Abstract
The recent arrival of large-scale cap analysis of gene expression (CAGE) data sets in mammals provides a wealth of quantitative information on coding and noncoding RNA polymerase II transcription start sites (TSS). Genome-wide CAGE studies reveal that a large fraction of TSS exhibit peaks where the vast majority of associated tags map to a particular location (∼45%), whereas other active regions contain a broader distribution of initiation events. The presence of a strong single peak suggests that transcription at these locations may be mediated by position-specific sequence features. We therefore propose a new model for single-peaked TSS based solely on known transcription factors (TFs) and their respective regions of positional enrichment. This probabilistic model leads to near-perfect classification results in cross-validation (auROC = 0.98), and performance in genomic scans demonstrates that TSS prediction with both high accuracy and spatial resolution is achievable for a specific but large subgroup of mammalian promoters. The interpretable model structure suggests a DNA code in which canonical sequence features such as TATA-box, Initiator, and GC content do play a significant role, but many additional TFs show distinct spatial biases with respect to TSS location and are important contributors to the accurate prediction of single-peak transcription initiation sites. The model structure also reveals that CAGE tag clusters distal from annotated gene starts have distinct characteristics compared to those close to gene 5′-ends. Using this high-resolution single-peak model, we predict TSS for ∼70% of mammalian microRNAs based on currently available data.
The transcription of genes to RNA is a fundamental step in the expression of information encoded in a genome. Animal genomes encode three RNA polymerases, and all protein-coding genes as well as regulated noncoding genes such as microRNAs (miRNAs) are transcribed by RNA polymerase II (Pol II). The precise mechanism and features by which the Pol II enzyme hones in on the location of the transcription start site(s) (TSS) to initiate transcription is still not completely resolved, in particular for complex genomes like those of mammals, where a comparatively small number of TSS are vastly outnumbered by the noncoding fraction of the genome. Rapidly accelerating technical advances in both hybridization-based and sequencing-based methods for high-throughput TSS identification (Sandelin et al. 2007) yield unprecedented opportunity for new insight into the mechanisms that guide transcription initiation by Pol II. In particular, the sequencing-based technology known as cap analysis of gene expression (CAGE) offers a unique advantage among high-throughput methods: the 5′-end sequencing of cap-selected cDNAs provides a count of the number of transcript starts (CAGE tags) that map to a particular location on the genome. CAGE tags therefore provide a view not only of where initiation events occur, but how they are distributed.
While it had been previously noted that some promoters do not show a preference for a single initiation site (Bucher and Trifonov 1986; Bucher 1990), transcription was largely viewed as a process that may begin at only a few particular locations per gene, perhaps with different frequency depending on tissue type and other cellular conditions. The recent CAGE studies that include >12 million 5′-ends of mouse and human transcripts have fundamentally altered our understanding of Pol II promoters (Sandelin et al. 2007), by demonstrating convincingly that initiation events are not limited to one or just a few single locations (Carninci et al. 2006). Rather, these events tend to cluster at different scales, and tag distributions over regions of frequent initiation (CAGE tag clusters) take on a variety of distinct shapes. Genome-wide detection of TSS using CAGE and other competing technologies thus strongly suggests that transcription can begin at millions of sites in the genome (Carninci et al. 2005, 2006; Kapranov et al. 2007), and that these sites have widely varying usage rates.
This CAGE tag information has been extensively analyzed by the RIKEN team to show that given experimental data on the tag frequency observed within an active promoter region, the relative transcription start site usage of each nucleotide within the region can be predicted with high accuracy using a first-order Markov model (Frith et al. 2008). TSS distributions for most promoters in this study were also found to be highly conserved between human and mouse, suggesting a mammalian “code” for transcription initiation. In particular, for ∼45% of mouse CAGE tag clusters that are supported by more than 100 tags, the cluster contains one or more strongly preferred regions of only a few nucleotides in width. The presence of a strong initiation event peak within these highly localized regions suggests that position-specific DNA sequence features may mediate transcription within a large subset of mammalian promoters. This finding motivates a fresh look at whether DNA-encoded transcription signals alone, only using TSS location and not tag frequencies, can predict the likelihood of transcriptional activity at any particular genomic location and serve as a complementary model to the positional Markov chain by Frith et al. (2008).
