Abstract
The human salivary microbiome may play a role in diseases of the oral cavity and interact with microbiomes from other parts of the human body (in particular, the intestinal tract), but little is known about normal variation in the salivary microbiome. We analyzed 14,115 partial (∼500 bp) 16S ribosomal RNA (rRNA) sequences from saliva samples from 120 healthy individuals (10 individuals from each of 12 worldwide locations). These sequences could be assigned to 101 known bacterial genera, of which 39 were not previously reported from the human oral cavity; phylogenetic analysis suggests that an additional 64 unknown genera are present. There is high diversity in the salivary microbiome within and between individuals, but little geographic structure. Overall, ∼13.5% of the total variance in the composition of genera is due to differences among individuals, which is remarkably similar to the fraction of the total variance in neutral genetic markers that can be attributed to differences among human populations. Investigation of some environmental variables revealed a significant association between the genetic distances among locations and the distance of each location from the equator. Further characterization of the enormous diversity revealed here in the human salivary microbiome will aid in elucidating the role it plays in human health and disease, and in the identification of potentially informative species for studies of human population history.
Molecular analyses have revealed enormous variation in the microbiome of diverse environments, including the human body (Eckburg et al. 2005; Dethlefsen et al. 2007; Grice et al. 2008; Oakley et al. 2008). These studies have revealed a wealth of not-yet-cultivated bacterial species, and several projects to study the human microbiome have been proposed and/or initiated (Turnbaugh et al. 2007; Mullard 2008). However, comparatively little attention has been paid to the human salivary microbiome, as most studies of the human oral cavity have focused on identifying bacteria that might be associated with diseases (Becker et al. 2002; Kumar et al. 2003; Diaz et al. 2006; Kilian et al. 2006; Machado de Oliveira et al. 2007; Faveri et al. 2008). More than 600 different species have been described from the human oral cavity (Fabian et al. 2008), but only limited information is available on the normal microflora of healthy individuals (Aas et al. 2005; Kang et al. 2006). A comprehensive understanding of the breadth of diversity in the human oral cavity microbiome is necessary to fully gauge the role of bacteria in diseases of the oral cavity (Aas et al. 2005; Paster et al. 2006; Fabian et al. 2008). Moreover, as the oral cavity is often the entry point of bacteria into the body, there may be important interactions between the saliva microbiome and other microbiomes in the human body, in particular, that of the intestinal tract (Schipper et al. 2007; Fabian et al. 2008).
A further reason for analyzing the human salivary microbiome is that since saliva is increasingly preferred in sampling humans as a source of DNA for epidemiologic and population genetic studies (Quinque et al. 2006; Hansen et al. 2007), it would be useful to identify bacterial taxa in saliva that may be able to provide insights into human population structure and migrations. When humans migrate, they take their bacteria with them, and it has previously been shown that patterns of variation in the stomach bacteria Helicobacter pylori correspond closely to migrations inferred from analyses of human DNA variation (Falush et al. 2003; Linz et al. 2007). Moreover, it was shown that two human ethnic groups, which could not be distinguished on the basis of human DNA markers, could be distinguished based on their patterns of H. pylori variation (Wirth et al. 2004). This result could reflect either genetic isolation of these two communities that is too recent to be reflected in the human DNA markers, or some cultural factor that influences H. pylori variation (such as diet) that differs between these communities; in either event, the H. pylori variation does provide insights into human populations that are not revealed by analysis of human DNA markers.
However, sampling H. pylori requires stomach biopsy material, so there would be obvious advantages if similarly informative bacterial species could be identified in saliva. The most informative bacterial species for this purpose should exhibit high levels of vertical versus horizontal transmission. Although this information remains largely unknown for bacteria in the oral cavity, there is some preliminary evidence from mother–child (Li et al. 2007) and twin (Corby et al. 2007) studies to suggest a significant role for vertical transmission. And even bacterial species that exhibit high levels of horizontal transfer may nonetheless provide evidence of recent contact between populations that would not be revealed by human DNA markers. Characterization of patterns of diversity in the human salivary microbiome thus may provide useful insights into human population structure and migration.
