Abstract
The human genome contains nearly 1.1 million Alu elements comprising roughly 11% of its total DNA content. Alu elements use a copy and paste retrotransposition mechanism that can result in de novo disease insertion alleles. There are nearly 900,000 old Alu elements from subfamilies S and J that appear to be almost completely inactive, and about 200,000 from subfamily Y or younger, which include a few thousand copies of the Ya5 subfamily which makes up the majority of current activity. Given the much higher copy number of the older Alu subfamilies, it is not known why all of the active Alu elements belong to the younger subfamilies. We present a systematic analysis evaluating the observed sequence variation in the different sections of an Alu element on retrotransposition. The length of the longest number of uninterrupted adenines in the A-tail, the degree of A-tail heterogeneity, the length of the 3′ unique end after the A-tail and before the RNA polymerase III terminator, and random mutations found in the right monomer all modulate the retrotransposition efficiency. These changes occur over different evolutionary time frames. The combined impact of sequence changes in all of these regions explains why young Alus are currently causing disease through retrotransposition, and the old Alus have lost their ability to retrotranspose. We present a predictive model to evaluate the retrotransposition capability of individual Alu elements and successfully applied it to identify the first putative source element for a disease-causing Alu insertion in a patient with cystic fibrosis.
Alu elements have the highest copy number of all of the human mobile elements, contributing nearly 11% of the genome with about 1.1 million copies (Lander et al. 2001). Alu elements are nonautonomous; requiring protein products from L1 elements to carry out the generally accepted target primed reverse transcription (TPRT) process necessary for their amplification (Boeke 1997; Batzer and Deininger 2002; Kajikawa and Okada 2002; Dewannieux et al. 2003; Ostertag et al. 2003; Kazazian 2004). Alu can be subdivided into several different subfamilies based on their specific diagnostic sequence positions (for reviews, see Batzer et al. 1993; Batzer and Deininger 2002). Alu started to amplify about 65 million years ago, with peak amplification occurring around 40 million years ago, prior to the divergence of the old and new world monkeys (Shen et al. 1991; Lander et al. 2001; Batzer and Deininger 2002). Activity of the old AluJ/S subfamilies declined while being replaced by the younger Y subfamily (∼100,000 copies) (Shen et al. 1991). Subsets of the Y subfamilies are the only known Alu elements currently active in the human genome, with variants of the Y, Ya, and Yb lineages currently dominating activity (Deininger and Batzer 1999; Hedges et al. 2004; Mills et al. 2007; Belancio et al. 2008). There are ∼900,000 older subfamily elements in the genome, predominately variants of the Alu S and J (Wang et al. 2006), and yet no de novo disease-associated insertions of these older elements have been found (Belancio et al. 2008).
Alu elements contribute significantly to human genetic instability; recent estimates calculate one new Alu insertion in every 20 live births (Cordaux et al. 2006), and at least one in every 1000 de novo genetic diseases are the result of an Alu insertion event (Deininger and Batzer 1999). There are at least 15 examples of Ya5 elements that have recently inserted causing disease (Belancio et al. 2008), despite there being only 3000 copies in the genome (Wang et al. 2006). In contrast, the older subfamilies have a 300-fold greater copy number than Ya5 while having no detectable amplification rate, suggesting that there must be at least a 4500-fold enrichment in activity per Ya5 copy relative to the old Alu subfamily members. This probably represents a minimal estimate as we have yet to see any AluJ or AluSx inserts generating disease alleles through insertional mutagenesis. To date, the exact reasons why these older Alu elements are “dead” or why only the younger Alu elements continue to amplify remain unclear. Surprisingly, Alu element insertions cause twice as much disease as L1 despite the fact that L1 is necessary for Alu activity (Belancio et al. 2008).
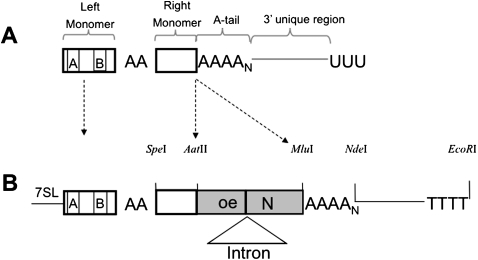
To further evaluate the reason older Alu elements are inactive, we looked at the different sequence components of an Alu element. Figure 1A shows a schematic of the basic structure of a transcript of a genomic Alu element. An Alu is a dimer of two nonidentical sequences ancestrally derived from the 7SL RNA gene separated by a middle A-rich region (Ullu and Tschudi 1984). The left monomer contains the internal RNA polymerase III (Pol III) promoter A and B boxes (Ullu and Tschudi 1984; Chu et al. 1995; Batzer and Deininger 2002). The basic Alu dimer is flanked by an adenosine (A) rich section (A-tail) at its 3′ end. Because an Alu element does not encode its own Pol III terminator, Alu transcripts will contain a unique 3′ end (Fig. 1A) derived from the genomic flanking region found between the end of the A-tail and the first downstream terminator sequence (usually –TTTT) present in the genomic flank. This sequence, which is effectively unique to each individual Alu, will be referred to as the 3′ unique region. Because the A-tail of the transcript is used for the priming during retrotransposition, the 3′ unique region associated with the parent (source) Alu is not included in the resulting insertion sequence. Therefore, it is not possible to unambiguously determine the 3′ unique region of the source Alu from examining the genomic sequences of new insertions. Nevertheless, the individual characteristics of this sequence may be important to the “parent” Alu's ability to retrotranspose.
Figure 1.
Schematic of Alu structure and transcript. (A) Representation of a typical transcript of a genomic Alu element with functional segments defined as the left and right monomers, A-tail, and 3′ unique region. (B) Schematic of the Alu base construct used in this study with 7SL promoter-enhancer sequence upstream of the Alu and the necessary restriction enzymes used for construct manipulation. The neomycin selection cassette in reverse orientation with a self-splicing intron that allows for the retrotransposition studies is introduced after the Alu and before the A-tail. The selection cassette contains a Pol III terminator (TTTT) at the 3′ end.
Because Alu elements amplify through a Pol III-directed transcript, initial studies focused on transcription as a factor controlling the Alu activity. However, analysis of Alu transcripts recovered from cultured cells demonstrates that the majority of the transcripts (66%) were derived from the older Alu subfamilies (Sinnett et al. 1992; Maraia et al. 1993) and <1% derived from AluYa5 elements (Shaikh et al. 1997), indicating that the RNA from older Alu elements was being made but was not undergoing retrotransposition. This expression of Alu subfamilies suggests that there is only about a sixfold difference in transcription per copy between older and younger Alus (Shaikh et al. 1997). Thus, transcriptional silencing does not appear to account for the lack of activity of the older Alu elements.
