A much weaker but significant antisilencing activity was associated with mouse major-satellite DNA.
The antisilencing activity of gamma-satellite DNA cannot be explained by direct or indirect up-regulation of transgene transcription, because tests for promoter or enhancer activity in gamma-satellite DNA were negative. However, antisilencing activity correlated with a specific chromatin structure in gamma-satellite DNA at the ectopic site in mouse chromosome 4, a chromatin state also seen at naturally occurring sites in human chromosomes. This result is consistent with the observation that gamma-satellite DNA adopts a transcriptionally permissive chromatin conformation in the cassette in MEL cells, but the vector DNA in the cassette retains a condensed chromatin structure. Because modification to heterochromatin is a directional activity, we propose that gamma-satellite DNA arrays are capable of arresting heterochromatin spreading from vector DNA. This hypothesis is consistent with the observation that enrichment for H3K9me3 decreases progressively and linearly along the chromosome from YAC/BAC vector DNA to the transgene.
Some antisilencing DNA sequences, also known as chromatin barriers, are a relatively rare and specialized class of DNA elements, which actively demarcate DNA regions with distinct chromatin structures or states (Sun and Elgin 1999). Chromatin barriers restrict heterochromatin to specific genomic regions and fall within a broader class of elements called insulators (Gaszner and Felsenfeld 2006; Valenzuela and Kamakaka 2006). One of the most well-characterized chromatin barrier elements was identified in fission yeast (Scott et al. 2007). The function of this element is to separate pericentromeric heterochromatin and centromeric chromatin. Deletion of this barrier results in propagation of pericentromeric heterochromatin beyond its normal boundary into centromeric chromatin, which leads to defective chromosome segregation during meiosis. Interestingly, this barrier activity is encoded by functional tRNA genes. In some cells, insulators establish separate domains that isolate transcription enhancer and promoter functions (Chung et al. 1997). Gamma-satellite DNA is unique with respect to previously described barrier or insulator elements, and we propose to call it a “heterochromatin-arresting repeat.”
In RL5 cells, the antisilencing and heterochromatin-arresting activities of gamma-satellite DNA require binding of Ikaros, a protein that regulates hematopoeiesis in lymphoid, myeloid, and erythroid lineages (Georgopoulos 2002). Alternative splicing of the primary transcript generates at least seven distinct isoforms of Ikaros that are expressed in a development-specific and cell type-specific manner (Molnár and Georgopoulos 1994; Sun and Elgin 1996; Georgopoulos 2002). In hematopoietic cells, Ikaros is associated with the NuRD histone deacetylase complex as well as with the SWI/SNF complex that activates gene expression (O'Neill et al. 2000). Ronni et al. (2007) recently showed that IK-H specifically recognizes bipartite 5′-GGGAA-3′ binding motifs and proposed that IK-H preferentially binds and promotes chromatin remodeling and activation of target genes in perichromatic heterochromatin. Therefore, binding of IK-H to multiple recognition sites in gamma-satellite DNA may play a role in preventing heterochromatin spreading in RL5 cells.
FISH and ChIP experiments revealed that the structure of gamma-satellite DNA varies in a cell type-specific manner. Thus, in human fibroblasts that do not express Ikaros, gamma-satellite DNA might be in a transcriptionally permissive or a compact heterochromatin-like state. This suggests that gamma-satellite DNA has the characteristic features and dynamic properties of facultative heterochromatin (i.e., euchromatic DNA that forms compact heterochromatin in a developmentally regulated manner). It is possible that proteins other than Ikaros modulate the function of gamma-satellite DNA in nonhematopoietic cells. Such proteins may be members of the Ikaros protein family (Perdomo et al. 2000, and references therein), replication proteins that modulate histone acetylation in euchromatic DNA regions (Fu et al. 2006), or the insulator protein CTCF (Ohlsson et al. 2001), whose recognition sites were found in gamma-satellite DNA by gel-shift assays (Supplemental Fig. S10). Future analysis is needed to determine which DNA-binding protein(s) modulate the chromatin structure of gamma-satellite DNA in human nonhematopoietic cells.