Here, we present the first description of patterns of global diversity in any human microbiome, based on analyses of partial 16S ribosomal RNA (rRNA) sequences from 120 individuals from diverse locations around the world. Our results provide a framework for further investigations of the ecological and cultural factors that may influence the composition of the human salivary microbiome and identify promising taxa for further characterization that may provide novel insights into human population structure, migration, and contact.
Results
Saliva samples were obtained from 120 individuals, consisting of 10 individuals from each of two locations from each of six geographic regions (Fig. 1). A highly variable portion of the 16S rRNA gene of ∼500 bp was amplified and cloned, and approximately 120 clones were sequenced from each individual; this number of clones is sufficient to capture most of the variation at the level of bacterial genera inferred from the partial 16S rRNA sequences (Supplemental Fig. S1). A total of 14,691 sequences were obtained, which were then analyzed for possible chimeras or other artifacts (Ashelford et al. 2006). We identified 576 potentially chimeric sequences, which represents ∼3.9% of the sequences. This is less than the average of 9% potentially chimeric sequences reported previously for full-length 16S rRNA clone libraries (Ashelford et al. 2006), probably because our partial 16S rRNA sequences provide a smaller target for jumping PCR or other processes that produce chimeric sequences. Excluding these 576 potentially chimeric sequences leaves 14,115 sequences for further analysis (Table 1); these sequences have been deposited in GenBank under accession numbers EU984515–EU998629. The sequences were analyzed in two ways: First, a genus was assigned to each sequence by comparison to the RDPII database (Cole et al. 2007), and the composition of genera across individuals was subsequently analyzed. Second, all sequences were used to generate a phylogenetic tree, which was subsequently analyzed.
Figure 1.
Map of the sampling locations. Ten saliva samples were obtained from each of the 12 sampling locations, for a total of 120 saliva samples analyzed.
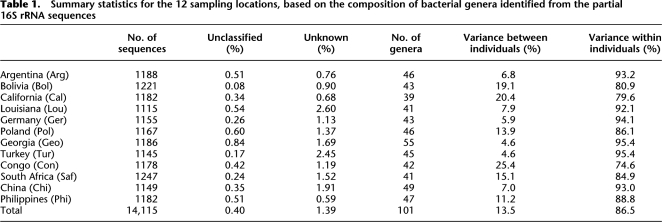
Table 1.
Summary statistics for the 12 sampling locations, based on the composition of bacterial genera identified from the partial 16S rRNA sequences
The distribution of bacterial genera assigned to the sequences from each individual (Supplemental Tables S1–S12) varied greatly. Overall, 196 sequences (1.4%) did not match any sequence in RDPII above the cutoff value of 90% similarity, while 57 sequences (0.4%) matched a sequence in RDPII for which the genus was unclassified. To estimate the number of genera that might be present among these unknown sequences, a phylogenetic tree was constructed (Supplemental Fig. S2). There are 64 clusters of sequences in this tree at the cutoff value of 90% similarity, indicating that there are an estimated 64 new genera among these sequences.
The number of genera identified in each individual ranged from six to 30 (Supplemental Tables S1–S12), while the number of genera per location ranged from 39 to 55 (Table 1), which is comparable to previous studies of the oral cavity microbiome (Aas et al. 2005; Paster et al. 2006). In total, 101 different genera were identified, 39 of which have not been previously reported from the human oral cavity (Supplemental Table S13). In combination with the unknown genera, we have therefore identified an estimated 103 previously undescribed bacterial genera from human saliva.
A few genera are quite frequent, while most are rare (Supplemental Fig. S3); 16 genera were observed only once, while the most frequent genus (Streptococcus) accounted for 22.7% of the sequences (Supplemental Table S13). In general, we identified 85%–95% of the genera that have previously been found in molecular surveys of the human oral cavity (Kumar et al. 2003; Aas et al. 2005; Diaz et al. 2006; Kang et al. 2006; Paster et al. 2006); the previously reported genera not found in this study include Eikenella, Lautropia, Synergistes, and Bacteroides.
The relative abundance of each genus in each individual appears qualitatively to be fairly uniform (Fig. 2). To quantitate differences in the distribution of genera, we apportioned the total variance within each location into between-individual and within-individual components (Table 1). California and the Congo showed the biggest differences between individuals, while Georgia and Turkey showed the smallest differences between individuals. Overall, ∼13.5% of the variation in the distribution of genera can be attributed to differences between individuals (i.e., any one individual has ∼86.5% of the variation that is observed when all locations are sampled).
