Abstract
Transcription factors (TFs) regulate the expression of genes through sequence-specific interactions with DNA-binding sites. However, despite recent progress in identifying in vivo TF binding sites by microarray readout of chromatin immunoprecipitation (ChIP-chip), nearly half of all known yeast TFs are of unknown DNA-binding specificities, and many additional predicted TFs remain uncharacterized. To address these gaps in our knowledge of yeast TFs and their cis regulatory sequences, we have determined high-resolution binding profiles for 89 known and predicted yeast TFs, over more than 2.3 million gapped and ungapped 8-bp sequences (“k-mers”). We report 50 new or significantly different direct DNA-binding site motifs for yeast DNA-binding proteins and motifs for eight proteins for which only a consensus sequence was previously known; in total, this corresponds to over a 50% increase in the number of yeast DNA-binding proteins with experimentally determined DNA-binding specificities. Among other novel regulators, we discovered proteins that bind the PAC (Polymerase A and C) motif (GATGAG) and regulate ribosomal RNA (rRNA) transcription and processing, core cellular processes that are constituent to ribosome biogenesis. In contrast to earlier data types, these comprehensive k-mer binding data permit us to consider the regulatory potential of genomic sequence at the individual word level. These k-mer data allowed us to reannotate in vivo TF binding targets as direct or indirect and to examine TFs' potential effects on gene expression in ∼1700 environmental and cellular conditions. These approaches could be adapted to identify TFs and cis regulatory elements in higher eukaryotes.
Transcription factors (TFs) are major regulators that control critical cellular processes and responses to environmental conditions. TFs control the expression of their target genes by binding to cis regulatory elements in a sequence-specific manner. Thus, TFs and their DNA-binding sites are of central importance for gene regulation, and intensive efforts have been invested in identifying TF binding sites.
S. cerevisiae is an important model organism in understanding fundamental biological pathways and transcriptional regulatory networks (Ideker et al. 2001; Harbison et al. 2004; MacIsaac et al. 2006; Workman et al. 2006), developing genome sequence analysis algorithms (Cliften et al. 2003; Kellis et al. 2003) that are subsequently applied to the genomes of higher organisms, and considering regulatory sequence evolution (Gasch et al. 2004; Tanay et al. 2005). However, even in S. cerevisiae, in which transcriptional regulation has been studied extensively both computationally (Hughes et al. 2000a; Cliften et al. 2003; Kellis et al. 2003; Beer and Tavazoie 2004) and experimentally using traditional and high-throughput genomic approaches (for review, see Bulyk 2006), the identities of the TFs that regulate major functional categories of genes or coexpressed genes remain unknown. For example, the PAC motif was identified nearly two decades ago (Dequard-Chablat et al. 1991) and computational studies have associated it with regulation of ribosomal RNA (rRNA) processing genes (Hughes et al. 2000a; Pilpel et al. 2001; Beer and Tavazoie 2004), but the trans factor(s) that bind this motif have remained unknown. A number of studies have utilized chromatin immunoprecipitation (ChIP) followed by microarray (ChIP-chip) (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001) to experimentally identify genomic regions occupied by TFs in vivo in the examined condition through either direct or indirect association with DNA; however, computational analysis of the binding data from those studies failed to yield sequence-specific binding motifs for half of the ∼200 known yeast TFs (Harbison et al. 2004; MacIsaac et al. 2006).
Most of the available ChIP-chip data on yeast TFs was obtained using array technology that provides TF binding data for entire intergenic regions. More recently, ChIP coupled with higher-density tiling arrays (Pokholok et al. 2005; Borneman et al. 2007) or with new generation sequencing technologies (ChIP-seq [Johnson et al. 2007], ChIP-paired-end diTag [ChIP-PET] [Wei et al. 2006]) has permitted higher resolution identification of in vivo TF binding sites. However, relatively few TFs have been examined by these higher resolution approaches, and the resulting data can still leave a challenge in distinguishing direct binding sites from those bound indirectly. Consequently, the functions and condition-specific regulatory roles of many known yeast TFs, even those with previously characterized binding specificities, are still not well understood, and many predicted TFs remain uncharacterized. Moreover, even well-studied TFs' DNA-binding preferences are often not sufficiently characterized to be able to accurately assess the consequences of nucleotide substitutions in their known binding sites, and a model of the binding preferences is typically generated based upon only a few dozen known binding sites.
Here we report high-resolution DNA-binding profiles and motifs for 89 TFs, utilizing protein-binding microarray (PBM) technology (Bulyk et al. 2001; Mukherjee et al. 2004) that covers all possible contiguous 8-mers and a large variety of gapped 8-mers (Berger et al. 2006). Briefly, custom-designed oligonucleotide arrays (Philippakis et al. 2008) are converted to double-stranded DNA arrays, incubated with GST-TF fusion protein in an in vitro-binding reaction, stained with fluorophore-conjugated anti-GST antibody, and scanned in a microarray scanner, followed by quantification of array signal intensities and sequence analysis (Berger et al. 2006). The resulting PBM data not only provide comprehensive DNA-binding preferences over all possible DNA-binding site variants, but also identify previously undiscovered DNA-binding proteins and their DNA-binding specificities, including two newly discovered TFs that bind to PAC sites and regulate rRNA processing genes. We predict the potential target genes, regulatory roles, and condition specificities of these TFs using their 8-mer binding profiles. While current PBM technology assesses the direct binding of protein to nucleosome-free DNA on the surface of arrays, PBM data on TFs' direct DNA-binding preferences in vitro are complementary to in vivo ChIP-based studies that provide information on both direct and indirect TF occupancy in particular conditions. We show that PBM data can be used to further interpret ChIP-chip data in order to distinguish likely direct versus indirect binding targets. Our extensive PBM k-mer data provide a valuable resource for future studies of transcriptional regulatory networks.
Results
DNA-binding specificity survey of known and predicted yeast transcription factors
To address major gaps in our knowledge of yeast TFs and their cis regulatory sequences, we have determined the comprehensive DNA-binding sequence specificities of 89 known and predicted yeast TFs using custom-designed universal protein-binding microarrays (PBMs) (Bulyk et al. 1999; Mukherjee et al. 2004; Berger et al. 2006). This data set constitutes the high-confidence data that we obtained from a DNA-binding specificity survey of 246 candidate TFs that we examined in this study (Supplemental Fig. S1, S2; Supplemental Table S1). Our survey included 157 proteins for which we did not obtain k-mer binding profiles that met our conservative acceptance criteria (Supplemental Methods). Our criteria for including candidate regulatory proteins in this survey were permissive and likely included proteins that do not bind DNA sequence specifically, if at all. Indeed, 14 of the proteins that did not yield motifs belong to structural classes for which there is no prior evidence for sequence-specific DNA-binding activity (bromodomain, PHD, Sir2, and Zf_CCCH), while another 58 had no identifiable DNA-binding domain and no prior evidence for DNA sequence-specific binding activity. Other factors, including 56 TFs that yielded motifs in previous analysis of ChIP-chip data (MacIsaac et al. 2006), may not have yielded motifs in our PBM survey as our criteria for acceptance of PBM-derived motifs may have been overly conservative. In addition, some TFs may not have been folded properly or may require protein partners, small molecule cofactors, post-translational modifications, particular buffer conditions, or a native chromatin context for sequence-specific DNA binding.