The younger, active subfamilies have a higher proportion of their members with long A-tails that then shrink rapidly in evolutionary terms (Roy-Engel et al. 2002; Odom et al. 2004). Although the bioinformatics made it seem possible that a primary factor differentiating older and younger Alu subfamilies was a threshold A-tail length (Roy-Engel et al. 2002), experimental studies with a tagged Alu in culture suggest that A-tail length only accounts for a modest difference in Alu activity between old and young subfamilies (Dewannieux and Heidmann 2005).
One possible explanation for subfamily differences in activity is that subfamily-specific sequence differences may result in altered interactions with the retrotransposition process (Sinnett et al. 1991). In addition, the older subfamily members have accumulated random mutations affecting the RNA secondary structures and/or the interactions with other components necessary for amplification (Sinnett et al. 1991; Alemán et al. 2000). For instance, mutations near the 5′ end of the Alu sequence have been found to alter binding ability to SRP9/14 causing inactivation of the Alu activity (Sarrowa et al. 1997; Bennett et al. 2008). In an effort to define Alu activity, Bennett and colleagues looked at the 280-bp “core” sequence of an Alu element (the left and right monomers) and determined that mutations within the primary sequence play a role in Alu activity levels, but that many of the old Alu subfamily members would be expected to remain active (Bennett et al. 2008). Thus, none of the existing data fully explain the minimum 4500-fold effect needed to explain the inactivity of the older Alu subfamilies. It seems that other differences between the RNA molecules generated by the old versus young subfamilies must be major contributors to relative Alu activity (see Fig. 1A for Alu components). We included evaluations of random mutations usually present in older Alus that lead to an increased A-tail heterogeneity and microsatellite formation (Arcot et al. 1995). In addition, because each Alu transcript has a unique 3′ end, this individual sequence may contribute major differences between individual elements contributing to the differences observed in Alu subfamily activity. In this manuscript, we present systematic analyses evaluating the impact of the observed sequence variation in the different sections of an Alu element on retrotransposition. We describe both a model of the evolutionary kinetics of Alu inactivation and a predictive model of the rules of Alu retrotransposition capability by estimating the effect of the different Alu sequence components.
Results
A-tail length
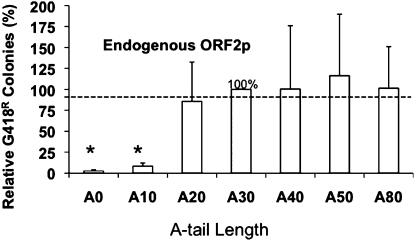
A previous report demonstrated that the A-tail length influences the retrotransposition efficiency of a tagged Alu element (Fig. 1B) driven by a cotransfected full-length L1 element (Dewannieux and Heidmann 2005), but the level of variation observed would not provide much explanation of the differential efficiency of genomic Ya5 amplification relative to Sx elements. Our own studies confirm this observation when using either a cotransfected wild-type L1 (JM101/L1.3 no tag; Wei et al. 2001) or an ORF2-only expression plasmid to drive Alu retrotransposition (data not shown). However, we were concerned that the increased expression of L1 proteins in transient transfection experiments might lead to unusually high concentrations of ORF2p and artificially augment its interaction with shorter A-tail Alus. Thus, we transfected only the tagged Alu construct into HeLa cells, which express a detectable level of endogenous L1 (Belancio et al. 2006), to determine whether the A-tail lengths (Table 1) worked the same at these endogenous L1 expression conditions (Fig. 2). These studies confirmed the relative influence of the A-tail, with only a modest benefit accruing once the A-tail exceeded 20 bases (Dewannieux and Heidmann 2005).
Table 1.
Sequence composition of Alu A-tail constructs
Figure 2.
The length of homogeneous A-tail affects Alu retrotransposition efficiency. The retrotransposition activity of Alu elements with homogenous A-tail of different lengths driven by the endogenous ORF2p present in HeLa cells was evaluated. Columns represent the mean G418R colonies (n = 3) normalized relative to the Alu with 30 homogenous As that was arbitrarily designated as 100% with the standard deviation shown as error bars. Results significantly different from the A30 control with P-values ≤ 0.0001 (Students paired t-test) are indicated by an asterisk (*).
A-tail heterogeneity
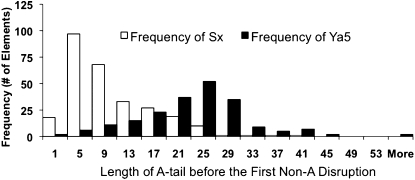
Because the older Alu elements also have less “perfect” A-tails than the newer elements, we hypothesized that disruption of the A-tails with other bases might serve the same role as shortening the A-tail. Figure 3 shows the distribution of uninterrupted A residues for a randomly chosen subset of AluSx (n = 276) and AluYa5 (n = 206) elements (see Supplemental Table 1S for exact A-tail sequences). Most AluSx elements have their longest uninterrupted A stretch significantly shorter than 20 bases and, therefore, might have little or no activity if homogeneous A-tails are required.
Figure 3.
Older Alu elements contain shorter homogenous A-stretches within the A-tail region than younger Alus. Data mining histogram that shows the frequency of Alu elements in different bin sizes (Sx n = 276; Ya5 n = 206)
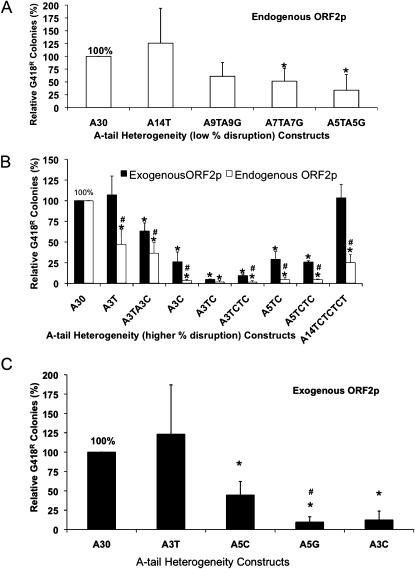
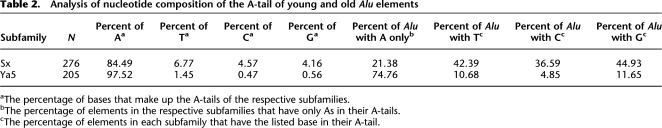
We introduced disruptions in the A-tail of our Alu-tagged construct starting with a uniform run of 30 As to evaluate whether they would lead to a decrease in retrotransposition (Table 1) and observed an inverse relationship between the amount of disruption and retrotransposition (Fig. 4A). These data demonstrate that interruptions influence the retrotransposition rate; however, the results are not simply the product of the longest stretch of uninterrupted As. Modest disruptions with non-A bases significantly affect retrotransposition (A7TA7G and A5TA5G, P < 0.03, Student's t-test), but it seems that any influence on retrotransposition insertion rate caused by this low level of disruption would be modest.