The idea to identify TSS with the help of positional sequence features is not a new one; computational approaches to identify the locations of Pol II promoters have a long history, and various models have been trained using different sets of sequence and structural features, with varying degrees of success and sometimes including positional preferences (Davuluri et al. 2001; Bajic et al. 2002; Down and Hubbard 2002; Ohler et al. 2002; Bajic and Seah 2003). As a successful example, the analysis of Drosophila sequences has, indeed, led to sets of positionally enriched sequence motifs (Ohler et al. 2002; Fitzgerald et al. 2006), and data subdivision according to promoter type leads to a significant improvement in modeling and classification success (Ohler 2006). Recent popular approaches applicable to mammals (Sonnenburg et al. 2006; Wang and Hannenhalli 2006; Goni et al. 2007; Zhao et al. 2007; Zhou et al. 2007; Abeel et al. 2008) are typically based on high-quality promoter data sets defined in the hand-curated Eukaryotic Promoter Database, EPD (Cavin Perier et al. 1998), or the Database of Transcription Start Sites, DBTSS (Suzuki et al. 2002). However, EPD and DBTSS are relatively small compared to the CAGE set, and the computational approaches, in general, did not use direct information on the distribution of initiation events occurring at each transcription start site or the surrounding region.
In light of the evidence coming from the CAGE tags, approaches that assume that all mammalian TSS are a homogeneous set sharing the same features may thus simply not be able to define Pol II promoters in the most appropriate way; the difficulties that have been traditionally observed are very suggestive of multiple underlying core promoter architectures. In mammals, a division of promoters based on the presence of so-called CpG islands in the TSS vicinity has been popular, and the recognition of promoters belonging to the CpG-poor group has been notoriously difficult. CpG islands are a by-product of mammalian DNA methylation that occurs at CpG dinucleotides and are defined as regions relatively rich in GC content in general and CpG dinucleotides in particular (Larsen et al. 1992). However, different architectures most likely go beyond the simple presence or absence of CpG islands, particularly given that CAGE analyses suggest that many layers of control by proximal and distal sequence elements influence TSS distribution, and given that there is no clear-cut association of TSS distribution types with CpG islands (Frith et al. 2008).
In this study, we explore in-depth how well we can computationally model the subset of promoters containing a strong TSS within a narrowly defined location. In particular, we examine whether the presence of known Pol II transcription factor (TF) binding sites alone is sufficient to predict the TSS location of promoters exhibiting a strong peak. We show that within this class of single-peak promoters, start sites for transcripts supported by current gene annotation can be predicted with astonishing accuracy using only DNA-binding affinity scores. We also observe that single-peak CAGE tag clusters not supported by current annotation constitute an overall different class. While the focus of our work is on the identification of the features defining the single-peak promoter class and not on a general-purpose promoter identification tool, we can apply these models for genome-wide scans and evaluate the resolution at which single-peak start sites for coding and noncoding transcripts can be predicted. Together, these results demonstrate that high-accuracy computational TSS prediction is achievable for a specific but large subgroup of mammalian promoters. Using this model to predict TSS of mammalian miRNAs at high accuracy and spatial resolution, we estimate that up to 70% of these miRNAs may have single-peak promoters.
Results
Transcription initiation can be accurately modeled by DNA-binding affinity
In order to investigate whether transcription initiation location at single-peak start locations could plausibly be encoded by DNA affinity for known TF binding elements, we first examined whether any of these elements exhibited strong localized enrichment within the immediate vicinity of CAGE-defined TSS locations. We reasoned that if the Pol II transcription machinery were guided by direct or indirect binding to a subcollection of such elements, binding would necessarily take on some degree of positional specificity with respect to the site of initiation. We began by examining a subset of CAGE single-peak locations that were also supported by UCSC Known and RefSeq gene annotation, the annotation-supported training set (see Methods). Using a standard log-likelihood TF binding site scanning technique and a collection of approximately 40 known TRANSFAC (Matys et al. 2003) and Jaspar (Sandelin et al. 2004) binding elements with positional enrichment reported in the literature, we identified a subset of 35 elements that exhibit marked enrichment within this data set. In particular, by incorporating a local background correction for dinucleotide frequency into our scanning method (see Methods), we could decouple specific local signal enrichment from broader enrichment arising because of the interplay between TF motif composition and background composition (Fig. 1). As a result, cumulative TF binding affinities for many elements resolved to display a sharp, highly localized signal (Fig. 2). We observed sharp enrichment signals in precisely the expected binding locations for canonical Pol II elements TATA and Initiator (Smale and Kadonaga 2003), along with sharp and broad regions of positional enrichment for more than 30 other elements (see Supplemental material for a complete list and positional enrichment plots).