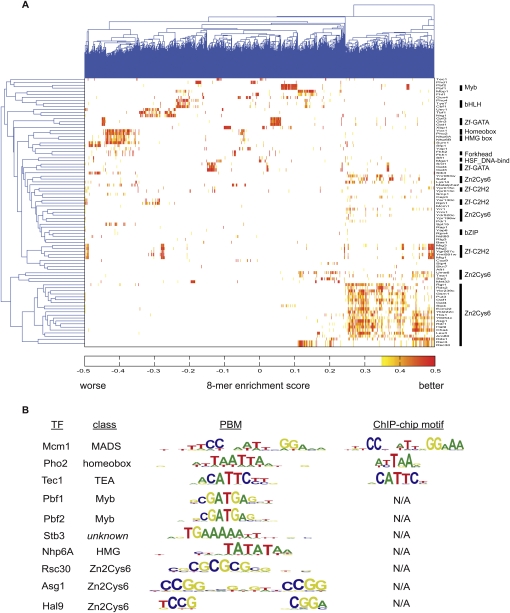
For each TF, from the PBM signal intensity data we calculated the relative sequence preferences, using an enrichment score (E-score) that ranges from −0.5 to 0.5 for each of more than 2.3 million gapped and ungapped 8-mers spanning the full affinity range from highest affinity to nonspecific sequences (Berger et al. 2006). These high-resolution k-mer binding profiles provide vastly more comprehensive binding specificity data than had been identified previously. In order to examine the landscape of sequence specificity across our entire data set, we performed two-dimensional clustering of the TFs' k-mer binding profiles (Fig. 1A; Supplemental Fig. S3). In general, the binding profiles of TFs of the same DNA-binding domain structural class were more similar to each other than to the profiles of TFs from different structural classes.
Figure 1.
PBM characterization of S. cerevisiae TF DNA-binding specificities. (A) Hierarchical clustering of PBM data over ungapped 8-mer E-scores determined for 89 yeast TFs. (B) Sequence logos for selected examples of newly discovered yeast TF DNA-binding site motifs.
Despite the fact that PBM k-mer data provide greater depth on the relative sequence preferences of TFs than do position weight matrices (PWMs) (Berger et al. 2008), in order to represent these DNA-binding specificities compactly, we constructed PWM-based motif representations using our Seed-and-Wobble algorithm (Berger et al. 2006) (Fig. 1B; Supplemental Fig. S4). For most previously characterized TFs, and in particular for the most well-known TFs, our PBM-derived motifs matched their previously known motifs (Supplemental Table S2). Consistent with prior knowledge of how proteins with a Zn2Cys6 (“Gal4-type”) DNA-binding domain can dimerize and interact with DNA (Reece and Ptashne 1993; Liang et al. 1996; Mamane et al. 1998), most TFs with a Zn2Cys6 DNA-binding domain have very similar half-site preferences, and appear to derive much of their specificities from the lengths of the degenerate spacers separating their half-sites. However, we found that not all Zn2Cys6 proteins bind CGG triplets; Rsc3 and Rsc30 appear to bind CGCGCGC and CGCGCGCGCG motifs, respectively.
We report experimentally determined DNA-binding site motifs for 30 known or predicted TFs lacking any prior motif data, and motifs for 11 additional TFs that have only consensus sequences reported in the literature. Among the 30 TFs we characterized that had no previously known DNA-binding specificities were 21 putative TFs, including many of unknown function. Interestingly, we observed sequence-specific DNA-binding preferences by the non-histone chromatin proteins Nhp6A (Fig. 1B) and Nhp6B (Supplemental Fig. S4), neither of which previously had been thought to have sequence-specific DNA-binding activity (Giavara et al. 2005). We confirmed the specificities of a subset of these novel TFs for their PBM-derived DNA-binding sequences by electrophoretic mobility shift assays (EMSAs) (Supplemental Fig. S5).
We also report direct DNA-binding site motifs for 20 TFs for which our PBM-derived motifs are substantially different (CompareACE score < 0.7; Supplemental Table S2) from the motifs computationally inferred from ChIP-chip data (Harbison et al. 2004; MacIsaac et al. 2006; Morozov and Siggia 2007) or from the literature, with our PBM-derived motifs being consistent with motifs of other TFs of the same DNA-binding domain structural classes. Three additional motifs are partial matches to previously derived motifs as the PBM motifs appear to be half-sites of apparently homodimeric ChIP-chip-derived motifs. Some of the ChIP-chip motifs that differ from PBM motifs appear to have captured a heterodimer. For example, the ChIP-chip-derived motif for Yox1 appears to contain not only a motif similar to our PBM-derived Yox1 motif, but also a MADS domain motif; of note, Yox1 is known to interact with the MADS domain protein Mcm1 (Pramila et al. 2002). All together we report new, direct DNA-binding specificities for 50 known or candidate TFs and for eight proteins for which only a (matching) consensus sequence was previously known; in total, this corresponds to over a 50% increase in the number of experimentally determined yeast DNA-binding site motifs (MacIsaac et al. 2006). Taking together our new PBM-derived motif data with prior motif data in the literature (MacIsaac et al. 2006; Morozov and Siggia 2007) or in the TRANSFAC database (Matys et al. 2003), experimentally determined DNA-binding site motif data are now available for 173 known or putative yeast TFs (Supplemental Table S3).
Analysis of in vivo binding site data with PBM k-mer data
Beyond looking at similarity between our PBM-derived motifs and literature motifs, we investigated the in vivo relevance of our k-mer data by comparing these data with ChIP-chip data. ChIP-chip experiments had previously been attempted for 70 of the 89 TFs in our data set as part of a large-scale survey (Harbison et al. 2004). We find that our k-mer binding data not only agree with the results of ChIP-chip for many TFs, but also aid in interpretation of the ChIP-chip data. Specifically, we used the k-mer binding data to calculate a predicted total TF occupancy score for each intergenic region (Supplemental Methods), ranked all of the intergenic regions by this score, and asked whether intergenic regions that scored well were enriched among the intergenic regions “bound” in vivo by ChIP-chip (P < 0.001) (Harbison et al. 2004).