Figure 4.
Increased A-tail heterogeneity reduces Alu retrotransposition capability. HeLa cells were transiently transfected with the corresponding Alu-tagged constructs driven by endogenously or exogenously supplied ORF2p. The relative activity of Alu elements with slightly disrupted A-tails (A) or extremely disrupted A-tails (B) is shown. The Alu with 30 homogenous As (A30) was arbitrarily selected as 100%; the asterisk (*) indicates a significant difference from A30, P < 0.05 (Students paired t-test). (C) The evaluation of how specific base disruptions in the A-tail affect retrotransposition rate (T >> C > G) under exogenous conditions of ORF2p is shown. (*) Asterisk indicates a significant difference between A5C, A5G, and A3C relative to A3T, P < 0.05, P < 0.01, and P < 0.01, respectively (Students paired t-test); # indicates a significant difference between A5C and A5G, P < 0.01 (Students paired t-test).
Higher amounts of disruption of the A-tails led to significantly different activity levels compared to the control when driven by either the exogenously supplied ORF2p (cotransfection of an expression plasmid) or the endogenous L1 expression (Fig. 4B). Under conditions with increased ORF2p expression, the relative activity of some of the disrupted A-tails also increased (Fig. 4B). For example, the A3C construct maintains only 5% activity under endogenous L1 expression relative to a perfect A-tail control (designated as 100%), while the same construct maintains 25% activity with ORF2p overexpression. Surprisingly, the A3T construct, although highly disrupted, can function almost as well as the control under these overexpressed ORF2p conditions, but decreases to ∼50% activity under endogenous conditions, indicating that overexpression of ORF2p may drive relatively inactive Alus in this assay system. Experiments using wild-type L1 (JM101/L1.3 no tag; Wei et al. 2001) as the driver (Supplemental Fig. 1S) showed an intermediate level of Alu activity between those observed when using the endogenous L1 and the exogenously supplied ORF2p conditions. In this case wild type is defined as the original L1.3 element (Sassaman et al. 1997) without any optimization or mutation of the L1.3 sequence that was cloned into an expression vector.
The distribution of altered bases relative to the length of uninterrupted As plays a role in Alu retrotransposition as both the A3TC and A14TCTCTCT constructs contain the same number of non-A bases but had significantly different capability driven under both conditions (Fig. 4B, P < 0.02 [endogenous], P < 0.001 [exogenous], paired Student's t-test). We observed a significant difference in activity of a 30-base A-tail disrupted with thymine residues when compared to a 30-base A-tail disrupted with cytosine residues in the exact same positions (Fig. 4B, P < 0.02 [endogenous], P < 0.01 [exogenous], Student's t-test), suggesting that the specific bases in the disruptions may also be important.
We further tested the relative influences of thymine (T), cytosine (C), and guanine (G) disruptions to the A-tail (Fig. 4C) and observed a general trend where T << C < G when it comes to disruptions in the A-tail influencing Alu activity. The A3T construct retrotransposed much better than the A5C and A5G constructs even though it contains more disruptions (P < 0.05 and P < 0.01, respectively, Student's t-test), and the A5C construct showed more colony-forming ability than the A5G construct (P < 0.01, Student's t-test).
Analysis of the same randomly chosen Sx and Ya5 elements used for the A-tail length study (Supplemental Table 1S) demonstrated the existence of considerable differences between the degree and nature of disruptions in the A-tails of the elements from these two subfamilies (Table 2). A-tails from the AluSx elements have over eight times as many C and G disruptions as the Ya5 elements, suggesting that these elements should have diminished retrotransposition capability. Additionally, ∼75% of the Ya5 elements have pure As in their tails, whereas only 21% of the Sx elements analyzed present no disruptions in their A-tails; 183 of the 276 Sx elements (66%) contain a C or G base, while only 32 of 206 (16%) of Ya5 A-tails contain either of these bases.
Table 2.
Analysis of nucleotide composition of the A-tail of young and old Alu elements
aThe percentage of bases that make up the A-tails of the respective subfamilies.
bThe percentage of elements in the respective subfamilies that have only As in their A-tails.
cThe percentage of elements in each subfamily that have the listed base in their A-tail.
We were uncertain whether the A-tail disruptions contributed to changes in RNA stability or whether the disruptions were contributing at a later stage in the insertion process. Real-time reverse transcriptase PCR (qRT-PCR) was performed on the whole cell RNA extracts from cells transfected with the A30, A3T, A5C, and A5G constructs. Our results indicate that the observed retrotransposition differences could not be attributed to variations in the RNA steady-state levels, indicating that the RNA is present at similar levels (data not shown).
3′ unique region
The most significant sequence difference between individual Alu transcripts is the downstream unique region located after the A-tail and before the transcription terminator (Fig. 1A). Previous studies suggested that these sequences may modestly influence Alu RNA levels (Alemán et al. 2000). We generated several constructs that contain 3′ unique regions of varying length ranging from 15 to 126 bp (Table 1). These sequences were selected as 3′ flanking sequences of de novo L1 insertions from cell culture experiments (Gilbert et al. 2005). The selected sequences include the nucleotides immediately downstream from the A-tail to the first set of four T residues that would be expected to act as a Pol III terminator. We would expect these L1 flanking regions to be similar to Alu flanking regions (Jurka 1997) and they would have been unlikely to have undergone any negative selection. The constructs were compared to a 30-base pure A-tail (A30) immediately followed by a terminator (A30-0 from Table 1) to determine the effect of the differing 3′ ends on Alu retrotransposition. The sequence found after the A-tail and prior to the terminator has a significantly detrimental effect on Alu retrotransposition rates whether under exogenous or endogenous conditions of ORF2p or L1 (Fig. 5A, P < 0.01 [exogenous], P < 0.03 [endogenous], Student's t-test). We see a strong decrease in Alu retrotransposition ability even with Alus containing little additional 3′ end sequence.
Figure 5.