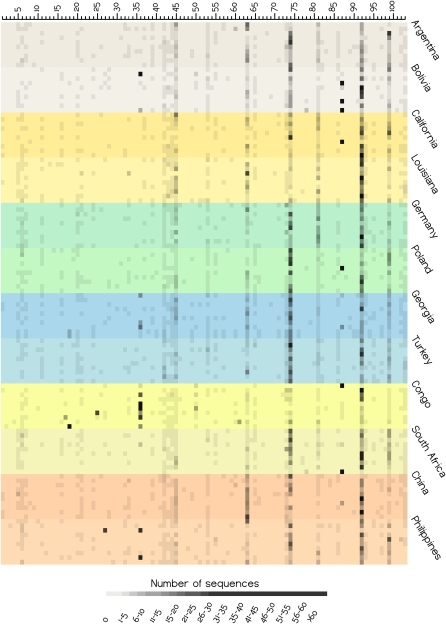
Figure 2.
Heat plot of the abundance of each bacterial genus in each individual, based on the partial 16S rRNA sequences. Each horizontal row corresponds to an individual saliva sample, with different colored shadings indicating the 10 individuals from each sampling location, and sampling locations from the same broad geographic region indicated by similar colors. Each column is a genus, with the numbers corresponding to the genus, as in Supplemental Table S13. The abundance of each genus (columns) is indicated by the grayscale value, according to the scale at the bottom of the plot.
The between-individual component of the total variation can be further subdivided into the amount of variation among individuals from the same location, and the amount of variation among individuals from different locations. The former estimate is 11.7%, and the latter is just 1.8%. Although both estimates are significantly different from zero (P < 0.001) based on a permutation analysis (Excoffier et al. 2005), this analysis indicates that differences in the composition of genera are larger among individuals from the same location than among individuals from different locations, and thus that differences in the composition of genera among individuals are not structured by geography.
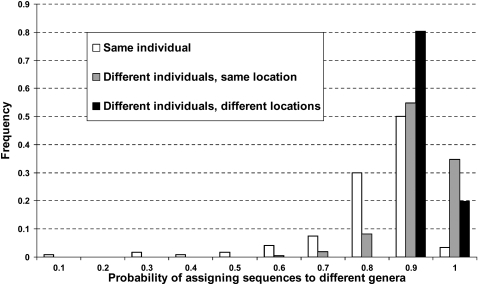
Another way to examine this question is to determine the probability that two sequences chosen at random will come from different genera, when the sequences are chosen from the same individual, from different individuals from the same location, or from different individuals from different locations (Fig. 3). The average probability that two sequences are from different genera when chosen at random from the same individual is 0.772, while for two sequences chosen from two different individuals from the same location, the corresponding probability is 0.875, which is significantly higher (Mann-Whitney test, P < 0.001). However, the average probability that two sequences are from different genera when chosen from two individuals from different locations is 0.892, which is not significantly different from the above probability of 0.875 for two sequences chosen from two individuals from the same location (Mann-Whitney test, P > 0.05). Thus, there is significantly more variation among sequences from different individuals than among sequences from the same individual, but there is not significantly more variation among individuals from different locations than among individuals from the same location.
Figure 3.
Frequency distribution of the probability that two sequences will be assigned to different genera when drawn from the same individual (values are for each of the 120 individuals); different individuals from the same location (values are for all pairwise comparisons of the 10 individuals from the same location, for all 12 locations); and different individuals from different locations (values are for all pairwise comparisons of the 12 locations).