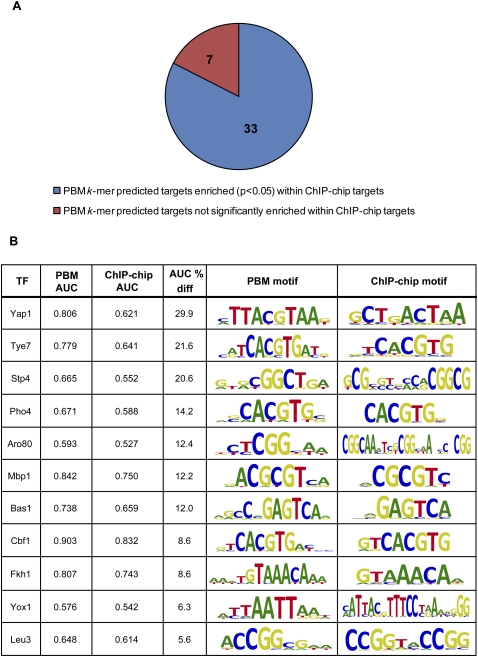
For 33 of the 40 TFs for which we had both PBM- and ChIP-chip-derived motifs (Harbison et al. 2004), we observed good agreement (AUC > 0.5, P < 0.05) between the ChIP-chip in vivo data and our scoring of intergenic regions by the PBM k-mer data (Fig. 2A). For 20 of these TFs, scoring of intergenic regions by the k-mer data yielded better agreement with the ChIP-chip binding data (Harbison et al. 2004) than did scoring using the ChIP-chip-derived motifs (Fig. 2B; Supplemental Methods; Supplemental Fig. S6). In contrast, for all but five (Mig1, Smp1, Mga1, Rph1, Mig3) of the 17 TFs that did not yield ChIP-chip motifs (MacIsaac et al. 2006), the k-mer-derived potential target genes were not enriched within the ChIP-chip bound regions (Harbison et al. 2004) (AUC > 0.5, P < 0.05); this suggests that the direct targets of those 12 TFs were not enriched in those ChIP-chip data sets (Supplemental Fig. S6).
Figure 2.
PBM k-mer binding profiles in most cases correspond well with ChIP-chip binding data. (A) For 33 of the 40 TFs for which we had both PBM- and ChIP-chip-derived motifs (Harbison et al. 2004), the PBM k-mer-derived potential targets were significantly enriched (AUC > 0.5, P < 0.05) among the ChIP-chip “bound” regions, showing good agreement between the ChIP-chip in vivo data and our scoring of genes based on the in vitro PBM k-mer data. (B) For 11 out of 40 TFs, intergenic regions scored by the PBM 8-mer data are more highly enriched (>5% improvement in AUC; all PBM AUC P-values are <0.05) among the ChIP-chip “bound” regions as compared with those scored by the ChIP-chip-derived motif.
The high-resolution nature of the PBM data presented an opportunity to reanalyze the ChIP-chip data (Harbison et al. 2004) and the subsequent improved regulatory map published by MacIsaac et al. (2006) for how well the in vivo binding data fit a model of direct TF–DNA binding. Previous computational analyses of those ChIP-chip data were performed using computationally inferred PWMs learned from those same ChIP-chip data (Harbison et al. 2004; MacIsaac et al. 2006; Tanay 2006). In contrast, the PBM data were generated by a direct biochemical approach, independent of the ChIP-chip experiments, and thus aid in annotation of direct targets.
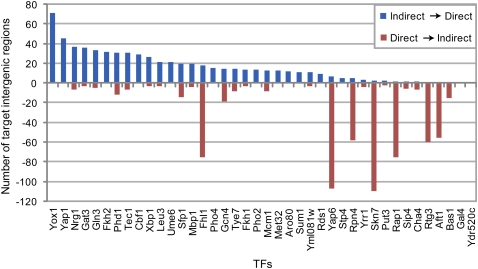
We reanalyzed the individual targets identified as “bound” in vivo to investigate whether they are likely to be bound directly by the TFs or indirectly via interaction with other proteins. Previously, MacIsaac et al. (2006) identified a set of high-confidence binding sites, which they defined as those containing motif matches that were bound by the corresponding factor at P < 0.001. We refer to those sites as that study's “direct” targets, and those bound by the corresponding factor at P < 0.001, but not identified by MacIsaac et al. (2006) as containing a motif match as their “indirect” targets. We scanned each of the “bound” intergenic regions for the presence of at least one k-mer at E > 0.45 (Berger et al. 2008), and annotated any such intergenic regions as “direct” targets according to k-mer matches and any bound intergenic regions not containing at least one k-mer at E > 0.45 as “indirect” targets. Using an E-score threshold rather than a continuous occupancy score in this analysis provides a consistent way to determine sets of target intergenic regions across TFs for comparison with the discrete sets of target intergenic regions reported in the ChIP-chip results (Harbison et al. 2004). This resulted in our reassignment of 682 intergenic regions that previously had been classified as direct targets according to the MacIsaac et al. (2006) regulatory map, rather as potentially being indirect targets (Fig. 3). Moreover, this suggests that 653 bound intergenic regions that previously had not been classified as high-confidence target sites, according to MacIsaac and colleagues, may actually be direct targets for one of these 40 TFs (Fig. 3). Using this same approach to analyze an additional 17 TFs that had at least 10 bound intergenic regions (P < 0.001) (Harbison et al. 2004), but for which MacIsaac et al. (2006) had not derived motifs from the ChIP-chip data, we annotated 279 out of a total of 852 binding instances as direct targets at E > 0.45 (Supplemental Table S4).
Figure 3.
Reclassification of TF occupancy at ChIP-chip “bound” (P < 0.001) intergenic regions as likely being due to direct DNA-binding sites versus indirect association of the TF with the DNA. Blue bars above the horizontal axis for each TF indicate the number of ChIP-chip bound intergenic regions that were previously called “indirect” (i.e., the regions do not contain a good match to the ChIP-chip motif as determined by MacIsaac et al. (2006) that are reclassified as potential “direct” TF targets by PBM data (i.e., the regions contain a PBM k-mer with an E-score > 0.45). Red bars below the axis indicate the number of intergenic regions previously annotated as “direct” targets by MacIsaac et al. (2006) that are reclassified as potential sites of indirect TF association according to the PBM data (i.e., the regions do not contain any k-mers with E-score > 0.45).