The length of the 3′ unique sequence drastically affects Alu retrotransposition capability. (A) Constructs with 30 homogenous As and variable 3′ unique region lengths (0–126 bp) were evaluated by transient transfection of HeLa cells. The activity of these Alu elements under both exogenous (black bars) and endogenous (white bars) conditions of ORF2p is shown. The A30-0, an Alu with 30 homogenous As immediately followed by a terminator (TTTTT), was arbitrarily selected as the 100%; the asterisk (*) indicates significant difference from A30-0, P < 0.01 exogenous, P < 0.03 endogenous (Students paired t-test). (B) Northern blot analysis of poly(A) selected RNA extracts was performed from cells transfected with the A30-0 control and the variable A30 3′ unique constructs. The unspliced (open arrowhead) and spliced (black arrow) neo-tagged Alu transcripts are indicated. The spliced Alu transcript from the variable length constructs were normalized to cyclophillin (C, loading control) and expressed relative to the A30-0 construct (designated as 1.00). The mean ± SD for the quantitation results for each construct are indicated below (n = 3). (C) Histogram of the length distribution of the 3′ unique sequence of young and old Alu elements. The distribution of the length of 3′ unique sequence (defined as the sequence between A-tail and the first four Ts in the 3′ genomic flank) of a subset of randomly selected AluSx (n = 289) and Ya5 (n = 227) families is shown as the frequency subdivided into bins of various sizes. (•) Alu elements containing a premature terminator within their internal dimeric sequence are included in this bin; Sx (n = 25), Ya5 (n = 1).
Northern blot analyses demonstrate only modest transcription differences between the different 3′ unique constructs and the A30-0 control (Fig. 5B). In this case, the observed transcription differences do not explain the huge decrease in activity, indicating that the observed effect of the 3′ unique sequence on Alu retrotransposition capability is largely independent of RNA stability. Relative to the A30-0 construct, the A30-126 construct maintains only 13% and 7% effectiveness under exogenous and endogenous ORF2p conditions, respectively, but it has 67% of the level of the A30-0's RNA level.
We analyzed the 3′ unique region (defined as the sequence between A-tail and the first four Ts in the 3′ genomic flank) in a random sample of AluSx (n = 289) and Ya5 (n = 227) subfamilies and observed no significant difference in length distribution (P = 0.69, Student's t-test; Fig. 5C). However, only 25% of the Sx elements had 3′ unique regions of 15 bases or less, indicating that close to 75% of these elements would be very limited in their retrotransposition capability, even if the other aspects of their 3′ regions were ideal. Upon further examination, roughly 10% of the Sx elements contain sequence mutations that generate a Pol III terminator within their sequences leading to a truncated Alu transcript devoid of an A-tail and 3′ unique sequence. When these premature terminating Alus are removed from the data set, only 17% of Sx elements present a 3′ unique region <15 bp. We observe a similar distribution with the Ya5 elements, where only 15% of them contain 15 or fewer bases between the end of the A-tail and the genomic Pol III terminator. However, most of the 15% of Ya5 elements that do have a short 3′ end would be expected to be active based on their A-tail homogeneity and length of continuous As.
Alu right monomer
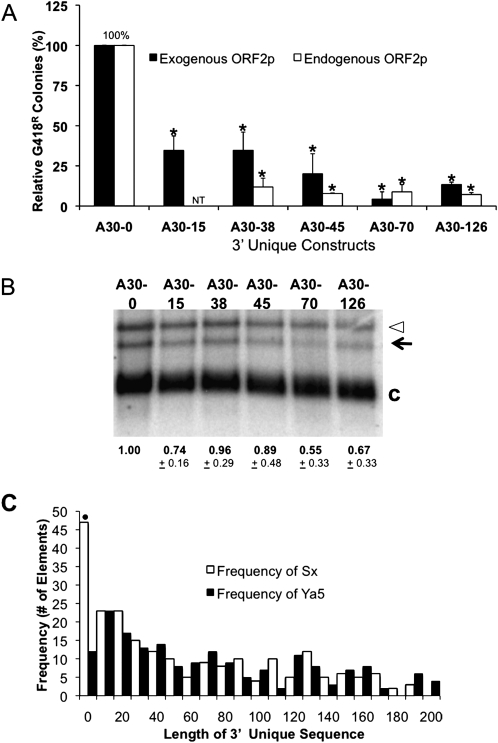
Older Alu elements, such as Sx subfamily members, have accumulated more sporadic mutations throughout their length, which led to the proposal that this may disrupt structure or interactions relative to consensus elements (Sinnett et al. 1991; Alemán et al. 2000). Because the promoter resides in the left half of the element (Fig. 1A), we could not measure the influence of mutations in this region independently from transcription influences. Therefore, we selected different right halves from several randomly chosen Alus to evaluate whether they influence the retrotransposition process. Nucleotide changes within the right monomer contribute sporadically to Alu activity (Fig. 6A; see Supplemental Table 2S for sequences). Some of the right monomer changes had no effect on Alu activity; however, others severely decreased Alu retrotransposition. These data demonstrate that a significant level of variation is likely to be tolerated, but certain mutations in the right monomer will affect retrotransposition, perhaps due to structural changes in the Alu RNA, which prevent its retrotransposition.
Figure 6.
Random right monomer mutations affect Alu retrotransposition capability. (A) Constructs of tagged Alu elements containing a randomly selected genomic AluSx right monomer with several mutations were evaluated by transient transfections in HeLa cells under exogenously supplied ORF2p conditions. The relative activity of these Alu elements is shown. The construct A30 that contains the consensus AluYa5 right monomer was designated as 100. (B) A Northern blot analysis of the steady-state level of poly(A) selected RNA from the constructs in the same order as A is shown. The unspliced (open arrowhead) and spliced (black arrow) neo-tagged Alu transcripts are indicated. The spliced Alu transcript from the variable length constructs were normalized to cyclophillin (C, loading control) and expressed relative to the A30 construct (designated as 1.00). The mean ± SD for the quantitation results for each construct are indicated below (n = 3).
The steady-state RNA levels of these constructs were evaluated by Northern blot analysis (Fig. 6B). The difference in retrotransposition ability is not primarily due to RNA stability of these constructs. This indicates that something other than RNA level is the contributing factor to retrotransposition in these random right constructs and our observations are in good agreement with other recent results (Bennett et al. 2008) supporting RNA structure as an important factor in the retrotransposition mechanism.