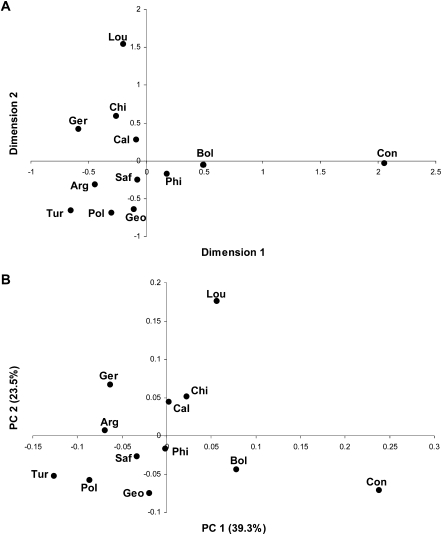
The genetic structure of human populations is strongly influenced by geography (Ramachandran et al. 2005; Li et al. 2008). However, a multidimensional-scaling (MDS) plot based on the differences in the composition of genera among locations does not indicate any strong influence of geography (Fig. 4A). The first dimension separates the Congo from the other locations, while the second dimension separates Louisiana from the other locations, and overall there is no grouping by geography. In agreement with the MDS plot, the distances among locations based on the composition of genera (Supplemental Table S14) are not correlated with the geographic distances among locations (Mantel test, r = −0.07, P = 0.65). Moreover, an MDS plot based on differences in the composition of genera among individuals, rather than locations, also does not reveal any geographic clustering (Supplemental Fig. S4).
Figure 4.
Graphical depictions of the relationships among sampling locations based on the partial 16S rRNA sequences. (A) MDS plot, based on linearized FST distances (Slatkin 1995) between each pair of locations, calculated from the genus composition. The location abbreviations are as in Table 1. The stress value is 0.03. (B) PCA plot, based on the UniFrac distances (Lozupone and Knight 2005); same abbreviations as in A. The percentage of the total variance explained by each PC is indicated in parentheses.
The above analyses are all based on classifying the genus of each sequence; to assess independently the geographic structure in the data without first classifying the sequences, we also carried out a UniFrac analysis (Lozupone and Knight 2005). We constructed a phylogenetic tree for the entire set of 14,115 sequences, which was then used to estimate the UniFrac distance (the fraction of the tree that is not shared in common) between each pair of locations. The PCA plot of the UniFrac distances (Fig. 4B) is quite similar to the MDS plot for the distances based on genus composition (Fig. 4A), and the UniFrac distances (Supplemental Table S14) are similarly not correlated with the geographic distances among locations (Mantel test, r = −0.14, P = 0.79). In agreement with the close similarity between the plots (Fig. 4), the UniFrac distances are highly and significantly correlated with the genus composition distances (Mantel test, r = 0.94, P < 0.001). Thus, the lack of any significant geographic structure to the human saliva microbiome is not a consequence of the procedure used to assign the genus of each sequence, but is an inherent property of the sequences retrieved from the saliva samples.
Although overall there is no significant geographic patterning with respect to the human salivary microbiome, specific genera do vary significantly in frequency among locations (Fig. 2; Supplemental Table S13). For example, the most extreme variation in frequency across locations is exhibited by Enterobacter, which accounts for 28% of the sequences obtained from the Congo (Supplemental Table S13) but was completely absent from California, China, Germany, Poland, and Turkey. Several other genera were observed at higher frequency in the Congo than elsewhere (Fig. 2; Supplemental Table S13), reinforcing the uniqueness of the salivary microbiome from the Congo (Fig. 4; Table 1). Another genus that occurs at relatively high frequency in several individuals from one particular location is Serratia (Bolivia). However, several genera that vary significantly in frequency (Supplemental Table S13) occur at high frequency only in one individual from a particular location (Fig. 2); more intensive sampling would be required to determine if these genera are truly characteristic of these particular locations.
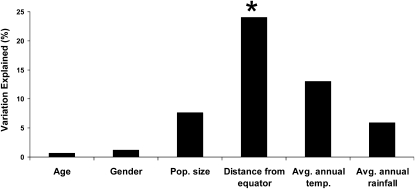
A prime question remains the identification of environmental and cultural factors that influence the human salivary microbiome. As a first step in this direction, we carried out a multivariate regression analysis (Anderson 2001, 2004) of associations between four factors (population size, distance from the equator, average annual temperature, and average annual rainfall) (Supplemental Table S15) and the UniFrac distances among the 12 locations. We also tested for associations between age and gender of the donor and the FST distances based on the composition of bacterial genera for each donor. The only significant association involved the distance of each location from the equator, which explained ∼24% of the variation in the UniFrac distances (Fig. 5). However, this apparently significant association should be viewed with caution. Many factors vary with distance from the equator (such as UV index), so further work will be required to ascertain what aspect(s) of the distance from the equator is responsible for this association. Moreover, the association is primarily driven by the Congo, which has high distances to other locations and is also closest to the equator; investigation of additional populations that are similarly close to the equator would be necessary to verify this association.
