Abstract
Hox genes control many aspects of embryonic development in metazoans. Previous analyses of this gene family revealed a surprising diversity in terms of gene number and organization between various animal species. In vertebrates, Hox genes are grouped into tightly organized clusters, claimed to be devoid of repetitive sequences. Here, we report the genomic organization of the four Hox loci present in the green anole lizard and show that they have massively accumulated retrotransposons, leading to gene clusters larger in size when compared to other vertebrates. In addition, similar repeats are present in many other development-related gene-containing regions, also thought to be refractory to such repetitive elements. Transposable elements are major sources of genetic variations, including alterations of gene expression, and hence this situation, so far unique among vertebrates, may have been associated with the evolution of the spectacular realm of morphological variations in the body plans of Squamata. Finally, sequence alignments highlight some divergent evolution in highly conserved DNA regions between vertebrate Hox clusters, which may coincide with the emergence of mammalian-specific features.
Hox genes encode homeodomain-containing transcription factors that play a central role in the specification of regional identities along the anterior-to-posterior body axis. In many animal species, they are characterized by their clustered organization, which generally coincides with the distribution of their expression domains along the developing body axis (spatial collinearity). In some taxa, in particular vertebrates, the physical order of genes also corresponds to the timing of their transcriptional activation (temporal collinearity) (e.g., Krumlauf 1994; Kmita and Duboule 2003). In the course of vertebrate evolution, Hox gene functions were coopted to accompany the development of several organs from multiple germ layers origins (Deschamps and van Nes 2005; Di-Poï et al. 2007; Zakany and Duboule 2007), making these genes of particular interest to study the genetic bases of evolutionary mechanisms.
Hox gene clusters have been used as a paradigm to study genome evolution (e.g., de Rosa et al. 1999; Ferrier and Holland 2002; Garcia-Fernandez 2005; Lemons and McGinnis 2006) ever since they were found conserved in vertebrates and invertebrates (Duboule and Dolle 1989; Graham et al. 1989). In the past few years, however, genomic analyses have revealed a surprising diversity in Hox gene number, genomic organization, and expression patterns between various metazoans. In several cases, indeed, clusters have been broken up, either partially or entirely, likely in conjunction with the implementation of a particular mode of development (discussed in Duboule 1994). Interestingly, all invertebrates and chordates investigated to date, which contain a single “intact” Hox gene cluster, including sea urchin and amphioxus, display rather large intergenic distances, which can vary considerably between species (Cameron et al. 2006; Amemiya et al. 2008). In contrast, jawed vertebrates (gnathostomes), which contain multiple Hox gene clusters as a result of successive genome duplication events (see Dehal and Boore 2005), exhibit an arguably more compact organization, with highly conserved distances between orthologous Hox genes (Duboule 2007). Vertebrate genomes generally contain four Hox clusters (HoxA, HoxB, HoxC, and HoxD), except for ray-finned fishes, which have encountered additional genomic duplications leading to the presence of seven clusters in zebrafish (see Hoegg and Meyer 2005) and as many as 13 in salmon (Mungpakdee et al. 2008). In contrast to the situation in protostomes, repetitive elements are strongly excluded from Hox clusters in chordates, albeit with different degrees of stringency. While few stretches of repeats are still found in the large intergenic regions in the amphioxus Hox cluster, such sequences are virtually absent from the vertebrate counterparts (Fried et al. 2004; Amemiya et al. 2008), suggesting that the elusive structural or functional constraint prohibiting the invasion of vertebrate Hox clusters by repetitive elements predated the dramatic size reduction (consolidation) of these loci in vertebrates. The nature of the(se) underlying constraint(s) has been associated with the general way vertebrates develop, with the necessity for an exact timing in the activation of these genes, the genomic cluster acting as a “clock” (Duboule 1994; Ferrier and Holland 2002). In this view, the introduction of any foreign piece of DNA into this interval would be detrimental to the implementation of this critical process. On the other hand, modifications of these collinear mechanisms may have been a rich source of genetic innovations accompanying the variations observed in vertebrate body plans (discussed in Gaunt 2000). Within amniotes, squamates (lizards, snakes) display an amazing realm of morphologies, suggesting that important modifications in the structure and/or regulation of the Hox system may have occurred. However, no comprehensive genomic information was so far available for any member of this large group. The whole genome sequence release of the green anole lizard (Anolis carolinensis) allowed us to investigate the genomic organization of Hox clusters in this group of animals and hence to see if any substantial differences exist with respect to other amniotes sequenced so far.
We have analyzed and annotated the four Hox gene clusters in the green anole, and we hereby report a massive accumulation of interspersed repeats, comprising non-long-terminal-repeat (non-LTR) retrotransposons. Such an accumulation, not yet observed for any vertebrate species, largely accounts for the increased size of these loci, when compared to other vertebrates' Hox clusters sequenced to date. We also show that similar repeats are present in range of developmental gene-containing regions, which were previously identified as being refractory to the invasion of repetitive elements too (Simons et al. 2006, 2007). Because transposable elements are a major source of genetic modifications, including the emergence of novel genes, the alteration of gene expression, and the genesis of major genomic rearrangements, their successful invasion into Hox clusters may have offered an ideal substrate for the evolution of phenotypic novelties. Finally, sequence alignments between Anolis and other vertebrate Hox clusters highlight common divergent regions in human and mouse that may be associated with the evolution of particular traits characterizing mammals.