These comparisons highlight the complementary nature of the in vivo ChIP-chip and in vitro PBM data and the value of an integrated analysis for an improved distinction of direct versus indirect binding events. For example, the previously inferred ChIP-chip motifs for Fhl1 and Sfp1 (MacIsaac et al. 2006) appear to actually be matches to the Rap1 binding-site motif instead of reflecting their own respective DNA-binding specificities. Thus, some intergenic regions annotated by MacIsaac and colleagues as direct targets of Fhl1 or Sfp1 may actually be indirectly associated with those factors by interactions with Rap1. For other TFs, the large numbers of reannotated sites may be due to sensitivity to the PBM E-score threshold used. Since some TFs might utilize lower affinity DNA-binding sites than others, the use of different thresholds for different TFs may provide a more accurate distinction between direct and indirect binding sites; reannotation results at different E-score thresholds are shown in Supplemental Figure S7. For TFs for which relatively many intergenic regions previously annotated as indirect targets were reannotated as direct targets, the ChIP-chip-derived motif may not have accurately reflected the binding specificities of those factors (e.g., see earlier discussion of Yox1). In other cases, the PBM data suggest that a wider diversity of sequences may be recognized by a factor than is represented by the ChIP-chip motif, and thus, that more targets contain these additional preferred sequences.
Effects of transcription factor perturbations on potential target genes
To begin to investigate the potential regulatory functions of each TF, we determined whether its top-ranked potential target genes, ranked according to the total occupancy score described above (Supplemental Methods; Supplemental Table S5), are over-represented for particular functional categories (Supplemental Table S6) (Tavazoie et al. 1999; Hughes et al. 2000a; Robinson et al. 2002). Function predictions using our k-mer binding data for previously characterized TFs generally were supported by the literature (Supplemental Fig. S8). However, DNA-binding specificity data alone cannot identify which particular binding-site sequence occurrences serve a regulatory role in vivo and which corresponding potential target genes are actually regulated by a TF.
Therefore, to further investigate the potential regulatory effects of these PBM-derived DNA-binding sites, we examined the effects of genetic perturbations (deletion, overexpression, or other gain-of-function or loss-of-function mutants) of the TFs on their PBM-derived potential target genes. For each gene expression microarray data set, we applied our CRACR algorithm (McCord et al. 2007) to determine whether potential target genes are significantly enriched among the up- or down-regulated genes in any of the gene expression arrays. In all, we considered 256 expression data sets for TF perturbation yeast strains that were available for 79 of the 89 TFs.
Significant effects of TF perturbations on their PBM-predicted target genes were found in many cases, and such support for the predicted target genes was found just as often for TFs with novel or significantly different motifs as for TFs with well-known motifs (Supplemental Table S7). Specifically, we found that the potential target genes of 44 of these 79 TFs were enriched at a conservative significance threshold (CRACR area statistic ≥ 0.095, P ≤ 5 × 10−4) for being either up- or down-regulated in a corresponding TF perturbation strain. Twenty of these 44 TFs correspond to those for which our PBM-derived motif is substantially different (CompareACE score < 0.7) from the motif computationally inferred (MacIsaac et al. 2006) from the ChIP-chip data (Harbison et al. 2004) or from the literature consensus sequence, and another 14 of these 44 TFs correspond to those for which we newly report direct DNA-binding site motifs. For example, the potential target genes of Ypr015c, a putative protein of unknown function, are significantly enriched (CRACR area statistic = 0.12, P = 6.1 × 10−10) among genes down-regulated in a YPR015C overexpression strain (Chua et al. 2006). More case examples for specific TFs are provided in the following sections. The lack of enrichment for the other 35 TFs might indicate that those TFs did not exert a strong regulatory effect in the profiled conditions, that there exist at least partially functionally redundant TFs, or that the gene expression effects are predominantly due to secondary effects of the perturbed TFs. Support for the potential target genes of several additional TFs came from expression data on perturbations of genes with which they exhibit genetic interactions (Supplemental Table S7). Further experiments will be needed to validate our predictions for those cases where in vivo TF binding data and gene expression data on TF deletion or mutant TF strains are not available.
Sequence-specific DNA binding by Rsc3 and Rsc30, components of the RSC chromatin remodeling complex
RSC is an essential 15-subunit ATP-dependent chromatin remodeling complex that repositions nucleosomes (Cairns et al. 1996, 1999). Two subunits of this RSC complex, Rsc3 and Rsc30, are Zn2Cys6 proteins that previously had been hypothesized to recognize specific sequences and thus help target this important regulatory complex to specific genes (Wilson et al. 2006). In line with this hypothesis, we observed sequence-specific binding by these factors in PBM experiments and determined novel DNA-binding site motifs for them (Fig. 1B; Supplemental Fig. S4).
ChIP-chip data on in vivo occupancy by RSC support the model that in vivo genomic binding of the RSC complex is conferred by the Rsc3/30 sequence preferences that we have identified. ChIP-chip data are available for the Rsc1 and Rsc2 isoforms of the RSC complex from separate ChIP experiments on five subunits, namely, Rsc1, Rsc2, Rsc3, Rsc8, and Sth1 (Ng et al. 2002). In all five of these data sets, we found that the intergenic regions that scored well by the k-mer data for Rsc3, and separately for Rsc30, are highly enriched among intergenic regions occupied in vivo (Supplemental Table S8). The potential target regions of Rsc3 and Rsc30 are also enriched among the intergenic regions occupied by Rsc9 in four different environmental conditions (Damelin et al. 2002).
Furthermore, analysis of gene expression microarray data revealed that the potential target genes of Rsc3 and Rsc30 are enriched among the genes that are differentially expressed when Rsc3 or Rsc30 were perturbed. In a RSC3 temperature-sensitive mutant strain (Angus-Hill et al. 2001) in which RSC3 itself is up-regulated, we found that Rsc3′s potential target genes are enriched among down-regulated genes (CRACR area statistic = 0.110, P = 1.41 × 10−6). In a RSC30 deletion strain (Angus-Hill et al. 2001), Rsc30's potential target genes are enriched among down-regulated genes (CRACR area statistic = 0.099, P = 1.70 × 10−4). Consistent with these findings, CRACR analysis of gene expression data performed on a RSC30 overexpressor strain (Chua et al. 2006), in which RSC3 is up-regulated, revealed that the potential target genes of Rsc3 are enriched (CRACR area statistic = 0.0986, P = 7.87 × 10−4) among the repressed genes. These data support the model that Rsc3 and Rsc30 may sometimes have opposite functions (Angus-Hill et al. 2001).
Finally, Rsc3 and Rsc30 are required for regulation of genes that regulate cell wall integrity (Angus-Hill et al. 2001). Consistent with these findings, we observed that the PBM-derived potential target genes of Rsc3 are highly enriched for various functional annotation terms pertaining to the cell wall and cell wall function (Supplemental Table S6). In addition, Ng et al. (2002) found that RSC targets several gene classes, including histones. Consistent with that result, we found that Rsc3's PBM-derived potential target genes are also highly enriched for various functional categories of genes pertaining to chromatin remodeling (Supplemental Table S6). Taken together, all of these analyses support the model that Rsc3 and Rsc30 target the RSC complex to relevant classes of genes through the sequence preferences discovered in our PBM experiments.