Figure 5.
Amount of variation explained (in percent) in the FST distances among individuals (for the age and gender of the donors), and in the UniFrac distances among locations (for the variables population size, distance from the equator, average annual temperature, and average annual rainfall) (Supplemental Table S15). There is a significant association between the UniFrac distances and the variable “Distance from Equator” (P = 0.006).
Discussion
The analyses of the human salivary microbiome presented here provide the first detailed investigation of global patterns of diversity in any human microbiome. There is high diversity in the human salivary microbiome; assignment of the partial 16S rRNA sequences generated in this study to bacterial genera via comparison to the RDPII database identified a total of 101 bacterial genera, including 39 that had not been previously described from the human oral cavity. Moreover, we estimate that an additional 64 unknown genera are represented in the sequences. Understanding the role of these known and unknown genera in the health of the human oral cavity and how they interact with the microbiomes of other parts of the human body (in particular, the intestinal tract) would be of considerable interest.
A potential criticism of this study is that there are many environmental and cultural factors (in particular, diet and hygiene) that were not controlled for during the sampling. Indeed, a limited investigation of some environmental factors revealed a significant association between the genetic distances among locations and the distance of the location from the equator (Fig. 5). However, we emphasize that a main conclusion of our study (discussed below) is that the human salivary microbiome does not vary greatly between different geographic locations. Therefore, if these uncontrolled environmental and/or cultural factors are having a significant impact on our results, then controlling for them would presumably result in even less geographic differentiation in the patterns of variation. That is, if (for example) differences in diet between locations are responsible for differences in the salivary microbiome composition, then controlling for diet would further decrease the already small differences between locations.
Another potential weakness is that possible temporal change in the salivary microbiome was not studied. In this respect, the sampling in this study should be regarded as “environmental,” that is, a single snapshot in time of the salivary microbiome diversity of an individual was captured, much as sampling the microbiome of hot springs or other environmental locations often use a single time point. To be sure, it would be of considerable interest to determine the extent of change in an individual's salivary microbiome over time and how it might be influenced by other factors. For example, what happens to the salivary microbiome if individuals change their diet? And what happens if an individual moves to a new location? Although these questions were not addressed in this study, the results of this study do provide a framework for further studies designed to answer such questions about the temporal stability of the human salivary microbiome.
This high diversity in the human salivary microbiome does not appear to be geographically structured (Fig. 2). While there is significantly more diversity in bacterial genera compared from different individuals than from the same individual, the diversity among individuals from the same location is nearly the same as the diversity among individuals from different locations (Fig. 3). Overall, the between-population component of the variance in the bacterial genera composition is ∼13.5% (Table 1), which means that any one individual contains on average 86.5% of the total variance observed when all individuals are sampled. This is remarkably similar to the amount of between-population variation typically observed among human populations for neutral genetic markers (Romualdi et al. 2002; Li et al. 2008). Thus, in this sense, the distribution of genera among individuals is behaving as a neutral genetic marker, if one equates the bacterial composition of individuals to the genetic composition of human populations.
It should be emphasized that this conclusion of an overall lack of geographic structure extends only to the pool of 16S rRNA sequences and the bacterial genera identified from them. Sequence variation within particular bacterial taxa may very well exhibit geographic structure that would provide novel insights into human population structure, relationships, and migrations, as has been previously observed for H. pylori (Falush et al. 2003; Linz et al. 2007). Multi-locus sequence typing (MLST) is a particularly promising approach (Urwin and Maiden 2003; Maiden 2006) that has shown geographic structure in some bacteria that was not evident in 16S rRNA comparisons (Margos et al. 2008). Additional characterization of coding sequence variation in particular taxa by MLST or other means would be highly desirable, and may very well reveal geographic structure that is not apparent in the overall composition of the salivary microbiome. For tracing human migrations, it would be most useful to focus on bacteria that are likely to be transferred primarily vertically (i.e., within families or households). However, even bacteria that are primarily transferred horizontally may provide useful markers of population contact that is too recent to be detected by analysis of human DNA polymorphisms. Perhaps the most useful taxa to start with would be those that are both widespread (i.e., found in many different locations) and frequent (i.e., found in multiple individuals from a location); these would include Streptococcus, Prevotella, Veillonella, Neisseria, Haemophilus, Rothia, Porphyromonas, and Fusobacterium (Fig. 2). Together, these eight genera account for >70% of the total number of sequences (Supplemental Table S13).