Results
Vertebrate Hox gene clusters
In order to better describe both the exact organization (e.g., in terms of gene number and sizes of intergenic regions) and the DNA content (in term of repetitive sequences) of vertebrate Hox gene clusters, we annotated newly available sequence data sets from various species, including mammals (mouse; Mus musculus), birds (chicken; Gallus gallus), amphibians (frog; Xenopus tropicalis), and reptiles (lizard; A. carolinensis). All four vertebrates showed a similar organization of Hox gene clusters, with comparable gene arrangement (Fig. 1), except for the Xenopus genome, which lacks two genes (Hoxb13 and Hoxd12). Furthermore, and in contrast to mammals, BLAST searches revealed the persistence of Hoxc3 in both the lizard and Xenopus HoxC clusters (Fig. 1C), similar to the situation observed in coelacanth, sharks, and some bony fishes (Hoegg and Meyer 2005).
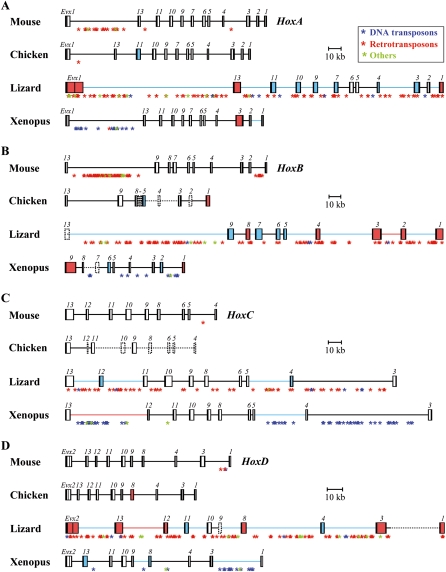
Figure 1.
Genomic organization of vertebrate Hox clusters. Schematic representation of: (A) HoxA, (B) HoxB, (C) HoxC, and (D) HoxD clusters in mouse, lizard, chicken, and Xenopus. The annotated relative sizes of predicted exons (black boxes), introns (white or colored boxes), and intergenic regions (thick lines) for both Hox and Evx genes allows for direct size comparisons between vertebrates. Gene names are shown above each box. Colors indicate either a 1.5-fold to threefold increase (blue), or a more than threefold increase (red), either in intronic (colored boxes) or intergenic (colored lines) distances. Gaps into the genomic sequences are indicated by dotted lines for intergenic regions and dotted boxes for coding regions. (C) Note the poor sequence coverage of the HoxC cluster in chicken and the presence of Hoxc3 in both lizard and Xenopus. The positions of interspersed repeats along the clusters are indicated by asterisks of different colors, reflecting the major classes of repetitive elements: (blue) DNA transposons; (red) LTR as well as non-LTR retrotransposons; (green) other conserved repeats.
A first survey of the annotated green anole Hox clusters immediately revealed an unexpected feature—while vertebrate Hox clusters are relatively homogeneous in size (the mouse, chicken, and Xenopus clusters are ∼100 kb large), all four lizard clusters were found substantially larger, from a factor of 1.5-fold (HoxA) to 2.5-fold (HoxD) (Fig. 1). When compared to other vertebrates, the Anole Hox genes are quite similar regarding the length of their protein-coding sequences, yet both intronic and intergenic distances are significantly larger in lizards. The relative increase in the length of these regions varies considerably within each cluster, with both the “anterior” (3′) and “posterior” (5′) extremities showing greater enlargement than more central parts of the gene clusters (Fig. 1), as previously noted for the related amphioxus locus (Amemiya et al. 2008). This is particularly prominent for the lizard HoxC cluster, which displays enlarged intergenic distances at both extremities of the cluster (the Hoxc13–Hoxc12 and the Hoxc5–Hoxc4 intergenic regions), as well as a long intergenic region of ∼80 kb between Hoxc4 and Hoxc3 (Fig. 1C). Interestingly, a similar structure was found for the Xenopus HoxC counterpart (Fig. 1C). In addition, some intronic sequences were also significantly enlarged in both lizard and Xenopus clusters, for example, within Hoxa3 and Hoxd13 (Fig. 1A,D).
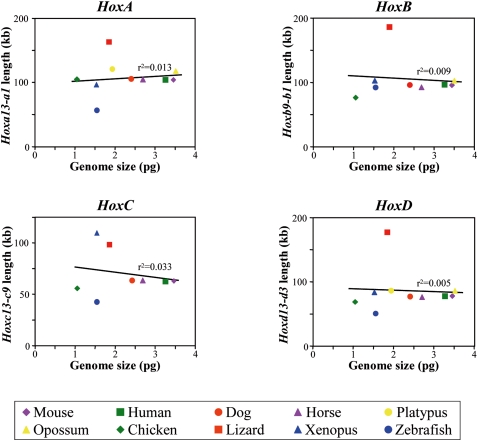
We examined the exact sizes of Hox gene clusters in other recently released vertebrate genomes, including those of the dog, horse, platypus, opossum, and zebrafish. As shown in Figure 2, lizard Hox gene clusters were found comparatively larger than those of all other vertebrate species investigated. In contrast to a previous study in which only a few species were considered (Santini et al. 2003), no correlation between vertebrate genome size and the extent of different Hox clusters was observed (HoxA: r = 0.11, P > 0.1; HoxB: r = 0.09, P > 0.1; HoxC: r = 0.18, P > 0.1; HoxD: r = 0.07, P > 0.1), even though such a relationship may have been anticipated from the case of birds (Fig. 2). We included in this size comparison some more distantly related vertebrates such as the coelacanth (Latimeria menadoensis) and the horn shark (Heterodontus francisci). Here again, despite the large genome size of the horn shark (Kim et al. 2000), the available genomic sequence covering parts of Hox gene clusters in these two species revealed a higher level of compaction, when compared to the lizard (Supplemental Fig. S1; Santini et al. 2003).