Two newly discovered PAC-binding factors associated with regulation of rRNA processing genes
The PAC and RRPE motifs are highly over-represented in the upstream regions of rRNA processing and transcription genes (Hughes et al. 2000a), and exhibit significant correlation with their expression (Pilpel et al. 2001). While an RRPE-binding factor, Stb3, has recently been described (Liko et al. 2007), the identity of a PAC-binding factor had remained unknown. Two of our novel TFs' DNA-binding site motifs are extremely good matches to the well-known PAC motif (Dequard-Chablat et al. 1991), and we now refer to these proteins of previously unknown function as “PAC-binding factor 1” (Pbf1; also known as Ybl054w) and “PAC-binding factor 2” (Pbf2; also known as Dot6, or Yer088c). By identifying Pbf1 and Pbf2 as PAC-binding factors, we have thus bridged a long-standing knowledge gap in the regulation of ribosome biogenesis. As expected given its recognition of the PAC motif, the genes predicted to have the highest occupancy by Pbf1 and Pbf2 are highly enriched for the Gene Ontology (GO) Biological Process terms “ribosome biogenesis” (P = 2.81 × 10−8 and 2.29 × 10−7, respectively, Bonferroni-corrected) and “rRNA processing” (P = 8.28 × 10−6 and 2.40 × 10−3, respectively, Bonferroni-corrected) (Supplemental Table S6).
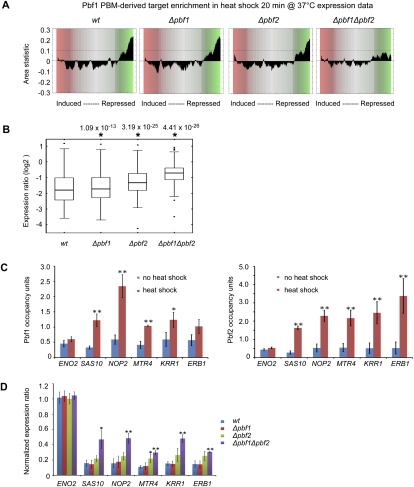
Experimental testing of these newly discovered TFs supports their involvement in regulation of rRNA processing genes. Consistent with prior studies of the association of the PAC motif with stress-induced changes in gene expression (Tavazoie et al. 1999; Causton et al. 2001; Pilpel et al. 2001), CRACR predicted that Pbf1 and Pbf2 regulate rRNA processing genes in a number of stress conditions in which ribosome biosynthesis is repressed, including in heat shock (CRACR P = 4.9 × 10−11 and 1.7 × 10−11, respectively). Therefore, we performed Affymetrix gene expression profiling of the single deletion mutants Δpbf1 and Δpbf2, the double deletion mutant Δpbf1Δpbf2, and the isogenic wild-type strain upon heat-shock treatment. We found that the potential target genes of Pbf1 and Pbf2 are significantly repressed (CRACR P < 10−12) during heat shock in wild-type and in the Δpbf1 and Δpbf2 single deletion strains, and that this repression is diminished in the Δpbf1Δpbf2 double deletion strain (Fig. 4A; Supplemental S9). Consistent with the literature (Gasch et al. 2000), we found that during heat shock, rRNA processing genes are down-regulated (Supplemental Table S9). Importantly, we found that rRNA processing genes (GO ID 0006364) that contain at least one k-mer at a conservative threshold of E ≥ 0.45 (Berger et al. 2008) for either Pbf1 or Pbf2 within 600 bp upstream of translational Start are significantly derepressed in Δpbf1 (P = 4.37 × 10−13, two-tailed paired Wilcoxon-Mann-Whitney test) and in Δpbf2 (P = 3.19 × 10−25, as above), and even more so in the Δpbf1Δpbf2 double deletion strain (P = 4.41 × 10−26, as above) (Fig. 4B).
Figure 4.
Pbf1 and Pbf2 regulate rRNA processing genes. (A) Predicted target genes of Pbf1 and Pbf2 are significantly repressed (CRACR P < 10−12) after 20 min heat shock (shift from 25°C to 37°C) in wild-type, Δpbf1, and Δpbf2 strains, but not in the Δpbf1Δpbf2 double deletion strain, in Affymetrix gene expression profiling of triplicate biological replicate cultures. (B) Box plots indicating expression changes of rRNA processing genes containing at least one k-mer at E ≥ 0.45 after 20 min heat shock in wild-type, Δpbf1, Δpbf2, and Δpbf1Δpbf2 strains, in the expression data from A. (C) Pbf1 and Pbf2 associate in vivo with the promoter regions of the rRNA processing genes SAS10, NOP2, MTR4, KRR1, and ERB1. ChIP-qPCR was performed on cells treated with 5-min heat shock, at predicted target sites in their upstream regions, and at a negative control region upstream of ENO2. Binding fold-enrichment was defined as the ratio of PCR product in “IP” versus “INPUT,” using an open reading frame free region on chromosome V as an internal normalization control. Error bars indicate 1 SD from triplicate biological replicate cultures (*P < 0.05; **P < 0.01; two-sided Student's t-test). (D) Expression ratio of rRNA processing genes after heat shock. RT-qPCR data were generated for either untreated yeast or yeast treated with 20-min heat shock. Gene expression was normalized relative to ACT1 as an internal normalization control. Error bars indicate 1 SD from triplicate biological replicate cultures (*P < 0.05; **P < 0.01; two-sided Student's t-test compared with wild type).
We used chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) to measure the association of Pbf1 and Pbf2 under heat shock to the promoter regions of five rRNA processing genes—SAS10, NOP2, MTR4, KRR1, and ERBI—which contain the PAC motif, and as a negative control we used ENO2, which is not involved in rRNA processing and does not contain a PAC site in its promoter region. We found that after heat shock, Pbf2 bound significantly (P < 0.01, Student's t-test) to all five of these rRNA processing genes' promoters, and not to ENO2 (Fig. 4C). Similarly, Pbf1 bound significantly to most of these promoters, although to a lesser degree. Quantitative RT-PCR data indicated that all five of these rRNA processing genes are normally repressed during heat shock, but that they are all significantly derepressed (P < 0.05, Student's t-test) in the Δpbf1Δpbf2 double deletion strain (Fig. 4D). The derepression appeared to be greater in Δpbf2 than in Δpbf1, but the derepression was statistically significant for only one of these five genes in the Δpbf2 single deletion mutant. Taking all of these data together, we conclude that both Pbf1 and Pbf2 coregulate rRNA processing genes upon heat shock, with Pbf2 apparently playing a greater role.