Alu source element identification
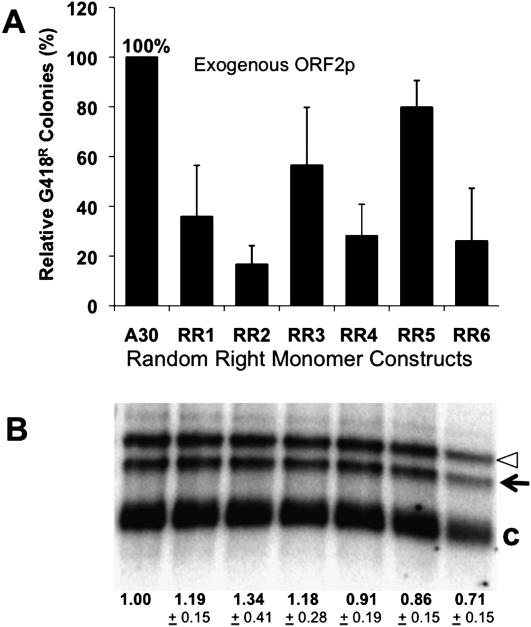
A recent study (Chen et al. 2008) determined that an Alu insertion in the Cftr gene was a direct cause of cystic fibrosis. When we performed a genomic search with the sequence of the disease inserted Alu, only one Alu in the genome shared 100% identity (Fig. 7). This candidate source Alu shares a T mutation in the A-tail with the sequence of the disease causing Alu, a highly unusual feature in new Alu inserts. Although this first T residue in the A-tail is shifted slightly in one sequence relative to the other, we have found that this type of slippage in A-tail lengths is extremely common between A-tails in different individuals (Roy-Engel et al. 2002) and, therefore, this is likely to represent a shared T residue with an A length polymorphism. Our data suggest that the T mutation in the A-tail would not be expected to disrupt retrotransposition activity. In addition, this putative source Alu contains all of the features predicted by our data of an active Alu: a reasonable A-tail length with little disruption (34 bp) and a very short 3′ unique region (0 bp) (Fig. 7). Collectively these observations, along with the data presented above, strongly suggest that we had identified the active “parent” or source Alu element responsible for the insertion into the Cftr gene. While it is impossible to demonstrate with absolute certainty, these observations likely represent the first identification of an active source Alu causing disease in humans, and furthermore, they demonstrate how the rules defined in these studies assist us in understanding the retrotransposition potential of individual Alu elements.
Figure 7.
Identification of the candidate source element for a disease-causing Alu insert. The sequence of the disease-causing Alu compared to the candidate source element found on chromosome 14 in the genome. The Alu sequences share 100% identity within the body of the element. (A) Alu insertion disrupting Cftr gene, exon 17b (Chen et al. 2008). Flanking sequence is lower case. Inserted Alu sequence is italicized. Bold text indicates the target site duplication formed during the retrotransposition process. The box outlines the thymine mutation that is inferred to have transferred to the progeny sequence during retrotransposition. Underlined portions within the direct repeat indicate the transcription terminator. (B) The parent locus of the Cftr Alu insertion. With the flanking sequence, Alu, target site duplication, thymine residue, and terminator labeled as in A.
Discussion
Our studies demonstrate why investigators have struggled for years to understand the pattern of evolutionary activity of Alu elements. The vast majority of Alu elements belong to older subfamilies that have shown very little activity in recent evolutionary times. Instead, modern Alu activity is dominated by a few small, young Alu subfamilies (Deininger and Batzer 1999). The master-gene hypothesis (Deininger and Batzer 1995) was developed to explain subfamily evolution of rodent ID repeats whose evolution is clearly controlled by the BC1 master locus, and by analogy the model was extended to Alu. This model relied primarily on the transcriptional regulation of one, or a very few loci. At the time, we felt that the only other reasonable alternative is that, due to a very limited number of active Alu elements at any one time, their evolution goes through an extreme bottleneck, allowing the subfamilies to drift in the manner observed. In fact, the truth is probably a mix of these ideas, with the likelihood that a relatively few loci maintain activity for a longer period of time (master loci), while some loci have limited activity as proposed by the stealth model (Han et al. 2005), with the vast majority of Alu elements being retrotranspositionally incompetent.
Transcription is still likely to be a major factor limiting the activity of many Alu elements. They are heavily influenced by flanking sequences at each new insertion locus (Chesnokov and Schmid 1996; Alemán et al. 2000; Roy et al. 2000; Ludwig et al. 2005), silenced by methylation (Englander et al. 1993; Liu and Schmid 1993; Liu et al. 1994), and the older Alus gradually accumulate mutations that incapacitate the internal promoter (Murphy et al. 1983). Because our studies utilize an Alu whose transcription is assisted by the strong 7SL RNA gene upstream region, we cannot address the influence of genomic sequence on individual Alu loci. It is likely that the majority of Alu elements are in relatively poor transcriptional environments that limit their activity. However, every transcriptional study carried out on Alu elements (Sinnett et al. 1992; Liu et al. 1994; Shaikh et al. 1997) demonstrates that many Alu loci from all subfamilies continue to be transcriptionally active. Thus, the only way to explain the minimum of 4500-fold amplification preference of young Alu families, like Ya5, relative to the older Alu families is through post-transcriptional regulatory factors. It was previously proposed that A-tail length might be a major factor in this regulation (Roy-Engel et al. 2002). However, experimental studies demonstrated that only very short A-tails are retrotranspositionally inactive, and therefore old subfamilies have sufficient A-tail length that we estimate this influence should only be about threefold (Dewannieux and Heidmann 2005). It would be reasonable to expect that interruptions in the A-tail that gradually accumulate through mutation of older Alu elements would also influence the ability of an A-tail to successfully participate in the predicted TPRT priming (Boeke 1997). Our studies confirm the negative impact of some A-tail disruptions. Surprisingly, A to T mutations in the A-tail had little impact on the retrotransposition efficiency. However, interruptions by C or G had significantly larger impacts. Thus, the influence of interruptions in the A-tail is complex. Clearly additional attributes beyond the length of the longest run of As contribute to TPRT priming efficiency. Table 2 shows that there are high levels of T residues within the A-tails early in Alu element evolution. This is somewhat surprising in that transversions are generally rarer than transitions. However, visual inspection of the Ya5 A-tails (Supplemental Table 1S) suggests that these T residues are less frequently the result of transversion mutations within the A-tail, and more from microsatellite-like amplifications from the A+T rich direct repeat region flanking the element. The AluSx elements contain higher levels of all base interruptions in their A-tails (Table 2), but the G and C mutations increase more proportionately, consistent with the accumulation of point mutations in the older A-tails. Thus, both the level of disruption and the higher incidence of G and C interruptions in the A-tails would jointly contribute to decrease activity from the Sx elements.
One of the most surprising influences on Alu activity levels is the distance between the A-tail at the 3′ end of the Alu and the Pol III terminator located randomly downstream. We find that having the terminator very close to the A-tail allows maximum activity, while having as few as 15 random bases between the A-tail and the four T residues that cause the termination can result in as much as an order of magnitude decrease in activity. Thus, we estimate that >90% of new Alu inserts have relatively poor insertion capability even if they insert in a genomic region that allows them to transcribe actively. While this does not help to explain the relative activity differences observed between the Ya5 and Sx subfamilies, it does help lead to relatively few Alus having high levels of activity which is likely to contribute to the overall pattern of Alu evolution.