Figure 2.
Relationships between genome size and the length of Hox gene clusters in vertebrates. To compare different lineages of vertebrate in the same data set, only the lengths of the indicated regions of the clusters are shown. The very low squared correlation coefficients (r 2) indicate that the respective sizes of Hox clusters are not correlated with genome size (see text for associated P-values).
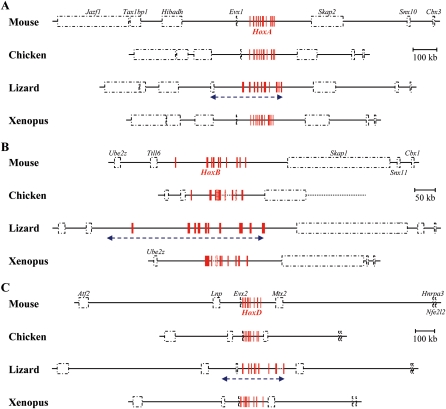
We assessed whether this size increase in the green anole was specific to Hox clusters or if it would affect the entire chromosomal neighborhood and analyzed large chromosomal regions surrounding Hox clusters, using genomic contigs assemblies available at the UCSC database. Interestingly, we found a significant positive correlation (P < 0.02) between genome size and the length of Hox cluster-flanking regions (as exemplified in Fig. 3) in diverse vertebrates (mouse, chicken, Xenopus, dog, horse, platypus, opossum, human) including lizard. The latter displayed flanking regions well in agreement with the size expectation, that is, within the 99% confidence interval inferred by the correlation. Altogether, the results indicate that the increase in intronic and intergenic distances observed in the lizard is clearly restricted to Hox clusters (and the associated Evx genes) (Figs. 1, 3), rather than being a general feature of these genomic regions in particular.
Figure 3.
Genomic organization of genes flanking the vertebrate Hox clusters. The correct relative size of the HoxA (A), HoxB (B), and HoxD (C) clusters (red boxes), as well as putative flanking genes (black dashed boxes) allow for direct size comparisons of large genomic regions between mouse, lizard, chicken and Xenopus. Gene names are shown either above or below each box for the mouse. (Arrows) The relative positions of the genomic regions that are increased in their sizes in the lizard genome. (Dotted lines) Gaps in the genomic sequences.
Distribution of interspersed repeats
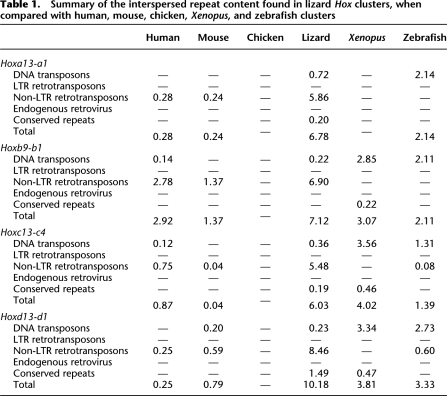
We next analyzed the Hox clusters for the presence of transposable and other repetitive elements. These elements represent a large fraction of all eukaryotic genomes, with a few exceptions, and they substantially contribute to the observed differences in genome size between various species (Feschotte and Pritham 2007). Sequence comparison between the entire human, mouse, chicken, lizard, Xenopus, and zebrafish Hox gene clusters revealed a frequency of interspersed repeats significantly higher in lizard than in other species (Table 1). The average content of interspersed repeats in lizard Hox clusters, as identified by the Censor software, was, indeed, 7.5%, excluding simple repeats. This is much higher than in human (1.1%), mouse (0.6%), chicken (none), Xenopus (2.7%), and zebrafish (2.2%). A comparable analysis of other available vertebrate genomes, including that of the opossum Monodelphis domestica, a genome that has been the target of heavy bombardment by transposable elements (Gentles et al. 2007), further confirmed that most vertebrate Hox gene clusters, in particular in mammals, are strongly refractory to invasion by repetitive elements (data not shown).
Table 1.
Summary of the interspersed repeat content found in lizard Hox clusters, when compared with human, mouse, chicken, Xenopus, and zebrafish clusters
Interestingly, within Hox gene clusters, both the distribution and nature of interspersed repeats differed greatly among vertebrate species. While the very few transposable elements found in the mouse and human Hox clusters predominantly include non-LTR retrotransposons, Xenopus and zebrafish Hox clusters mostly contain DNA transposons belonging to different families (Table 1). Surprisingly, only a few simple or conserved repeats were identified in lizard Hox gene clusters (Table 1), where the predominant type of interspersed repeat consists of two families of non-LTR retrotransposons, previously identified in reptile genomes: Penelope-like elements (PLEs) and Sauria short interspersed elements (Sauria SINEs). The accumulation of these two elements is largely responsible for the observed dot plots profile, showing multiple small units repeated along lizard Hox gene clusters (Supplemental Fig. S2).