Figure 1.
The effect of local dinucleotide background frequency correction on cumulative TF scores within several kilobases of the TSS. (Left panels) VDR (vitamin D receptor) is typical of a relatively GC-rich motif that shows score enrichment in the TSS vicinity partly due to an increasingly GC-rich background near many TSS, and partly due to a sharp locally enriched signal, which can often be difficult to distinguish as a separate entity. (Right panels) TEF (thyrotrophic embryonic factor) is typical of a relatively AT-rich motif that shows depletion in the TSS vicinity for the same reason. The local background correction decouples specific local signal enrichment from broader enrichment arising because of the interplay between TF motif composition and background composition. TRANSFAC ID for each binding element is displayed in plot titles.
Figure 2.
Regions of positional enrichment with respect to TSS for TF-binding elements TATA, OCT1, and YY1. TRANSFAC ID for each binding element is displayed in plot titles. Plots display cumulative score (summed over the annotation-supported training set TSS regions) for each element as a function of position with respect to TSS. Colors show the region subdivisions diagrammed in Figure 3A. (Red and gray) Flanking regions.
This observation suggests that many of these elements may play a guiding role in initiation for at least some single-peak TSS locations and that their regions of positional enrichment reflect the locations in which they are most likely to do so. To test this hypothesis, we asked whether a model based on this group of sharp and broad regions of enrichment could accurately predict the probability that any given genomic location is a single-peak TSS. In order to allow such a model to distinguish locations of high binding affinity that are the most predictive of a single-peak initiation site, we divided the regions of enrichment into several subwindows and flanking regions as shown in Figure 3A. A cumulative score that approximates affinity for the relevant binding element was computed over each subwindow and flanking region, and this procedure was performed for all locally enriched binding elements to construct the scoring features for a particular location (see Methods). Additionally, GC content in a surrounding 200-nt region is computed. We sought to understand whether a model based on these features could distinguish single-peak TSS locations not only from gene-poor regions of DNA, but also from nearby upstream locations and coding sequence, as both may lie in regions that are CpG-rich and/or proximal to TF binding sites. We therefore computed scoring features for each TSS in the annotation-supported training set (positive examples), and for negative examples selected from the immediate upstream regions of these TSS as well as from annotated coding sequence (CDS) (Fig. 3B). Positive examples, negative intergenic examples, and negative CDS examples are selected in a 1:20:1 ratio (see Methods).
Figure 3.
(A) For each example (location) considered, features are generated by adding up affinity scores for each TF within its region of enrichment. The lower portion of the diagram illustrates how this is done in detail: Each region is divided into five overlapping subwindows covering the region of enrichment, plus two flanking subwindows. Positive log-likelihood scores are summed over all positions in each subwindow, generating seven features for each TF. Additionally, GC content within a 100-nt region on either side of the location is also computed as a feature. The intuition behind this setup is to allow a trained model to select which elements and regions are most predictive of a TSS. (B) Training data sets are constructed from positive examples (the TSS locations themselves) and two types of negative examples: intergenic locations drawn at random from the immediate upstream regions of the TSS locations, and coding sequence examples drawn at random from annotated CDS regions on the mouse genome. Twenty intergenic locations are drawn from each immediate upstream region, and CDS locations are drawn in a 1:1 ratio with positive examples (to comprise ∼5% of the negative data set).