In conclusion, our results provide a framework for further studies of human microbiome diversity and for investigations of the environmental, dietary, and genetic factors that influence an individual's salivary microbiome. These, in turn, will lead to a better understanding of the role of the salivary microbiome in diseases of the oral cavity and how the salivary microbiome interacts with the intestinal and other microflora of the human body. In addition, further characterization of some of the taxa identified in this study by MLST or other means should provide useful markers for studies of human population history and/or recent population contact.
Methods
Samples and DNA extraction
Saliva samples from 10 individuals from each of 12 locations (two locations from each of six geographic regions) (Fig. 1) were collected with informed consent. The criteria for selection of individuals were that individuals should not be from the same household, and they should not have been away from the sampled location within the past two months. Although dental health was not examined in detail, all individuals were in good health, and no individual was suffering from any obvious dental disease at the time of sampling.
Two milliliters of saliva was collected from each individual into a tube containing an equal volume of lysis buffer (Quinque et al. 2006). Samples were stored at ambient temperature and transported to the laboratory (MPI-EVA, Leipzig, Germany), and DNA was extracted as described previously (Quinque et al. 2006).
PCR amplification of the microbial 16S rRNA gene
We designed primers to amplify an informative segment of the microbial 16S rRNA gene that would be of optimal size for cloning and DNA sequence analysis. We downloaded and aligned ∼100,000 complete microbial 16S rRNA sequences from the Ribosomal Database Project II (RDPII) database (Cole et al. 2007) and searched for highly conserved segments that flank highly variable segments of the gene. The gene fragment we amplified corresponds to nucleotide positions 2,769,690 to 2,770,259 in the Escherichia coli genome (Blattner et al. 1997), with an amplicon length of 570 bp. The primer sequences used in this study are: forward primer, 5′-CTACGTGCCAGCAGCCGCGG-3′; and reverse primer, 5′-CTCACGACACGAGCTGACGA-3′. PCR amplification was carried out in a 50-μL total volume, including 38 μL of ddH2O, 5 μL of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl at pH 9.0, 1.0% Triton X-100), 3 μL of 25 mM MgCl2, 1 μL of 10 mM dNTPs (10 mM each dATP, dTTP, dGTP, dCTP), 1 μL of 20 μM forward primer, 1 μL of 20 μM reverse primer, 0.5 μL of Taq polymerase (Applied Biosystems), and 20 ng of template DNA, for 35 cycles using the following conditions: denaturation step for 1 min at 95°C, annealing step for 1 min at 62°C, and extension step for 1 min at 72°C.
Cloning and sequencing of amplicons
Amplicons were cloned using the TOPO TA cloning kit (Invitrogen), following the protocol recommended by the supplier. Colony PCR was carried out using the M13 forward and reverse primers supplied with the cloning kit in a 50-μL total volume that included the same reagents described above, to which a small amount of cells were added with a sterile pipette tip, and the following thermocycling conditions: an initial incubation for 5 min at 95°C to lyse cells and denature the DNA, followed by 30 cycles of 1 min at 95°C, 1.5 min at 54°C, and 1 min at 72°C, followed by a final incubation for 5 min at 72°C. The forward stands of colony PCR products with appropriate inserts were sequenced using the 16S rRNA PCR primers and the Big Dye DNA Sequencing Kit (Perkin-Elmer), following the protocol recommended by the supplier, and an ABI 3730 automated DNA sequencer. Sequence quality was assessed via the software package DNASTAR (DNASTAR Inc.).
Data analysis
Chimeric or other potentially artifactual sequences were identified with the Mallard program (Ashelford et al. 2006), using an iterative procedure and a quantile value of 95% as the cutoff line for determining outliers. This procedure identified 576 potentially chimeric sequences, which were excluded from further analyses, leaving 14,115 sequences.