In the corresponding murine loci, a high density of interspersed repeats is scored only within the unusually large intergenic region between Hoxb13 and Hoxb9, as well as outside the HoxA cluster, between Evx1 and Hoxa13 (Fig. 1A,B). The Xenopus Hox clusters contain significantly more transposable elements and conserved repeats (satellites) than mammals, in particular in some extended DNA regions like the Hoxc13–Hoxc12, Hoxc5–Hoxc4, Hoxc4–Hoxc3, and Hoxd3–Hoxd1 intergenic regions (Fig. 1C,D). Strikingly, while the chicken Hox clusters are apparently devoid of repetitive elements, lizard Hox clusters, which have the closest evolutionary distance to birds, massively accumulated retrotransposons in almost all intergenic regions, as well as in some intronic sequences (Fig. 1; Supplemental Fig. S2). Once the sequences related to interspersed repeats were deleted from the lizard Hox clusters, the average lengths of both the introns and intergenic regions were nevertheless still larger than in other vertebrate species, and dot plot profiles showed the persistence of small repeat units (data not shown). By using further nucleotide BLAST searches on the green anole genome, we confirmed the presence of new repetitive DNA elements along Hox clusters (data not shown), which are either not yet indexed in the Repbase library of vertebrate repeat sequences or too degenerated to be detected by Censor and RepeatMasker programs.
Regarding nucleotide content, the lizard Hox genomic regions shared some features with the amphibians and fish counterparts, with a prevalence of AT-rich regions. However, no significant correlation between GC content and the length of Hox gene clusters was identified among vertebrates, including lizards (data not shown). Altogether, we conclude that the Anole Hox gene clusters have accumulated interspersed repeats including non-LTR retrotransposable elements, and hence such DNA elements are not excluded from these specific genomic loci, as is the case in other vertebrates. This important difference largely accounts for the significant increase in the size of the lizard Hox gene clusters when compared to other vertebrates.
Comparison of development-related genes in vertebrates
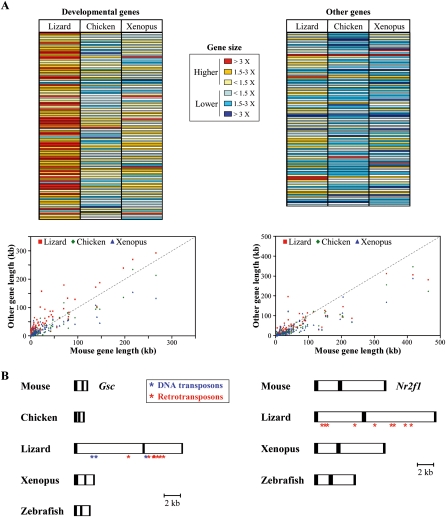
Besides Hox gene clusters, the longest vertebrate transposon-free regions identified to date are associated with development-related genes, usually encoding transcription factors such as members of the Pax, Fox, Sox, Six, and Tbx gene families (Simons et al. 2006, 2007). Noteworthy, these regions are well conserved among the vertebrate species used in our analysis (Simons et al. 2007). We thus assessed whether or not the observed tolerance for repeated elements in lizard Hox clusters is also observed within these regions, by comparing the distribution of gene sizes for more than a hundred development-related genes in mouse, lizard, chicken, and Xenopus. As controls, flanking genes with no known function during development were analyzed (Supplemental Table S1). The relative lengths of several development-related genes is significantly higher in lizards, when compared to mouse orthologous genes (Fig. 4A), with one-third of the genes being increased by at least threefold (Fig. 4A, left panels; average increase of threefold), whereas control genes display comparable average lengths in lizard and mammals (Fig. 4A, right panels).
Figure 4.
Distribution of gene sizes (lengths), either for genes with a known developmental-specific function, or without. (A, top panel) The relative lengths of developmental-specific genes and their non-developmental-specific flanking genes in lizard, chicken, and Xenopus are compared with their mouse counterparts and expressed as a ratio to mouse gene size. In order to compare exact orthologous genomic regions between vertebrates, the analysis includes both full-length and portions of genes. (Increasing red color intensity) An increasing gene size, as compared with the mouse size reference; (blue color intensity) a reduction in gene size. (Bottom panels) The distribution of developmental-specific (left) and nondevelopmental-specific (right) gene sizes between the mouse and other vertebrates. (B) Comparison of Gsc (left) and Nr2f1 (right) gene structures in some vertebrates. The correct relative sizes of predicted exons (black boxes) and introns (white boxes) allows for direct size comparisons between the various vertebrates. The locations of intronic transposons (blue asterisks) and non-LTR retrotransposons (red asterisks) within lizard genes are indicated.
Similar comparisons were made with the chicken and Xenopus genomes and revealed that the average gene length, within both “categories” of genes, is similar or, if anything, slightly decreased when compared to the situation in mice (Fig. 4A). Therefore, the lizard-specific expansion in the size of transcription units, observed in Hox genes clusters, also occurs at other genomic regions with known functions during development. We searched these regions for the presence of transposable—or any other repetitive—elements, which may have caused this general elongation, focusing on those genes recently identified in other vertebrate species to reside into evolutionarily conserved, transposon-free regions (Simons et al. 2006, 2007). Sequence comparisons of both the Gsc and Nr2f1 genes, between mouse, chicken, lizard, Xenopus, and zebrafish revealed the presence of interspersed repeats only in the lizard loci, in agreement with the observed increase in the size of these two genes in this species (Fig. 4B). Much like Hox gene clusters, the majority of these repeats included non-LTR retrotransposons, as well as some transposons. Their presence in these specific genomic regions of the lizard, whereas totally absent from the syntenic regions of all other vertebrate species analyzed to date, indicate that transposon-resistant genomic regions have been generally maintained in vertebrates, with the exception of lizard.