At present, it is not known how the chromatin structure in endogenous gamma-satellite DNA in human chromosomes is regulated. Nevertheless, structural conservation of gamma-satellite DNA in pericentromeric regions of most chromosomes in humans and nonhuman primates suggests that gamma satellite may play a structural and/or functional role in the centromere. We propose that the primary role of human gamma-satellite DNA may be to prevent spreading of pericentric heterochromatin into chromosomal arms; as such, it resembles a barrier element. Gamma-satellite DNA may also play a role in separating specific domains of chromatin/heterochromatin. Further analysis of gamma-satellite and other DNA repeats may provide insight into how heterochromatic and euchromatic domains are established and maintained in the human centromere as well as other regions of the human genome.
Methods
Construction of synthetic DNA repeats and vectors
Construction of synthetic DNA repeats by rolling-circle amplification and recombinational cloning in yeast was previously described (Ebersole et al. 2005). Source of human alpha- and gamma-satellite DNA repeats as well as corresponding primers are described in Supplemental Tables S5 through S8. A sequence of the 3-mer of mouse major-satellite DNA used for the construction of arrays is ∼97% identical to mouse gamma-satellite DNA, clone 6A (GI:193675), and is available from GenBank (accession no. EF028077). Construction of the pYB targeting cassette is described in Supplemental Figure S2 and Supplemental Table S7. A modified Cre expression plasmid was constructed using iCre, a mammalian codon optimized Cre variant, the kind gift of R. Sprengel (Max-Planck Institute for Medical Research, Heidelberg, Germany). The pCpG-iCre expression plasmid, with no CpG sites in promoter or vector sequences and MAR elements flanking the iCre gene, generally gave improved efficiency in recovery of targeted recombinants in the mouse MEL/RL5 cells. The pCAGGS-FLP vector was purchased from GeneBridges.
Cell culture and transformation
Mouse MEL cells were grown in DMEM (Invitrogen) with 10% FBS (Hyclone) at 37°C in 7.5% CO2. The cells were maintained in 700 μg/mL Hygromycin B (Invitrogen). Electroporation was performed as follows: The pYB reporter gene construct (100 μg) and the iCre expression plasmid (50 μg) in ∼100 μL TE were added to ∼5 × 106 cells in 700 μL of PBS in a 0.4-cm gap cuvette and then coelectroporated at 0.3 kV, 960 μF into MEL cells. Selection for loss of the HYTK gene with 10 μM Gancyclovir was begun ∼48 h post electroporation. Gancyclovir-resistant clones were expanded and tested for targeted recombination by PCR (Supplemental Table S9). Clones were subject to FACS analysis and Southern blotting for further analysis of genomic organization as appropriate.
FACS analysis
FACS analysis of eGFP expression was performed on a FACSCalibur instrument (BD Biosciences) under the control of CellQuest acquisition software and analyzed statistically with FlowJo software (Feng et al. 2005). A minimum of 4 × 104 cells were analyzed for each cell line.
Excision of repeats and the YAC/BAC vector sequence
Excision was performed by transfection of ∼5 × 106 cells of each clone with 50 μg pCAGGS-FLPe vector (Gene Bridges). Forty-eight hours after transfection, cells were selected with 500 μg/mL puromycin for 7 d. Then, cells were distributed into 96-well plates in 10 μM Gancyclovir. Selection of the excised clone was confirmed by the changing the eGFP gene expression level and by PCR (Supplemental Table S9).
ChIP assay
ChIP assay was carried out using ChIP assay kit (Upstate) according to the manufacturer's instructions. Briefly, 6 × 106 cells were fixed with 1% formaldehyde for 10 min at 37°C. After serial washings, cells were resuspended to 1 × 106 cells per 200 μL of SDS Lysis Buffer and sonicated for 30 sec with subsequent 30-sec intervals for 16 min at 4°C using a Bioruptor (Cosmo Bio). Immunoprecipitated DNA (IP-DNA) with lysine 4-dimethylated and trimethylated histone H3 (H3K4me3 and H3K4me3), lysine 9-trimethylated histone H3 (H3K9me3) (Upstate), antibodies IK-H specific for the largest Ikaros isoform, antibodies IK-CTS that recognize all isoforms (Ronni et al. 2007), and a mixture of nine anti-CTCF mouse monoclonal antibodies was quantitated by real-time PCR using the icycler IQ (Bio-Rad). The sequences of primers are listed in Supplemental Table S10.