These 14,115 sequences were then searched against the RDPII database (Cole et al. 2007), using the online program SeqMatch (Wang et al. 2007) and a threshold setting for the similarity score of 90%, to assign a genus to each sequence. The appropriate similarity score for defining bacterial genera is a matter of debate, generally ranging from 90% to 95%; we chose a value at the low end of the range, in accordance with some previous studies (Pei et al. 2004; Gao et al. 2007), in order to minimize falsely designating new taxa (at the possible expense of mis-assigning some taxa). In the event of multiple matches to sequences in the database, the genus was assigned according to the highest similarity score. Comparing the sequences in a similar fashion to GenBank resulted in a highly similar assignment of genera, except that more sequences were classified as unknown (data not shown). Supplemental Tables S1–S12 give the frequency of each genus for each individual, as well as the frequency of unclassified sequences (those that matched a sequence in RDPII, but the genus of the matching sequence has not been determined) and unknown sequences (those that did not match any sequence in RDPII above the 90% threshold). The apportionment of variation based on the frequency distribution of genera within and between individuals and locations, and linearized FST distances (Slatkin 1995) were calculated with Arlequin 3.1 (Excoffier et al. 2005); for these analyses, each genus was considered equivalent to an allele at a single haploid locus. Multidimensional scaling (MDS) analysis was carried out using STATISTICA 7.1 (StatSoft, Inc.).
We also compared the differences among sequences from different individuals and locations, without first assigning sequences to genera, using the UniFrac method (Lozupone and Knight 2005; Lozupone et al. 2006). To implement this method, we first carried out a multiple alignment of all of the sequences using ClustalW2 (Larkin et al. 2007). A matrix of the sequence differences between each pair of sequences was then determined using the F84 model (Kishino and Hasegawa 1989) as implemented in the DNADIST program in PHYLIP 3.67 (Felsenstein 1989). A UPGMA tree was then constructed from the pairwise distance matrix, using the NEIGHBOR program in PHYLIP, and the UniFrac distance metric (Lozupone et al. 2006) was calculated between each pair of locations with the online resource (http://bmf2.colorado.edu/unifrac/). These distances were used as input for principal coordinates analysis; a neighbor-joining tree resulted in UniFrac distances that were nearly identical to the distances based on the UPGMA tree (Mantel test, r = 0.97, P < 0.001), and the PC plot based on the neighbor-joining distances was virtually identical to that based on UPGMA distances (data not shown). Multivariate regression analysis (Anderson 2001; McArdle and Anderson 2001) for associations between the genetic distance matrices and potential predictor variables of interest was carried out with the DISTLM program (Anderson 2004); this method treats the entire distance matrix as the response variable and uses a permutation method to test for associations with potential explanatory variables, in a regression analysis framework.
Acknowledgments
We thank all individuals who kindly donated a sample for this study; Rebecca Atencia, Jarek Bryk, Heather Buckner, Bryan Buckner, Daniel Corach, Anne Fischer, Michel Halbwax, Janet Kelso, Marina Nagveradze, Samra Sardas, Beth Trachtenberg, and Guido Valverde for assistance with sample collections; Susanne Gutsmuths and Anna Schönberg for technical assistance; and Micah Hamady for assistance with the UniFrac distance calculations. Funding was provided by the Max Planck Society.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. EU984515–EU998629.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084616.108.