Sequence comparison within vertebrate Hox clusters
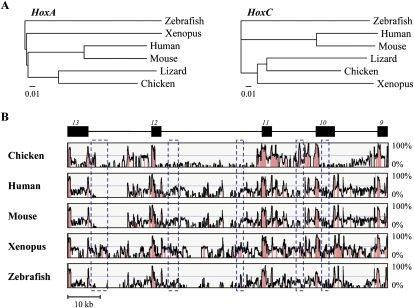
We looked at the phylogenetic relationships between these atypical lizard Hox clusters and those of other vertebrates, by considering each cluster separately, in mouse, human, chicken, lizard, Xenopus, and zebrafish. Phylogenetic trees were produced based on global genomic sequence alignments of annotated Hox loci, using both neighbor-joining and maximum likelihood methods. As expected, the alignments between HoxA, HoxB, or HoxD clusters generated phylogenetic trees well in agreement with the common view of vertebrate phylogeny. In marked contrast, however, alignments of the various HoxC clusters produced a clearly aberrant tree, where amphibians (Xenopus) were positioned as the sister group to birds (chicken) plus squamates (anole) (Fig. 5A).
Figure 5.
Sequence comparison of vertebrate HoxA and HoxC clusters. (A) Phylogenetic tree of some vertebrate HoxA (left) and HoxC (right) clusters, using the neighbor-joining method and the zebrafish Hox cluster as outgroup. To include the chicken sequence in this study, only the posterior part of the HoxC cluster (from Hoxc13 to Hoxc9) was used. Branch lengths are proportional to the number of nucleotide substitutions per site. (B) Global sequence alignment of lizard, chicken, human, mouse, Xenopus, and zebrafish HoxC clusters. Sequence comparisons were carried out using the indicated lizard HoxC cluster region (from Hoxc13 to Hoxc9, common to all vertebrates) as a reference sequence. Exons of the respective Hox genes are indicated above the black boxes. Nucleotidic homologies relative to the lizard sequence are given by histogram peaks. (Red peaks) Significant nucleotide homology. (Blue dashed boxes) Conserved elements between lizard and Xenopus (with >60% identity), yet not recognizable in mammals.
We compared the Anolis genomic sequences with orthologous regions in other species using methods of multiple global sequence alignments, including MLAGAN and TBA. The strongest regions of nucleotide homology were found in both exons of each Hox gene. However, additional evolutionarily conserved non-coding sequences were also detected, in all clusters, in both the intergenic and intronic regions, which are known to contain regulatory elements (Fig. 5B; Supplemental Fig. S3). Both the number and length of sequence matches expectedly decreased with increasing evolutionary distance, in the HoxA, HoxB, and HoxD clusters, and several regions showed high conservation between lizard, chicken, and mammals, whereas it was absent from Xenopus and zebrafish (Supplemental Fig. S3). However, consistent with the disturbed phylogenetic tree produced with the HoxC clusters, the alignment of this locus in vertebrates, from Hoxc13 to Hoxc9, identified several non-coding regions showing >60% identity between lizard and Xenopus, yet undetectable in mammals (Fig. 5B). While these sequences were most likely conserved in birds, their formal identification was made difficult owing to the poor sequence coverage (or wrong contig assembly) of the chicken HoxC cluster.
Remarkably, these conserved sequences were distributed within intergenic regions, all along the HoxC cluster, including at its two extremities, which were comparably larger in size between the lizard and Xenopus (see Fig. 1C; Supplemental Fig. S4). In addition, most of these motives were also well conserved between the lizard and the distantly related coelacanth, indicating that these sequences were probably lost from the mammalian genomes (Supplemental Fig. S4). Likewise, sequence conservation within exons of several HoxC genes was significantly higher between lizard and Xenopus than between lizard and mammals (data not shown), further suggesting that the mammalian HoxC cluster underwent divergent evolution. The aberrant phylogenetic tree generated by comparing the HoxC clusters thus likely derives from a wrong positioning of the mammalian counterpart, too deep in the phylogeny, rather than as a misplacement of the Xenopus HoxC cluster itself.
Discussion
Accumulation of retrotransposons in lizard Hox gene clusters
The annotation of lizard Hox gene clusters revealed the presence of four distinct loci, with 40 Hox genes, a situation comparable to other amniotes (39 and 38 genes in mammals and Xenopus, respectively). The general organization of the gene clusters (gene order and transcriptional orientation) were also conserved, as well as the presence of microRNAs in some intergenic regions, proposed to play important roles in regulating the expression of adjacent genes in other amniotes (Tanzer et al. 2005; data not shown). However, the overall sizes of the green anole Hox clusters were found systematically larger than those of any of the vertebrate counterparts sequenced to date (except for the Xenopus HoxC; see below). This increase in size is almost entirely due to the accumulation of non-LTR retrotransposons, that is, genetic elements usually excluded from these genomic loci.
In vertebrates, Hox gene clusters are not the only loci devoid of foreign genetic elements, and a robust correlation exists between this particular property, on the one hand, and the presence of transcription units of key importance for known developmental processes, on the other hand (Simons et al. 2007). We looked at some other genomic loci of this kind in the green anole genome and demonstrate that here again, in marked contrast to the situation observed in other amniotes, interspersed repeats are well tolerated. This observation indicates that transposon-free genomic regions have not been equally maintained in all vertebrate lineages, and hence it suggests that drastically different constraints must exist, even within amniotes, to either tolerate or exclude such genetic elements from particular loci. As previously suggested by whole vertebrate genome studies, the nature of the few interspersed repeats that invaded Hox clusters differed greatly both within and between vertebrate lineages.