Prediction of transcription factor condition-specific regulatory roles
Condition-specific binding-site usage is a vital aspect of TF function (Simon et al. 2001; Harbison et al. 2004). While ChIP-chip provides a “snapshot” of what genomic regions are occupied in vivo by a TF either directly or indirectly in the particular examined condition(s), PBM data provide information on the inherent, direct DNA-binding preferences of a protein. Therefore, we applied our CRACR algorithm (McCord et al. 2007) to the potential target genes of these 89 S. cerevisiae TFs in order to generate specific hypotheses about the condition-specific binding-site utilization and functions of these TFs, considering the environmental and cellular conditions represented in 1693 publicly available microarray gene expression data sets (Supplemental Tables S10, S11). Some of these CRACR predictions have been experimentally validated. Using ChIP-qPCR, we validated our CRACR predictions of condition-specific Rap1 binding site usage after diamide treatment (Supplemental Fig. S10). In addition, CRACR predicted that Pbf1 and Pbf2 coregulate rRNA processing genes in heat shock (Fig. 4; Supplemental Table S10).
The CRACR results from analysis of 1693 conditions suggested condition-specific regulation by additional previously uncharacterized TFs in conditions including sporulation, carbohydrate metabolism, and stress. The predicted conditions could be used to direct future in vivo experiments in cases where previous experiments may have failed because the yeast were grown in conditions in which the TF of interest does not bind its target sites. For example, a prior Tbs1 ChIP-chip experiment (Harbison et al. 2004) was performed on yeast grown only in YPD rich media conditions in the absence of stress, while CRACR suggests that Tbs1 regulates its target genes in sodium chloride and heat-shock stress conditions.
Identification of potential coregulatory transcription factors
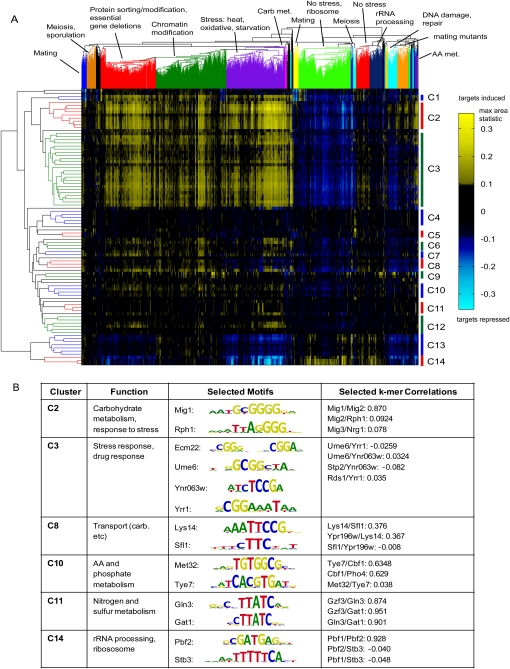
To identify sets of TFs that may exhibit similar regulatory effects on their target genes over various conditions, we performed two-dimensional hierarchical clustering of the 89 TFs according to their CRACR statistics across all 1693 microarray expression data sets (Fig. 5A; Supplemental Fig. S11). This CRACR clustering analysis provides different information from clustering the TFs' own gene expression patterns, as it identifies the conditions in which the TFs' potential target genes are differentially expressed rather than when the TFs themselves are differentially expressed. This clustering analysis also provides different information than an analysis of overlap in the sets of intergenic regions bound in ChIP-chip (Harbison et al. 2004), where binding events could involve either direct DNA binding or indirect DNA associations by the TFs.
Figure 5.
Analysis of TFs' regulatory associations and coregulatory factors. (A) Two-dimensional hierarchical clustering of 89 TFs (rows) according to their CRACR statistics across 1693 expression conditions (columns). (B) Examples of predicted coregulatory TFs from A with distinct motifs, and their 8-mer binding profile correlations. Clusters annotations are derived from the literature and functional predictions from this study. A high-resolution heatmap with full labeling is available in Supplemental Fig. S11, S12.
Expression conditions in general segregated according to general functional categories, and various sets of TFs have similar regulatory associations across conditions. The largest pattern in the CRACR clustering heatmap includes numerous TFs generally involved in stress response that appear to regulate their genes in response to various stress conditions. Using Pbf1, Pbf2, and Stb3 as a guide because of their DNA-binding site motifs' known coregulatory associations (Hughes et al. 2000a; Pilpel et al. 2001; Beer and Tavazoie 2004), we considered all TF clusters with equal or greater CRACR profile similarity as that of the Pbf1/Pbf2/Stb3 cluster, as groups of putative coregulatory TFs (Fig. 5B; Supplemental Fig. S12). At this threshold, 14 of 18 TFs predicted to be involved in drug response according to over-represented annotation terms among their potential target genes (Supplemental Table S6) clustered together in cluster C3 (Supplemental Fig. S12); these include the novel candidate drug response TFs Asg1 and Ykl222c as well as the known drug response TF Yrr1. While some clusters, such as C11, identified coregulatory TFs that have very similar k-mer binding profiles, other clusters identified coregulatory TFs with distinct motifs (Fig. 5B; Supplemental Fig. S12). Such hypotheses about potential coregulation can be used to direct future in vivo experimentation on yeast gene regulatory networks.
Discussion
This study provides both comprehensive binding specificity data for many known and newly identified TFs and a framework for identifying all yeast DNA-binding proteins and predicting their regulatory roles. The use of experimentally determined, comprehensive k-mer binding data to identify candidate cis regulatory elements and to predict candidate target genes is novel not only in the depth of experimentally measured TF binding specificity data, but also in tactical approaches for scoring genomic sequence for its regulatory potential. We expect that these yeast data will serve as a valuable model system for developing new, k-mer-based approaches for modeling transcriptional regulatory networks. Since PBM data correlate with DNA-binding affinities (Berger et al. 2006), in the future, the methods and data presented here could be adapted to explore differences in the usage of binding sites of different affinities between TFs or in different cellular or environmental conditions. Finally, although many challenges exist in the identification of cis regulatory elements in higher eukaryotes because of their frequently more distant regulatory regions and greater combinatorial regulatory mechanisms, the approaches described in this study could be integrated with additional strategies, such as phylogenetic footprinting, nucleosome occupancy data, high-resolution ChIP-chip, or ChIP-seq, in order to identify trans regulatory factors, to predict their cis regulatory elements and target genes, and to distinguish their directly bound and indirectly associated regulatory sites in genomes.
Methods
Cloning, expression, and purification of S. cerevisiae TFs
We cloned the 245 full-length ORFs and 99 DBDs into Gateway-compatible Entry and Destination vectors, pDONR201 or pDONR221, and pDEST-GST (Braun et al. 2002), as described previously (Hu et al. 2007). A separate, partially-redundant set of 118 DBDs were a generous gift from Tim Hughes (University of Toronto, Ontario, Canada) and Jason Lieb (University of North Carolina, Chapel Hill). Thus, our final clone collection includes 245 ORFs as full-length constructs, and 208 as DBDs. We did not pursue cloning of DBDs if the Pfam-annotated DBDs spanned >80% of the full-length protein, or in some cases, if careful manual inspection indicated that these were not likely DBDs based on their descriptions (e.g., bromodomain).