The most difficult aspect to assess Alu sequence variation is the influence of random mutations throughout the Alu element on the activity. Previous studies have shown that random mutations in the right monomer can have a modest effect on the RNA level of Alu elements (Alemán et al. 2000), that deletion of the right half decreases Alu activity by an order of magnitude (Dewannieux et al. 2003), and that deviation from Alu subfamily consensus sequences, particularly those >10% divergence, affects Alu retrotransposition efficiency (Bennett et al. 2008). In addition, previous studies altering the SRP9/14 binding motif in SINE RNAs showed a profound influence on activity (Sarrowa et al. 1997; Dewannieux et al. 2003; Bennett et al. 2008). Some of the influence might include the specific bases that define the subfamilies themselves. However, subfamily diagnostic mutations have at most a two- to threefold influence on Alu activity (Bennett et al. 2008; B. Wagstaff and A. Roy-Engel, unpubl.). Our data assessed random mutations in the right half of the Alu RNA structure because of the complexity of dissecting the individual contribution of transcriptional and post-transcriptional influences that might be expected from mutating the left half of the Alu structure. Several of the random right halves selected from old, Sx, Alu subfamily members negatively impacted insertion efficiency while others did not. These influences were post-transcriptional as we did not alter the promoter and saw only minimal variation in RNA levels.
We believe that it requires a combination of all of the factors described above to explain the relative inactivity of old Alu subfamilies and that these factors contribute differentially to silencing Alu elements at different stages of their evolution. For example, previous studies suggested about a sixfold advantage in transcription per Alu copy on average between the old and new elements (Shaikh et al. 1997). If we combine this with a roughly threefold influence of subfamily mutations and a 10-fold influence of A-tail length and heterogeneity, we have the potential for 180-fold regulation. This is still ∼20-fold short of explaining the full regulation, but a large portion of this is likely to be explained by the influence of random mutations throughout the Alu element (Bennett et al. 2008). The activity may also not be a direct product of the factors measured here. There may be synergistic or more complicated interactions between different factors, such as A-tail length, and the level and distribution of heterogeneity.
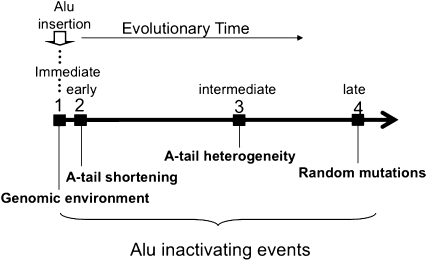
Figure 8 presents a relative timeline on the implementation of these various levels of control. Thus, when a new Alu insert occurs, it is likely that its new surrounding DNA results in either a poor transcriptional environment (which includes epigenetic regulation) or a long 3′ end that limits activity post-transcriptionally. Those Alu elements that are active in their new insertion site will then be subject to rapid shortening of their A-tail (Roy-Engel et al. 2002), which may also decrease activity. The A-tail accumulates variation relatively rapidly because of its microsatellite nature and therefore results in further inactivation of elements. Eventually, over many millions of years, the whole Alu element will accumulate sufficient mutations to either silence its promoter or alter its post-transcriptional activities to make it permanently silent.
Figure 8.
Time frame of events influencing Alu element activity. (1) De novo Alu insertion acquiring new 5′ transcription regulators and 3′ unique sequence which dramatically affect future retrotransposition efficiency. In addition, epigenetic changes will impact transcriptional capability of the Alu insert. (2) Alu A-tail length shortening occurs rapidly after insertion to squelch future activity. (3) The quick introduction of heterogeneity in the A-tail through microsatellite expansion causing an additional reduction in retrotransposition efficiency. (4) Slower and random accumulation of random mutations throughout the element including mutations to the A and B box reducing Alu transcription efficiency, mutations in the A-tail increasing heterogeneity, and mutations throughout the left and right monomers reducing overall identity with a consensus sequence possibly affecting RNA stability and structure.
There are still factors that we are unable to model in our studies with respect to individual loci, such as the influence of flanking sequences on Pol III transcription, or the possibility that very specific 3′ unique sequences from a very limited number of Alu elements contribute in an unpredicted manner. Therefore, it is not possible to fully predict which Alu elements are active or, for that matter, how many. Our data demonstrate that the majority of new Alu insertions are probably not very active upon insertion and this may help explain the low numbers active that could result in the pattern of evolution observed. In addition, because Alu elements contribute to genetic instability and disease, it is possible that there is genetic selection against the most active elements. This leaves the possibility that an Alu element that inserts into a favorable genomic environment (both for transcription and 3′ unique region) may undergo negative selection. This would lead to either its selective elimination from the population or rapid changes, such as A-tail length or heterogeneity that eliminates its activity. Thus, even though certain characteristics between old and young Alu subfamily members may look similar, it may be that all of the features necessary for activity are present only among a small group of the very youngest elements, those for which selection has not had sufficient time to remove from the population.
The finding that the most likely source element for a recent Alu insertion in the Cftr locus (Chen et al. 2008) was predicted based on its sequence similarity and shared T in the A-rich region (Fig. 7) also conforms to our rules of active elements having short 3′ ends and long, relatively perfect A-tails. This represents the first identification of a likely source element for a disease-causing Alu element and illustrates how our increasing understanding of the factors influencing Alu element activity can help us predict the potential strength of individual Alu elements and potentially predict likely source elements for other Alu insertions causing disease.
In a number of our studies, the level of expression of ORF2p driving the tagged Alu insertion (Wallace et al. 2008) appeared to have a significant influence. Most studies using this Alu retrotransposition assay make use of either overexpression of an L1 or simply of ORF2 from L1. We were able to obtain measurable levels of Alu activity in HeLa cells from the endogenous expression of L1 in these cells. Although our data did not see a significant influence on Alu activity due to A-tail length (Fig. 2) based on ORF2p expression levels, we saw consistently stronger influences from A-tail interruptions, the length of 3′ unique regions, as well as from random mutations in the right half, when we used only endogenous levels of L1 to drive the Alu. We envision that at high levels of ORF2p expression even relatively poor Alu substrates can form active insertion complexes. However, at lower ORF2p levels, other cellular RNAs, or even endogenous Alus, compete for the ORF2p and it is less available for the less favorable Alu elements. Even HeLa cells may have higher levels of L1 expression than typical normal cells (Perepelitsa-Belancio and Deininger 2003; Belancio et al. 2006), and therefore it is possible that our measurements of the various factors that influence Alu activity are underestimates of the potential influence each has in normal cells.