The prominent types of repeats identified in lizard Hox clusters consist of two non-LTR retrotransposon families previously identified in reptiles: the PLEs and Sauria SINEs. PLEs are a widespread, yet poorly studied, class of transposable elements characterized by an endonuclease domain as well as an unusual but active reverse transcriptase domain with similarity to telomerases (Evgen'ev and Arkhipova 2005). Sauria SINEs are non-viral tRNA-derived repetitive sequences that are widespread in lizards and snakes (Piskurek et al. 2006). These elements contain RNA polymerase III–specific internal promoter sequences whose activity can be enhanced by upstream genomic sequences. Such elements are frequently used by host genomes to achieve important roles during organogenesis, including the formation of transcriptional boundary elements or the production of microRNAs (Belancio et al. 2008). In the case of the lizard Hox clusters, however, only some of these transposable elements could, in principle, exert an active function, as many of these are either truncated or heavily rearranged.
Impact on the body plans in Squamata?
Among vertebrates, two large groups of animals display unusually high adaptive capacities and concurrent major variations in their body plans: Teleostei and Squamata. In both cases, interestingly, the structure of the Hox gene clusters differs significantly from the prototypic situation described for birds, amphibians, and mammals. In the case of teleostean fishes, morphological flexibility was associated with the additional genome duplication(s), which would have provided novel opportunities to evolve highly adaptive traits (Meyer 1998), perhaps controlled by small and partial Hox subclusters coopted to achieve such functions (Duboule 2007). In the case of Squamata, we now report that the green anole Hox clusters are full of repeated sequences. While genomic sequences of others species of lizards and snakes are required to associate this property with this group of animal in general, the presence of such elements in lizard Hox clusters suggests that they may be a rich source of regulatory variations, concurrent with the morphological versatility of Squamates. The massive accumulation of interspersed repeats between Hox transcription units may have modified some regulatory properties, thus opening the possibilities for substantial variations in both their timing and places of transcription (Cohn and Tickle 1999). Transposable elements have been shown to influence transcriptional control mechanisms (Feschotte and Pritham 2007), as well as to regulate epigenetic modifications of heterochromatin when inserted either within or nearby. Their insertion into specific introns may disrupt regulatory elements identified there in several vertebrate Hox genes (Brown and Taylor 1994), and longer intronic sequences have been shown to considerably reduce the transcriptional elongation of the targeted genes (Castillo-Davis et al. 2002). Repeat insertions can also influence post-transcriptional events by perturbing splicing and/or RNA editing of coding genes (Gazave et al. 2007). Finally, the reverse transcriptase activity from retroelements may play a functional role in early embryo development by perturbing proliferation and differentiation programs (Sinibaldi-Vallebona et al. 2006). Altogether, the presence of so many repeats within the lizard Hox gene clusters makes it doubtful that the precise regulatory mechanisms described for Hox genes in other amniotes will be similarly implemented. For example, it was shown that the general transcriptional outcome of the HoxD cluster in developing digits is a function of the number of transcription units present there (Montavon et al. 2008), a situation that would be drastically affected by the integration of retrotransposons between the relevant genes.
An alternative (and not exclusive) view is that retrotransposons may be tolerated within Hox clusters in the green anole because of the disappearance of a major structural or regulatory constraint. For example, retrotransposons may increase internal recombinations, a process selected against in other vertebrate Hox gene clusters, owing to the requirement for coordinated regulation in cis. Lizards may have lost a component necessary for such illegitimate recombinations, and hence these sequences become tolerated since they no longer represent a danger for the structural integrity of the clusters. On the other hand, the release of a regulatory, rather than structural, constraint may have favored the accumulation of interspersed repeats, as was proposed for the case of the Hox and ParaHox genes in Ciona intestinalis (Ferrier and Holland 2002). In this explanatory framework, however, the fact that such elements would have been tolerated without impacting too much on local gene regulation is difficult to reconcile with genetic data obtained in mice, either when additional promoters were introduced into the cluster (Rijli et al. 1994; Herault et al. 1999) or when the respective distances between transcription units and their regulatory elements were modified (Tarchini and Duboule 2006). Consistently, whenever repeats are observed in mammalian Hox clusters, they lie at positions of minimal functional and regulatory impact. For example, retrotransposons are found between Hoxd1 and Hoxd3, in the mouse HoxD cluster, that is, between two genes showing unusually different regulations (Zakany et al. 2001). Also, repeats are found between Hoxb9 and Hoxb13, an exceptionally large region devoid of any Hox gene. In addition, sporadic retrotransposons are found in the largest intergenic regions of the HoxA and HoxC clusters. Interestingly, The Xenopus Hox clusters tend to better tolerate transposons (mostly DNA transposons) than their mammalian or avian counterparts. Here again, however, repeats are grouped either at the extremities of the clusters or within large intergenic regions. The presence of multiple DNA transposons at both extremities of the Xenopus HoxC cluster contributes to its exceptionally large size, larger, in fact, than the lizard counterpart. In addition, alignments of vertebrate HoxC cluster sequences identified particular coding and non-coding sequences that were not (or much less) conserved in mammals, suggesting that the mammalian HoxC cluster underwent divergent evolution. In this context, it is intriguing that some of the global functions attributed to HoxC cluster genes are associated with ear-marked mammalian features, such as hair follicles (Godwin and Capecchi 1998) or mammary glands development (Garcia-Gasca and Spyropoulos 2000), raising the possibility that this particular cluster played a prominent role in accompanying the emergence of mammals.