Protein production
For all 245 full-length TF and 208 DBD clones, we performed high-throughput overexpression in E. coli cultures and subsequent affinity purification using glutathione resin in 96-well plates, essentially as described previously (Hu et al. 2007). For each purified protein we performed Western blots to assess quality and to approximate its concentration. Overall, this resulted in 246 nonredundant TFs.
Protein-binding microarrays (PBMs)
We constructed microarrays covering all 10-bp sequence variants (Berger et al. 2006; Philippakis et al. 2008) by converting high-density single-stranded oligonucleotide arrays to double-stranded DNA arrays (Berger et al. 2006; Berger and Bulyk 2009). Using these universal arrays, we measured the relative preferences of a TF for all possible contiguous 8-mers, as well as gapped 8-mers spanning up to 10 total positions. The new array designs we developed for this study also included typically 32-fold redundancy for all non-palindromic 8-mers consisting of two 4-bp half-sites separated by spacers from 1 to 12 bp in length; we added this array design feature to allow us to capture the sequence preferences of TFs with long or gapped recognition motifs, such as members of the Zn2Cys6 structural class (i.e., “Gal4-type” motifs).
Identification of DNA-binding specificities
Every nonpalindromic 8-mer occurs on at least 32 spots in each chamber of our universal PBM. Because of this redundancy, we are able to provide a robust estimate of the relative preference of a TF for every contiguous and gapped 8-mer that is covered on our array. Here, for each 8-mer, we provide the median normalized signal intensity and a rank-based statistical enrichment score (E-score). Median normalized signal intensity refers to the median normalized signal intensity for the set of probes containing a match to each 8-mer (usually ∼32 probes). Our E-score is a rank-based, nonparametric statistical measure that has been described previously in detail (Berger et al. 2006) and ranges from +0.5 (most favored) to −0.5 (most disfavored). We applied our “Seed-and-Wobble” algorithm to derive position weight matrices (PWMs) from universal array PBM data (Berger et al. 2006; Berger and Bulyk 2009). All PBM data are publicly available via the UniPROBE database (Newburger and Bulyk 2009).
Comparison of PBM motifs
We used CompareACE (Hughes et al. 2000a) to compare our 89 PBM-derived motifs against a list of 4282 PWMs for previously published motifs. We required a minimum CompareACE motif similarity score of 0.7 to consider motifs as matching.
Identification and scoring of potential target genes using PBM k-mer data
A predicted total occupancy score for a given TF was calculated for the upstream promoter region of each gene by summing the background-subtracted median PBM signal intensities for each overlapping 8-mer, considering all those 8-mers at E ≥ 0.35, over the sequence up to 600 bp upstream of translation start. The median value of the median intensities over all 8-mers was used as a measure of the background signal and was subtracted from each individual 8-mer's intensity before summation. For most analyses, the total occupancy score was used to rank genes according to their likelihood of being TF target genes. The top 200 scoring genes were considered for analysis of functional category enrichment among a TF's potential target genes. We utilized our CRACR algorithm essentially as described previously (McCord et al. 2007), except here, genes were first ranked by the predicted total occupancy of the sequence up to 600 bp upstream of its translational start site by a TF as described above.
Analysis of ChIP-chip data
All yeast intergenic regions (IGRs) were scored using PBM 8-mer data as described above for each TF, except that in this case the entire IGR length was scored, rather than only 600 bp upstream of translation start. This enabled a direct comparison between scores derived from PBM data and those measured from ChIP-chip experiments, in which the entire IGR was included on the array. Target IGR sets for each TF were defined as IGRs bound by the TF in a ChIP-chip experiment at P < 0.001 in any experimental condition, as reported by the authors of that study (Harbison et al. 2004). TFs were excluded from the analysis if fewer than 10 IGRs were bound at P < 0.001 in the ChIP-chip data. An area under the receiver operating characteristic (ROC) curve (AUC statistic) was then calculated by comparing the PBM-derived ranks of IGRs within the ChIP-chip “bound” IGRs (foreground set, or “class 1”) to the ranks of the rest of the yeast IGRs (background set, or “class 0”). For comparison, ChIP-chip-derived motifs, if available, were used to rank the IGRs as well. ScanACE (Roth et al. 1998; Hughes et al. 2000b) was used to score ChIP-chip motif matches in all yeast IGRs at a threshold of 2 SD below the mean motif score. If multiple matches occurred within an IGR, these scores were summed to obtain a final score for each IGR. The resulting ChIP-chip IGR ranking was then used to calculate an AUC statistic comparing the ChIP-chip-derived ranks for ChIP-chip target IGRs versus background IGRs.
EMSAs
Sixty-nucleotide EMSA probes were designed such that the 5′ 40-nt sequence corresponds to a putative target intergenic region in the yeast genome and contains the predicted DNA-binding site, and the next 20 nt corresponds to a common priming sequence at the 3′ end that can anneal to a universal biotinylated primer. Primer extensions reactions were performed in order to convert the single-stranded probes to double-stranded probes. Approximately 5 nM DNA probe and ∼0.2 μM protein were used in each reaction.
Yeast strains and growth conditions
BY4741, Δpbf1, and Δpbf2 were purchased from Open Biosystems. The Δpbf1Δpbf2 double deletion mutant was generated by replacing PBF2 with URA3 by homologous recombination in the Δpbf1 background. PCR epitope tagging was used to generate yeast strains with a 3xHA (hemagglutinin) N-terminal epitope tag using plasmid pMPY-3xHA. All yeast were grown in standard yeast YPD medium if not otherwise specified.
Chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR)
We carried out chromatin immunoprecipitation as described previously (Aparicio et al. 2005) with minor modifications. Three independent cultures were grown in parallel in order to carry out triplicate biological replicates for ChIP assays. Cells were then subjected to heat-shock treatment, i.e., growth temperature shifted from 25°C to 37°C, for 5 min, prior to fixation with 1% formaldehyde for 20 min. qPCRs were performed using iQ SYBR Green SuperMix (Bio-Rad) on an iCycler real-time PCR thermocycler.
Gene expression profiling and quantitative RT-PCR (RT-qPCR)
Three independent cultures of the BY4741, Δpbf1, Δpbf2, and Δpbf1Δpbf2 strains were grown in parallel in order to carry out triplicate biological replicates. Cells were then subjected to heat-shock treatment, i.e., growth temperature shifted from 25°C to 37°C, for 20 min, and subsequently spun down and flash-frozen at −80°C. RNA was extracted and purified using Qiagen RNeasy Mini kit with DNase I treatment. Gene expression profiling was performed using Affymetrix Yeast Genome 2.0 GeneChip oligonucleotide arrays essentially according to Affymetrix protocols. Microarray data were analyzed as described previously (Choe et al. 2005). We imposed a false discovery rate (FDR) of 0.0001 as the cut-off value to identify differentially expressed genes. Microarray data were deposited into the GEO database under accession number GSE13684. GO term enrichment analysis was performed by applying FuncAssociate (Berriz et al. 2003) on lists of differentially expressed genes ordered by their expression ratio. RT-qPCR reactions were performed essentially as described above.