Methods
Plasmid constructs
All of the constructs used in this study are based on the AluYa5-neo TET retrotransposition cassette provided by T. Heidmann (Dewannieux et al. 2003). Modifications (shown in Fig. 1B) via the Quick Change mutagenesis kit (Stratagene) were made to introduce a MluI site between the SV40 promoter in the reverse orientation and the A-tail of the Alu (see Supplemental Table 3S for primers), and the modified retrotransposition cassette was moved into pZero-Zeo (Invitrogen). Complementary oligonucleotides were synthesized (IDT) with the appropriate ends to introduce the desired 3′ sequence (Supplemental Table 3S) into the MluI–EcoRI site to generate the different A-tail length and heterogeneity constructs.
The 3′ unique region constructs were built into the modified 30 pure A pZero vector from the heterogeneity study. Oligonucleotides were synthesized that contained a 30-base A-tail to introduce an NdeI site after the 30 As (Fig. 1B; Supplemental Table 3S). These oligonucleotides contained overlaps that allowed them to be cloned into the digested pZero backbone between the MluI site and the EcoRI site. This vector, pZero7SL AluNdeIneo TET, was confirmed by DNA sequencing. The 3′-15, 3′-38, and 3′-45 constructs were built by annealing NdeI and EcoRI containing oligonucleotides and inserting them into pZero. The 3′-70 and -126 unique end sequences were isolated from HeLa genomic DNA by specific PCR amplification of the selected genomic locations. The primers were designed so that they would have NdeI and EcoRI overhangs so they could be inserted into the compatible locations of the pZero-NdeI vector (Supplemental Table 3S).
The random right monomer constructs were generated in the Alu retrotransposition cassette within the Topo-TA 2.1 vector that had the kanamycin resistance cassette removed by digestion with MscI and RsrII followed by Mung Bean Nuclease treatment and religated with T4 DNA ligase. A SpeI site was introduced using site-directed mutagenesis into the middle A-rich region of the Alu making p7SL AluSpeIneo TET. Randomly selected Alu right monomers from the genome, isolated by PCR (Supplemental Table 3S), were cloned into this vector using the SpeI and AatII sites found at the 3′ end of the Alu element (Fig. 1B). All constructs were confirmed by sequencing (TGen) to ensure proper subcloning. Plasmids were isolated and purified by double-banding in CsCl.
Transfection
500,000 or 100,000 HeLa cells were seeded onto a 75-cm2 cell culture flask the day before the transfection to evaluate Alu retrotransposition driven by endogenous L1 or by ORF2 or L1 expression plasmids, respectively. Transfections were carried out using Lipofectamine and Plus Reagents according to the manufacturer's protocol (Invitrogen). For L1 or ORF2 complemented retrotransposition assays, 2 μg of Alu-containing plasmid were transfected with 1 μg of the appropriate driver plasmid per flask. For assays using endogenous L1 expression, only 2 μg of Alu retrotransposition plasmid were added to the transfection solution. The transfection solution was left on the cells for 3 h with DMEM (GIBCO) without serum; after this incubation period, the medium was removed and MEM (GIBCO) supplemented with 10% FBS (Atlanta Biologicals), and 1× sodium pyruvate, 1× non-essential amino acids, and Pen/Strep (GIBCO) were added to these cells. After 24 h, G418 selection medium with 400 μg/mL Geneticin (Invitrogen) was added and the cells were grown for 2 wk. Medium was changed on the cells every three days, and after 2 wk, colonies were fixed and stained with crystal violet prior to automated colony counting (Colcount, Oxford Optronics) or manual counting. Each experiment contained three flasks for each construct, and each experiment was repeated three times to obtain the relative number of colonies compared to an Alu with a 30-base pure A-tail.
Data mining
In order to assess A-tail length and heterogeneity of genomic Alus, 276 elements from the AluSx and 206 elements from the AluYa5 families were randomly sampled from the RepeatMasker annotation of build 36.1 (hg18) of the human genome and extracted, along with 5000 bp of flanking sequence, using in-house Perl scripts. The 3′ Flank studies came from this same data set but included 289 AluSx elements from chromosomes 1, 2, and 3 plus the 227 total AluYa5 elements. The boundaries of the A-tail were defined by first identifying the target site duplications flanking the Alu insertion, then subsequently defining the end of the tail as the first non-A base in the second (3′) TSD site (Roy-Engel et al. 2002). The initiation of the A-tail was defined by the first A-nucleotide subsequent to the conserved 3′ Alu consensus motif. Nucleotide composition was calculated using the previously described boundaries. The length of uninterrupted As in the A-tail was determined by counting from the initiation of the tail until the first non-A nucleotide. GC content of the A-tail, and the distance after the A-tail and before the genomic terminator were also assessed.
Northern blot analyses
RNA extraction and poly(A) selection were performed 48 h post-transfection as previously described (Perepelitsa-Belancio and Deininger 2003). Briefly, total RNA was extracted from two 75-cm2 cell culture flasks transfected with the Alu construct of interest using the TRIzol Reagent (Invitrogen) following the protocol supplied by the manufacturer. Poly(A) selection was performed using the PolyATract mRNA isolation system III (Promega) following the manufacturer's protocol. The poly(A) RNA was separated by electrophoresis in a 2% agarose-formaldehyde gel and transferred to a Hybond-N nylon membrane (Amersham Biosciences). The RNA was cross-linked to the membrane using a UV-light (GS Gene linker, BioRad) and pre-hybridized in 5× SSC, 5× Denhardt's, 1% SDS, and 100 μg/mL herring sperm DNA for at least 6 h at 60°C. A riboprobe complementary to the neomycin gene was used. A DNA template was amplified by PCR using the following primers T7neo(−): 5′-TAATACGACTCACTATAAGGACGAGGCAGCG-3′ and Neo northern(+): 5′-GAAGAACTCGTCAAGAAGG-3′. The isolated PCR product was used as a DNA template to generate a 32P-UTP (MP Biomedicals) labeled single strand-specific RNA probe using the MAXIscript T7 kit (Ambion) following the manufacturer's recommended protocol. We generated a riboprobe for cyclophillin (Ambion) to use as loading control. The radiolabeled probe was purified by filtration through a NucAway Spin column (Ambion). Hybridization with the probe (final concentration of 4–12 × 106 cpm/mL) was carried out overnight in the hybridization solution consisting of 30% formamide, 1× Denhardt's solution, 1% SDS, 1 M NaCl, 100 μg/mL salmon sperm DNA, 100 μg/mL yeast tRNA at 60°C. The membranes were washed twice for 15 min with a high stringency wash buffer (0.1× SSC, 0.1% SDS) at 60°C. The results of the Northern blot assays were evaluated using a Typhoon PhosphorImager (Amersham Biosciences) and quantitated with the ImageQuant software.