Methods
Annotation of Hox clusters
Genomic sequences from mouse Mus musculus (genome assembly 37), human Homo sapiens (assembly 36.1), dog Canis familiaris (assembly v.2.0), horse Equus caballus (assembly EquCab1), platypus Ornithorhynchus anatinus (assembly v.5.0.1), opossum Monodelphis domestica (assembly momDom4), chicken Gallus gallus (assembly v.2.1), frog Xenopus tropicalis (assembly v.4.1), zebrafish Danio rerio (assembly Zv.7), and lizard Anolis carolinensis (AnoCar1.0 assembly at 6.8× coverage) were extracted from the UCSC, VISTA, and Ensembl genome browsers. The annotations of the different Hox clusters as well as flanking genomic regions in chicken, Xenopus, and lizard were generated from sequence alignments of the orthologous regions with other vertebrate species available at the UCSC genome browser, using recent human and mouse assemblies as reference. Lizard Hoxc3 was identified by BLAST searches, using the coelacanth (Latimeria menadoensis) nucleotidic sequence of the HoxC cluster available at NCBI (AC151571, from Hoxc9 to Hoxc1), and putative coding regions were predicted using the GenScan program (http://genes.mit.edu/GENSCAN.html).
Owing to the poor sequence coverage of some Hox clusters in the chicken genome project, especially for HoxC, the recent genomic reconstruction and annotation was used as well (Richardson et al. 2007). For most vertebrate HoxC clusters, no flanking gene could be identified because of poor sequence coverage. To further validate exon boundaries and exon–intron organization of Hox clusters in non-mammalian species, the coding sequences of individual Hox genes were confirmed using nucleotide BLAST searches at NCBI and global sequence alignment at VISTA. Similar analysis procedures were performed to compare the size of all other developmental and non-developmental vertebrate genes in mouse, lizard, chicken, and Xenopus. Estimated vertebrate genome size values are based on genome sequence assemblies found at the Ensembl genome browser.
Identification of interspersed repeats
Interspersed repeats were identified and classified using both Censor (http://www.girinst.org/censor/index.php) and RepeatMasker (http://www.repeatmasker.org/) programs, which scan genomic sequences for regions of significant homology with the Repbase library of vertebrate repeat sequences (Kohany et al. 2006). For more accurate detection of transposable elements, Censor analysis was performed using nucleotide (BLASTN) and translated nucleotide (TBLASTN) sequences and default parameters. Fragments of repetitive elements were only selected if they exhibited at least 75% similarity over 75% of their lengths, and they were grouped according to major classes (DNA transposon, LTR or non-LTR retrotransposon, endogenous retrovirus and conserved repeat). Multiple copies of additional repetitive elements (length from 100 bp to 1 kb), probably not yet indexed in the Repbase library, were identified by nucleotide BLAST searches on the lizard genome at UCSC and Ensembl.
Multiple alignments and phylogenetic analysis
For multiple global alignments of genomic sequences, we used both MLAGAN (VISTA server: http://genome.lbl.gov/vista/index.shtml) and TBA programs (Mulan server: http://mulan.dcode.org/), which display different degrees of specificity and sensitivity (Margulies and Birney 2008). The default settings for the analysis visualization were a window of 100 bp and a minimum sequence identity of 50%. Phylogenetic analyses of nucleotide sequences common to all vertebrate species were performed based on global genomic sequence alignments using the neighbor-joining method implemented in VISTA and ClustalW with default parameters. To obtain more accurate estimates of the phylogeny, we also analyzed the data set by maximum likelihood using the PhyML algorithm with the GTR substitution model (Guindon and Gascuel 2003). Outputs were displayed using the TreeView application (Page 1996).
Acknowledgments
We thank the Broad Institute at MIT and Harvard for releasing lizard and horse genome data ahead of publication, and J. Woltering for sharing information. This work was supported by funds from the Canton de Genève, the Louis-Jeantet Foundation, the Claraz Foundation, the Swiss National Research Fund, the National Research Center (NCCR) “Frontiers in Genetics,” and the EU programs “Cells into Organs” and “Crescendo.”
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.087932.108.