Acknowledgments
We thank G. Badis, T.R. Hughes, and J.D. Lieb for sharing 118 N-term GST fusion DNA-binding domain expression clones; G. Berriz for assistance with FuncAssociate; F. Winston, I. Laprade, and X. Li for technical assistance with yeast experiments; J.-A. Kwon and J. Love for technical assistance with Affymetrix gene expression profiling experiments; T. Gurbich for technical assistance with PBMs; and K. Struhl for helpful discussion. Plasmid pMPY-3xHA was a generous gift from K. Struhl. We thank S. Gisselbrecht, T. Siggers, and A. Dudley for critical reading of the manuscript. We also thank anonymous reviewers for helpful comments. This work was funded by grants R01 HG003985 and R01 HG003420 from NIH/NHGRI to M.L.B. M.F.B. and R.P.M. were supported in part by National Science Foundation Graduate Research Fellowships.
Footnotes
[Supplemental material is available online at www.genome.org and at http://thebrain.bwh.harvard.edu/. Gene expression microarray data have been submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under accession no. GSE13684. Protein-binding microarray data are available at http://thebrain.bwh.harvard.edu/ and in the UniPROBE database, http://thebrain.bwh.harvard.edu/uniprobe/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.090233.108.
References
- Angus-Hill M.L., Schlichter A., Roberts D., Erdjument-Bromage H., Tempst P., Cairns B.R. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Aparicio O., Geisberg J., Sekinger E., Yang A., Moqtaderi Z., Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. 2005 doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- Beer M.A., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Berger M., Bulyk M. Universal protein binding microarrays for the comprehensive characterization of the DNA binding specificities of transcription factors. Nat. Protocols. 2009 doi: 10.1038/nprot.2008.195. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., III, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Badis G., Gehrke A.R., Talukder S., Philippakis A.A., Pena-Castillo L., Alleyne T.M., Mnaimneh S., Botvinnik O.B., Chan E.T., et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz G.F., King O.D., Bryant B., Sander C., Roth F.P. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Borneman A.R., Zhang Z.D., Rozowsky J., Seringhaus M.R., Gerstein M., Snyder M. Transcription factor binding site identification in yeast: A comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct. Integr. Genomics. 2007;7:335–345. doi: 10.1007/s10142-007-0054-7. [DOI] [PubMed] [Google Scholar]
- Braun P., Hu Y., Shen B., Halleck A., Koundinya M., Harlow E., LaBaer J. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 2002;99:2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L. DNA microarray technologies for measuring protein–DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L., Gentalen E., Lockhart D.J., Church G.M. Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- Bulyk M.L., Huang X., Choo Y., Church G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., Kornberg R.D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns B.R., Schlichter A., Erdjument-Bromage H., Tempst P., Kornberg R.D., Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.E., Boutros M., Michelson A.M., Church G.M., Halfon M.S. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G., Morris Q.D., Sopko R., Robinson M.D., Ryan O., Chan E.T., Frey B.J., Andrews B.J., Boone C., Hughes T.R. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Damelin M., Simon I., Moy T.I., Wilson B., Komili S., Tempst P., Roth F.P., Young R.A., Cairns B.R., Silver P.A. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III) J. Biol. Chem. 1991;266:15300–15307. [PubMed] [Google Scholar]
- Gasch A., Spellman P., Kao C., Carmel-Harel O., Eisen M., Storz G., Botstein D., Brown P. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Moses A.M., Chiang D.Y., Fraser H.B., Berardini M., Eisen M.B. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavara S., Kosmidou E., Hande M.P., Bianchi M.E., Morgan A., d'Adda di Fagagna F, Jackson S.P. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr. Biol. 2005;15:68–72. doi: 10.1016/j.cub.2004.12.065. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T., Zhu C., Berger M., Camargo A., Kelley F., McCarron S., Jepson D., et al. Approaching a complete repository of sequence-verified, protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., Estep P.W., Tavazoie S., Church G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000a;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Hughes T., Marton M., Jones A., Roberts C., Stoughton R., Armour C., Bennett H., Coffey E., Dai H., He Y., et al. Functional discovery via a compendium of expression profiles. Cell. 2000b;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Liang S.D., Marmorstein R., Harrison S.C., Ptashne M. DNA sequence preferences of GAL4 and PPR1: How a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb J.D., Liu X., Botstein D., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Liko D., Slattery M.G., Heideman W. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:26623–26628. doi: 10.1074/jbc.M704762200. [DOI] [PubMed] [Google Scholar]
- MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y., Hellauer K., Rochon M.H., Turcotte B. A linker region of the yeast zinc cluster protein leu3p specifies binding to everted repeat DNA. J. Biol. Chem. 1998;273:18556–18561. doi: 10.1074/jbc.273.29.18556. [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord R.P., Berger M.F., Philippakis A.A., Bulyk M.L. Inferring condition-specific transcription factor function from DNA binding and gene expression data. Mol. Syst. Biol. 2007;3:100. doi: 10.1038/msb4100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov A.V., Siggia E.D. Connecting protein structure with predictions of regulatory sites. Proc. Natl. Acad. Sci. 2007;104:7068–7073. doi: 10.1073/pnas.0701356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger D.E., Bulyk M.L. UniPROBE: An online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Robert F., Young R.A., Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis A.A., Qureshi A.M., Berger M.F., Bulyk M.L. Design of compact, universal DNA microarrays for protein binding microarray experiments. J. Comput. Biol. 2008;15:655–665. doi: 10.1089/cmb.2007.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y., Sudarsanam P., Church G.M. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Pramila T., Miles S., GuhaThakurta D., Jemiolo D., Breeden L.L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes & Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R.J., Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robinson M., Grigull J., Mohammad N., Hughes T. FunSpec: A web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth F.P., Hughes J.D., Estep P.W., Church G.M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Simon I., Barnett J., Hannett N., Harbison C., Rinaldi N., Volkert T., Wyrick J., Zeitlinger J., Gifford D., Jaakkola T., et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A., Regev A., Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J., Campbell M., Cho R., Church G. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wilson B., Erdjument-Bromage H., Tempst P., Cairns B.R. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics. 2006;172:795–809. doi: 10.1534/genetics.105.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C.T., Mak H.C., McCuine S., Tagne J.B., Agarwal M., Ozier O., Begley T.J., Samson L.D., Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]