Real-time reverse transcriptase PCR
Total RNA extraction was performed 24 h post-transfection from 75-cm2 cell culture flasks transfected with the A-tail heterogeneity constructs as described above. cDNA was generated from the extracted RNA samples using a commercially available reverse transcription system (Promega) and evaluated by real-time PCR using Platinum SYBR Green Kit (Invitrogen) in a Bio-Rad IQ5 Real-Time PCR Detection System following the manufacturers' protocols. The following primers were used: neomycin selection cassette of the Alu construct: 5′-CCTCGGCCTCTGAGCTATTC-3′ and 5′-AGTCCCTTCCCGCTTCAGTGACAAC-3′, and GAPDH: 5′-GAAATCCCATCACCATCTTCCAGG-3′ and 5′-GAGCCCCAGCCTTCTCCATG-3′ (West et al. 2004). Quantitation of the RNA from each heterogeneity construct was performed relative to the A30 control.
Acknowledgments
This publication was made possible by Grants Number P20RR020152 (P.L.D., A.M.R.-E.), R01GM45668 and NSF EPSCOR grant (P.L.D.) and R01GM079709A (A.M.R.-E.) from the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Competitive Advantage Funds (2006) from the Louisiana Cancer Research Consortium (LCRC) were also awarded to A.M.R.-E.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089789.108.
References
- Alemán C., Roy-Engel A.M., Shaikh T.H., Deininger P.L. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–4761. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcot S.S., Wang Z., Weber J.L., Deininger P.L., Batzer M.A. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer M.A., Schmid C.W., Deininger P.L. Evolutionary analyses of repetitive DNA sequences. Methods Enzymol. 1993;224:213–232. doi: 10.1016/0076-6879(93)24017-o. [DOI] [PubMed] [Google Scholar]
- Belancio V.P., Hedges D.J., Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V.P., Hedges D.J., Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D. LINEs and Alus—The polyA connection. Nat. Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Chen J.M., Masson E., Macek M., Jr, Raguenes O., Piskackova T., Fercot B., Fila L., Cooper D.N., Audrezet M.P., Ferec C. Detection of two Alu insertions in the Cftr gene. J. Cyst. Fibros. 2008;7:37–43. doi: 10.1016/j.jcf.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Schmid C.W. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J. Mol. Evol. 1996;42:30–36. doi: 10.1007/BF00163208. [DOI] [PubMed] [Google Scholar]
- Chu W.M., Liu W.M., Schmid C.W. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23:1750–1757. doi: 10.1093/nar/23.10.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Hedges D.J., Herke S.W., Batzer M.A. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A. SINE master genes and population biology. In: Maraia R.J., editor. The impact of short interspersed elements (SINEs) on the host genome. RG Landes; Georgetown, TX: 1995. pp. 43–60. [Google Scholar]
- Deininger P.L., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Englander E.W., Wolffe A.P., Howard B.H. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J. Biol. Chem. 1993;268:19565–19573. [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Xing J., Wang H., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A. Under the genomic radar: The stealth model of Alu amplification. Genome Res. 2005;15:655–664. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges D.J., Callinan P.A., Cordaux R., Xing J., Barnes E., Batzer M.A. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M., Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu W.M., Schmid C.W. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.M., Maraia R.J., Rubin C.M., Schmid C.W. Alu transcripts: Cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A., Rozhdestvensky T.S., Kuryshev V.Y., Schmitz J., Brosius J. An unusual primate locus that attracted two independent Alu insertions and facilitates their transcription. J. Mol. Biol. 2005;350:200–214. doi: 10.1016/j.jmb.2005.03.058. [DOI] [PubMed] [Google Scholar]
- Maraia R.J., Driscoll C.T., Bilyeu T., Hsu K., Darlington G.J. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol. Cell. Biol. 1993;13:4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.E., Bennett E.A., Iskow R.C., Devine S.E. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Murphy D., Brickell P.M., Latchman D.S., Willison K., Rigby P.W.J. Transcripts regulated during normal embryonic development and oncogenic transformation share a repetitive element. Cell. 1983;35:865–871. doi: 10.1016/0092-8674(83)90119-8. [DOI] [PubMed] [Google Scholar]
- Odom G.L., Robichaux J.L., Deininger P.L. Predicting mammalian SINE subfamily activity from A-tail length. Mol. Biol. Evol. 2004;21:2140–2148. doi: 10.1093/molbev/msh225. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V., Deininger P.L. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Roy A.M., West N.C., Rao A., Adhikari P., Alemán C., Barnes A.P., Deininger P.L. Upstream flanking sequences and transcription of SINEs. J. Mol. Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- Roy-Engel A.M., Salem A.H., Oyeniran O.O., Deininger L., Hedges D.J., Kilroy G.E., Batzer M.A., Deininger P.L. Active Alu element “A-Tails”: Size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrowa J., Chang D.Y., Maraia R.J. The decline in human Alu retroposition was accompanied by an asymmetric decrease in SRP9/14 binding to dimeric Alu RNA and increased expression of small cytoplasmic Alu RNA. Mol. Cell. Biol. 1997;17:1144–1151. doi: 10.1128/mcb.17.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Roy A.M., Kim J., Batzer M.A., Deininger P.L. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J. Mol. Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- Shen M.R., Batzer M.A., Deininger P.L. Evolution of the master Alu gene(s) J. Mol. Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Richer C., Deragon J.M., Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J. Biol. Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- Sinnett D., Richer C., Deragon J.M., Labuda D. Alu RNA transcripts in human embryonal carcinoma cells. Model of post-transcriptional selection of master sequences. J. Mol. Biol. 1992;226:689–706. doi: 10.1016/0022-2836(92)90626-u. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Wallace N., Wagstaff B.J., Deininger P.L., Roy-Engel A.M. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Song L., Gonder M.K., Azrak S., Ray D.A., Batzer M.A., Tishkoff S.A., Liang P. Whole genome computational comparative genomics: A fruitful approach for ascertaining Alu insertion polymorphisms. Gene. 2006;365:11–20. doi: 10.1016/j.gene.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: Cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.B., Kapatos G., O'Farrell C., Gonzalez-de-Chavez F., Chiu K., Farrer M.J., Maidment N.T. N-myc regulates parkin expression. J. Biol. Chem. 2004;279:28896–28902. doi: 10.1074/jbc.M400126200. [DOI] [PubMed] [Google Scholar]