References
- Amemiya C.T., Prohaska S.J., Hill-Force A., Cook A., Wasserscheid J., Ferrier D.E., Pascual-Anaya J., Garcia-Fernandez J., Dewar K., Stadler P.F. The amphioxus Hox cluster: Characterization, comparative genomics, and evolution. J. Exp. Zoolog. B Mol. Dev. Evol. 2008;310:465–477. doi: 10.1002/jez.b.21213. [DOI] [PubMed] [Google Scholar]
- Belancio V.P., Hedges D.J., Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Brown W.M., Taylor G.R. The 5′-sequence of the murine Hox-b3 (Hox-2.7) gene and its intron contain multiple transcription-regulatory elements. Int. J. Biochem. 1994;26:1403–1409. doi: 10.1016/0020-711x(94)90184-8. [DOI] [PubMed] [Google Scholar]
- Cameron R.A., Rowen L., Nesbitt R., Bloom S., Rast J.P., Berney K., Arenas-Mena C., Martinez P., Lucas S., Richardson P.M., et al. Unusual gene order and organization of the sea urchin hox cluster. J. Exp. Zoolog. B Mol. Dev. Evol. 2006;306:45–58. doi: 10.1002/jez.b.21070. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis C.I., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Cohn M.J., Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- Dehal P., Boore J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa R., Grenier J.K., Andreeva T., Cook C.E., Adoutte A., Akam M., Carroll S.B., Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Deschamps J., van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Di-Poï N., Zakany J., Duboule D. Distinct roles and regulations for Hoxd genes in metanephric kidney development. PLoS Genet. 2007;3:e232. doi: 10.1371/journal.pgen.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994;1994:135–142. [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Duboule D., Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen'ev M.B., Arkhipova I.R. Penelope-like elements—A new class of retroelements: Distribution, function and possible evolutionary significance. Cytogenet. Genome Res. 2005;110:510–521. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- Ferrier D.E., Holland P.W. Ciona intestinalis ParaHox genes: Evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 2002;24:412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- Feschotte C., Pritham E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried C., Prohaska S.J., Stadler P.F. Exclusion of repetitive DNA elements from gnathostome Hox clusters. J. Exp. Zoolog. B Mol. Dev. Evol. 2004;302:165–173. doi: 10.1002/jez.b.20007. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J. Hox, ParaHox, ProtoHox: Facts and guesses. Heredity. 2005;94:145–152. doi: 10.1038/sj.hdy.6800621. [DOI] [PubMed] [Google Scholar]
- Garcia-Gasca A., Spyropoulos D.D. Differential mammary morphogenesis along the anteroposterior axis in Hoxc6 gene targeted mice. Dev. Dyn. 2000;219:261–276. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1048>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gaunt S.J. Evolutionary shifts of vertebrate structures and Hox expression up and down the axial series of segments: A consideration of possible mechanisms. Int. J. Dev. Biol. 2000;44:109–117. [PubMed] [Google Scholar]
- Gazave E., Marques-Bonet T., Fernando O., Charlesworth B., Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Wakefield M.J., Kohany O., Gu W., Batzer M.A., Pollock D.D., Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin A.R., Capecchi M.R. Hoxc13 mutant mice lack external hair. Genes & Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A., Papalopulu N., Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Herault Y., Beckers J., Gerard M., Duboule D. Hox gene expression in limbs: Colinearity by opposite regulatory controls. Dev. Biol. 1999;208:157–165. doi: 10.1006/dbio.1998.9179. [DOI] [PubMed] [Google Scholar]
- Hoegg S., Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kim C.B., Amemiya C., Bailey W., Kawasaki K., Mezey J., Miller W., Minoshima S., Shimizu N., Wagner G., Ruddle F. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl. Acad. Sci. 2000;97:1655–1660. doi: 10.1073/pnas.030539697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M., Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Kohany O., Gentles A.J., Hankus L., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lemons D., McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Birney E. Approaches to comparative sequence analysis: Towards a functional view of vertebrate genomes. Nat. Rev. Genet. 2008;9:303–313. doi: 10.1038/nrg2185. [DOI] [PubMed] [Google Scholar]
- Meyer A. Hox gene variation and evolution. Nature. 1998;391:225–228. doi: 10.1038/34530. [DOI] [PubMed] [Google Scholar]
- Montavon T., Le Garrec J.F., Kerszberg M., Duboule D. Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes & Dev. 2008;22:346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungpakdee S., Seo H.C., Angotzi A.R., Dong X., Akalin A., Chourrout D. Differential evolution of the thirteen atlantic salmon Hox clusters. Mol. Biol. Evol. 2008;25:1333–1343. doi: 10.1093/molbev/msn097. [DOI] [PubMed] [Google Scholar]
- Page R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Piskurek O., Austin C.C., Okada N. Sauria SINEs: Novel short interspersed retroposable elements that are widespread in reptile genomes. J. Mol. Evol. 2006;62:630–644. doi: 10.1007/s00239-005-0201-5. [DOI] [PubMed] [Google Scholar]
- Richardson M.K., Crooijmans R.P., Groenen M.A. Sequencing and genomic annotation of the chicken (Gallus gallus) Hox clusters, and mapping of evolutionarily conserved regions. Cytogenet. Genome Res. 2007;117:110–119. doi: 10.1159/000103171. [DOI] [PubMed] [Google Scholar]
- Rijli F.M., Dolle P., Fraulob V., LeMeur M., Chambon P. Insertion of a targeting construct in a Hoxd-10 allele can influence the control of Hoxd-9 expression. Dev. Dyn. 1994;201:366–377. doi: 10.1002/aja.1002010408. [DOI] [PubMed] [Google Scholar]
- Santini S., Boore J.L., Meyer A. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 2003;13:1111–1122. doi: 10.1101/gr.700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C., Pheasant M., Makunin I.V., Mattick J.S. Transposon-free regions in mammalian genomes. Genome Res. 2006;16:164–172. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C., Makunin I.V., Pheasant M., Mattick J.S. Maintenance of transposon-free regions throughout vertebrate evolution. BMC Genomics. 2007;8:470. doi: 10.1186/1471-2164-8-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi-Vallebona P., Lavia P., Garaci E., Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosomes Cancer. 2006;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- Tanzer A., Amemiya C.T., Kim C.B., Stadler P.F. Evolution of microRNAs located within Hox gene clusters. J. Exp. Zoolog. B Mol. Dev. Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- Tarchini B., Duboule D. Control of Hoxd genes' collinearity during early limb development. Dev. Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Zakany J., Duboule D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zakany J., Kmita M., Alarcon P., de la Pompa J.L., Duboule D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell. 2001;106:207–217. doi: 10.1016/s0092-8674(01)00436-6. [DOI] [PubMed] [Google Scholar]