Luciferase reporter assays
The reporter constructs were transfected with Fugene 6 reagent according to the manufacturer's protocol (Roche) into mouse MEL or NIH3T3 cells grown to 30%–50% confluence in a 12-well plate (Corning) using 1 μg of pGL2 reporter plasmids and 0.1 μg of the internal transfection efficiency control plasmid expressing Renilla luciferase according to the protocol (Promega). Two days after the transfection cells were lysed, and luciferase activity was measured in a luminometer using a Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla expression.
Enhancer blocking assay
Enhancer blocking assays were performed as previously described (Chung et al. 1997). Two micrograms of AatII linearized constructs were electroporated into K562 cells. After 1 d, cells were plated in soft tissue culture agar (Sigma) with 750 μg /mL geneticine (Invitrogen). Colonies were counted after 2–3 wk of selection.
Immunofluorescence (IF)-FISH
Chromatin fibers from human cells were prepared according to published protocols (Sullivan and Karpen 2004). Briefly, cells were centrifuged onto glass microscope slides; lysed for 15 min in lysis buffer containing 25 mM Tris HCl, 0.5 M NaCl, 1% Triton X-100, 0.5 M Urea; and fixed in 4% paraformaldehyde (in PBS) before antibody detection. Histone modifications were detected using primary antibodies recognizing H3K4me2 (ab7766, Abcam) and H3K4me3 (ab1012 or ab8580, Abcam) and anti-rabbit or anti-mouse secondary antibodies conjugated to Alexa Fluor 488 (Molecular Probes, Invitrogen). Gamma-satellite probes were labeled by nick translation with either biotin-16-dUTP (Roche) or Alexa Fluor 568-dUTP (Molecular Probes, Invitrogen). Biotin-labeled probes were detected with Cy3-avidin (Jackson Immunoresearch) or Alexa Fluor 594 streptavidin (Molecular Probes). Fibers were counterstained with 5–10 μg/mL DAPI diluted in Vectashield (Vector Labs) and imaged using an Olympus IX71 inverted microscope as part of the Deltavision RT Imaging System (Applied Precision).
ChIP-PCR for histone modification enrichment at gamma-satellite DNA in human dermal fibroblasts
Native chromatin containing oligonucleosomes was isolated from cultured cells and prepared by micrococcal nuclease digestion as described (Lam et al. 2006). Twenty-five micrograms of digested chromatin (per IP reaction) was immunoprecipitated with antibodies recognizing lysine-specific histone modifications H3K4me2 (ab776) and H3K4me3 (ab8580), purchased from Abcam.
IP-DNA was used for semiquantitative PCR and/or quantitative PCR (QPCR). Primers that amplified gamma-satellite specific for human X chromosome were published previously (Spence et al. 2002); 0.5 μL of IP-DNA was used in each PCR reaction done in duplicate. Relative enrichment for histone antibodies at a given genomic position (query) was calculated using the following formula: relative enrichment = [(IP-Mock)/(Input-Mock)]Query/[(IP-Mock)/(Input-Mock)]Normalizer. Enrichment was normalized to enrichment at a STS marker located near ZXDA, the most proximal human gene on Xp located in euchromatin just outside of the centromere region.
Acknowledgments
We thank Eric Bouhassira for providing the RL5 and RL4 cell lines, Rolf Sprengel for the pBlue.iCRE plasmid, and Gary Felsenfeld for HS4 insulator plasmids. We thank Megumi Nakano for fruitful discussions and help with some experiments. This research was supported by the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are http://www.genome.org/cgi/doi/10.1101/gr.086496.108.