We then performed 10-fold cross-validation over the annotation-supported training set using L1-regularized logistic regression (Koh et al. 2007). We optimized the L1 regularization parameter over the validation set of each partition and estimated performance over an independent test set within the partition (see Methods). The optimal L1 parameter on each partition determines how many features are removed from the model in such a way that the best classification performance is achieved. Classification performance is measured by the area under the ROC curve (auROC). We found that performance was remarkably high, with a test auROC averaging 0.98 over all partitions (Fig. 4). For a baseline performance comparison on exactly the same feature set, we also performed cross-validation over the same data partitions with an empirical näive Bayes classifier. For performance comparison with a more elaborate generative model, we retrained the generalized hidden Markov model (HMM) defined in the McPromoter classifier (Ohler et al. 2000). Figure 4 compares cross-validation performance outcome for these three models. We found that L1-regularized logistic regression outperforms the McPromoter HMM, which outperforms näive Bayes, and, in fact, we consistently observed this performance relationship between the three model types on all subsequent CAGE data sets examined. We defined a final annotation-supported model by training on the entire annotation-supported training set using the average of optimal L1 parameters from cross-validation. We then tested the annotation-supported model on a completely separate test set composed of annotation-supported single-peak TSS and 100,000 randomly selected genomic locations (see Methods). Supplemental Figure 1 illustrates the outcome with two conferring performance measures, auROC and auPRC (area under the precision-recall curve). The model again performed remarkably, with a near-perfect auROC of 0.99.
Figure 4.
Tenfold cross-validation performance comparisons for the annotation-supported model. (Left) The plot compares the performance of two additional classifiers, a näive Bayes classifier and McPromoter's HMM classifier. (Right) The plot compares CpG-island and non-CpG-island models. ROC curves with threshold averaging are displayed in both plots, along with the average area under the curve (auROC). Positive examples are the experimentally supported CAGE single-peak TSS locations, while negative examples are selected from intergenic and coding regions (see Fig. 3B).
We observed that our annotation-supported data set contains ∼40% of single-TSS that are not located in CpG islands. We therefore also divided the data set according to previous convention, with one group of TSS in CpG islands and the other group not in CpG islands (the CpG-island and non-CpG-island training sets). Using the same regions of enrichment as for the model trained on the full set but retraining the L1-regularized logistic regression classifier and performing a corresponding cross-validation under this data division, we observed an average auROC of 0.99 and 0.96, respectively, for the CpG-island and non-CpG-island cases (Fig. 4). This shows that the annotation-supported set contains a large fraction of non-CpG-island TSS that it classifies almost as successfully as CpG-island TSS. In total, the outcome suggests that the annotation-supported model provides an internally consistent, high-resolution binding affinity-based code for the majority of single-peak promoters and does not need to be resolved into CpG-rich and CpG-poor TSS. This is markedly different from previous reports, which consistently reported significantly poorer performance on non-CpG island promoters (Wang et al. 2007; Zhao et al. 2007).
Test set scans demonstrate the model's ability to identify TSS locations with high precision
In order to understand how the annotation-supported model performs over contiguous genomic regions, we scanned 8-kb regions surrounding all TSS in the annotation-supported test set. We observe that within a reasonable range of classifier cutoffs, the annotation-supported model picks up single-peak TSS with very high resolution. The ability of the annotation-supported model to accurately identify start sites both within and outside of CpG islands is also confirmed. Figure 5 displays the percentage of TSS hit by a probability peak as a function of the number of additional peaks (hits) observed and examines how well these TSS-containing peaks approximate actual TSS location. Results for the annotation-supported model are displayed as solid dotted curves in Figure 5. About 70% of the TSS were hit within 10 nt at thresholds allowing very few additional hits, even in this difficult set of TSS proximal genomic regions. The average distance to peak center for probability peaks containing a TSS is well within 20 nt at thresholds where ∼90% of TSS are contained by these peaks.
Figure 5.
Performance on scans of the annotation-supported test set. (Left) The case in which a TSS is considered to be a hit if a probability peak contains the TSS. The curve represented by each symbol type shows the percentage of TSS hit as a function of the number of additional hits per kilobase. (Right) Each curve displays the average distance to the center of the probability peak computed over all of the peaks containing a TSS. At each threshold value (color), the plots give a comparative view of how many additional peaks are being called versus how well the TSS-containing peaks approximate actual TSS location.