References
- Aas J., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Anderson M.J. DISTLM v.5: A FORTRAN computer program to calculate a distance-based multivariate analysis for a linear model. Department of Statistics, University of Auckland; New Zealand: 2004. [Google Scholar]
- Ashelford K.E., Chuzhanova N.A., Fry J.C., Jones A.J., Weightman A.J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.R., Paster B.J., Leys E.J., Moeschberger M.L., Kenyon S.G., Galvin J.L., Boches S.K., Dewhirst F.E., Griffen A.L. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cole J.R., Chai B., Farris R.J., Wang Q., Kulam-Syed-Mohideen A.S., McGarrell D.M., Bandela A.M., Cardenas E., Garrity G.M., Tiedje J.M. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby P.M., Bretz W.A., Hart T.C., Schork N.J., Wessel J., Lyons-Weiler J., Paster B.J. Heritability of oral microbial species in caries-active and caries-free twins. Twin Res. Hum. Genet. 2007;10:821–828. doi: 10.1375/twin.10.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., McFall-Ngai M., Relman D.A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P.I., Chalmers N.I., Rickard A.H., Kong C., Milburn C.L., Palmer R.J., Kolenbrander P.E. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fabian T.K., Fejerdy P., Csermely P. Salivary genomics, transcriptomics, and proteomics: The emerging concept of the oral ecosystem and their use in the early diagnosis of cancer and other diseases. Cur. Genomics. 2008;9:11–21. doi: 10.2174/138920208783884900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Wirth T., Linz B., Pritchard J.K., Stephens M., Kidd M., Blaser M.J., Graham D.Y., Vacher S., Perez-Perez G.I., et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Faveri M., Mayer M.P.A., Feres M., de Figueiredo L.C., Dewhirst F.E., Paster B.J. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 2008;23:112–118. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Gao Z., Tseng C., Pei Z., Blaser M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Kong H.H., Renaud G., Young A.C., Bouffard G.G., Blakesley R.W., Wolfsberg T.G., Turner M.L., Segre J.A. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.V., Simonsen M.K., Nielsen F.C., Hundrup Y.A. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: Comparison of the response rate and quality of genomic DNA. Cancer Epidemiol. Biomarkers Prev. 2007;16:2072–2076. doi: 10.1158/1055-9965.EPI-07-0611. [DOI] [PubMed] [Google Scholar]
- Kang J.G., Kim S.H., Ahn T.Y. Bacterial diversity in the human saliva from different ages. J. Microbiol. 2006;44:572–576. [PubMed] [Google Scholar]
- Kilian M., Frandsen E.V.G., Haubek H., Poulsen K. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000. 2006;42:158–179. doi: 10.1111/j.1600-0757.2006.00159.x. [DOI] [PubMed] [Google Scholar]
- Kishino H., Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kumar P.S., Griffen A.L., Barton J.A., Paster B.J., Moeschberger M.L., Leys E.J. New bacterial species associated with chronic periodontitis. J. Dent. Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li Y., Ismail A.I., Ge Y., Tellez M., Sohn W. Similarity of bacterial populations in saliva from African-American mother–child dyads. J. Clin. Microbiol. 2007;45:3082–3085. doi: 10.1128/JCM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Linz B., Balloux F., Moodley Y., Manica A., Liu H., Roumagnac P., Falush D., Stamer C., Prugnolle F., van der Merwe S.W., et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Hamady M., Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado de Oliveira J.C., Siqueira J.F., Rocas I.N., Baumgartner J.C., Xia T., Peixoto R.S., Rosado A.S. Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol. Immunol. 2007;22:14–18. doi: 10.1111/j.1399-302X.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- Maiden M.C.J. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- Margos G., Gatewood A.G., Aanensen D.M., Hanincova K., Terekhova D., Vollmer S.A., Cornet M., Piesman J., Donaqhy M., Bormane A., et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. 2008;105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle B.H., Anderson M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Mullard A. Microbiology: The inside story. Nature. 2008;453:578–580. doi: 10.1038/453578a. [DOI] [PubMed] [Google Scholar]
- Oakley B.B., Fiedler T.L., Marrazzo J.M., Fredricks D.N. The diversity of human vaginal bacterial communities and their association with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B.J., Olsen I., Aas J., Dewhirst F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Pei Z., Bini E.J., Yang L., Zhou M., Francois F., Blaser M.J. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinque D., Kittler R., Kayser M., Stoneking M., Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Anal. Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Rosemann C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi C., Balding D., Nasidze I.S., Risch G., Robichaux M., Sherry S.T., Stoneking M., Batzer M.A., Barbujani G. Patterns of human diversity, within and among continents, inferred from biallelic DNA polymorphisms. Genome Res. 2002;12:602–612. doi: 10.1101/gr.214902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper R.G., Silletti E., Vingerhoeds M.H. Saliva as research material: Biochemical, physicochemical and practical aspects. Arch. Oral Biol. 2007;52:1114–1135. doi: 10.1016/j.archoralbio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin R., Maiden M.C. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003;11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Wang X., Linz B., Novick R.P., Lum J.K., Blaser M., Morelli G., Falush D., Achtman M. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc. Natl. Acad. Sci. 2004;101:4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]