References
- Basu J., Willard H.F. Artificial and engineered chromosomes: Non-integrating vectors for gene therapy. Trends Mol. Med. 2005;11:251–258. doi: 10.1016/j.molmed.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Chen Z.Y., He C.Y., Meuse L., Kay M.A. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H.A. The centromere. Oxford University Press; New York: 1997. [Google Scholar]
- Chung J.H., Whiteley M., Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Bell A.C., Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole T., Okamoto Y., Noskov V.N., Kouprina N., Kim J.-H., Leem S.-H., Barrett J., Masumoto H., Larionov V. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.Q., Warin R., Li T., Olivier E., Besse A., Lobell A., Fu H., Lin C.M., Aladjem M.I., Bouhassira E.E. The human beta-globin locus control region can silence as well as activate gene expression. Mol. Cell. Biol. 2005;25:3864–3874. doi: 10.1128/MCB.25.10.3864-3874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Wang L., Lin C.M., Singhania S., Bouhassira E.E., Aladjem M.I. Preventing gene silencing with human replicators. Nat. Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- Gaszner M., Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and Ikaros. Nat. Rev. Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova E.M., Morse III H.C., Ohlsson R., Lobanenkov V.V. The novel Boris + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Sem. Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- Lam A.L., Boivin C.D., Bonney C.F., Rudd M.K., Sullivan B.A. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl. Acad. Sci. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Lee C., Critcher R., Zhang J.G., Mills W., Farr C.J. Distribution of gamma-satellite DNA on the human X and Y chromosomes suggests that it is not required for mitotic centromere function. Chromosoma. 2000;109:381–389. doi: 10.1007/s004120000095. [DOI] [PubMed] [Google Scholar]
- Lin C.C., Sasi R., Lee Y.S., Court D. Isolation and identification of a novel tandemly repeated DNA sequence in the centromeric region of human chromosome 8. Chromosoma. 1993;102:333–339. doi: 10.1007/BF00661276. [DOI] [PubMed] [Google Scholar]
- Molnár A., Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Yoshida T., Georgopoulos K. Ikaros and chromatin regulation in early hematopoiesis. Curr. Opin. Immunol. 2007;19:116–122. doi: 10.1016/j.coi.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Ohlsson R., Renkawitz R., Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Okada T., Ohzeki J.-I., Nakano M., Yoda Y., Brinkley W.R., Larionov V., Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- O'Neill D.W., Schoetz S.S., Lopez R.A., Castle M., Rabinowitz L., Shor E., Krawchuk D., Goll M.G., Renz M., Seelig H.P., et al. An Ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo J., Holmes M., Chong B., Crossley M. Eos and Pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J. Biol. Chem. 2000;275:38347–38354. doi: 10.1074/jbc.M005457200. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Kubicek K., Mechtler R.J., O'Sullivan A.A., Derijck L., Perez-Burgos A., Kohlmaier S., Opravil M., Tachibana Y., Shinnkai Y., et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Ronni T., Payne K.J., Ho S., Bradley M.N., Dorsam G., Dovat S. Human Ikaros function in activated T cells is regulated by coordinated expression of its largest isoforms. J. Biol. Chem. 2007;282:2538–2547. doi: 10.1074/jbc.M605627200. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Sullivan B.A. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 2006;7:301–313. doi: 10.1146/annurev.genom.7.080505.115613. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Dunn J.M., Bird C.P., Ross M.T., Viggiano L., Rocchi M., Willard H.F., Green E.D. Progressive proximal expansion of the primate X chromosome centromere. Proc. Natl. Acad. Sci. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.C., White C.V., Willard H.F. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One. 2007;10:e1099. doi: 10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.M., Critcher R., Ebersole T.A., Valdivia M.M., Earnshaw W.C., Fukagawa T., Farr C.J. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Karpen G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F.L., Elgin S.C. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Kasai K., Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J. Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindle S.C., Kim H.G., Klug C.A. Mutation of CpGs in the murine stem cell viral retroviral vector long terminal repeat represses silencing in embryonic stem cells. J. Biol. Chem. 2004;279:34–41. doi: 10.1074/jbc.M309128200. [DOI] [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R.T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Vissel B., Nagy A., Choo K.H. A satellite III sequence shared by human chromosomes 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet. Cell Genet. 1992;61:81–86. doi: 10.1159/000133374. [DOI] [PubMed] [Google Scholar]