To place these results in context, we also scanned this same set of 8-kb test regions using three additional programs: (1) the retrained version of McPromoter; (2) ARTS (Sonnenburg et al. 2006), a support vector machine (SVM)-based TSS prediction program designed for high-performance genome-wide scanning; and (3) CoreBoost (Zhao et al. 2007), a decision-tree-based program intended for high-resolution prediction in shorter regions known to contain a TSS, for example, regions preidentified by a chromatin immunoprecipitation with microarray hybridization (ChIP-chip) experiment. Results are displayed in Figure 5 for comparison with the annotation-supported model. We observe that the annotation-supported model outperforms ARTS and CoreBoost in sensitivity/specificity and spatial resolution, although the trade-offs between these two programs are apparent. ARTS achieves nearly the same performance in calling the TSS peaks as the annotation-supported model and does a bit better than CoreBoost in this sense, but at a cost of much lower spatial resolution. Interestingly, the retrained McPromoter algorithm calls a slightly higher percentage of TSS per additional kilobase hit when low thresholds are considered, but produces such wide probability peaks that spatial resolution is by far the lowest of the programs considered. Encouragingly, we also observe that additional peaks called by the annotation-supported model in the region of the TSS frequently agree with the annotated starts of mRNAs, ESTs, and other CAGE tag clusters. Figure 6 displays a typical example of an annotation-supported model scan.
Figure 6.
At the top, the UCSC custom track displays probability output from a representative scan over the region of a test set TSS using the annotation-supported model (this particular example shows CAGE tag cluster T17F00727F78). The model calls out highly probable single-peak start regions with surprising accuracy, often indicating additional possible single-peak starts in locations that are supported by mRNA transcripts from GenBank.
TF affinity-based code enables high-resolution genomic scans of coding and noncoding sequence
Having observed that the annotation-supported model can delineate single-peak TSS with excellent spatial resolution in gene-proximal promoter regions, we explored the model's output when scanning on a chromosome-wide scale. We applied the annotation-supported model to mouse chromosome 16 (chr 16), selected for its high degree of synteny with human chromosome 21 (a historical gold standard of comparison for genome-wide promoter prediction). By removing the relatively few TSS regions on mouse chr 16 contained in the annotation-supported test set, this chromosome provides an ∼100-Mb body of sequence that has not previously been seen by either the annotation-supported model or by other TSS prediction programs evaluated here. As McPromoter was less successful than the other predictors and CoreBoost is intended to only scan small regions, we limited ourselves here to comparing our approach to the ARTS predictor.
We obtained single-nucleotide resolution predictions for both ARTS and our model and performed a comparison for these two programs over RefSeq genes and over CAGE start sites supported by 10 or more tags following the genome-wide performance comparison strategy in Sonnenburg et al. (2006) (see Methods). In brief, chr 16 is divided into equal-sized chunks, and the prediction having the largest value within each chunk is computed for each program. For RefSeq genes, the comparison is implemented just as described in Sonnenburg et al. (2006): positive chunks are defined as those that contain a RefSeq start, while negative chunks are all non-positive chunks containing any downstream portion of a RefSeq gene. For CAGE starts, full-length transcripts are not available, so all non-positive chunks are considered as negatives. Figure 7 and Supplemental Figure 2 display the results for 50-nt and 500-nt chunks, respectively. While both RefSeq and CAGE sets contain all types of promoters, it is striking that performance as defined by auROC is not vastly different. ARTS clearly picks up less putative start sites downstream from annotated RefSeq gene starts and calls less very-high-probability additional chunks with respect to the CAGE set although auROC values on this set are nearly identical. As chunk size becomes smaller, the annotation-supported model consistently improves its auROC performance relative to ARTS across different types of data sets. These trends are not difficult to reconcile given the nature of the output signals observed in test scans. Smaller chunks allow the annotation-supported model to “home in” on single-peak promoters and to distinguish between more alternative high-probability start regions than ARTS does, particularly for the RefSeq gene set.
Figure 7.
Output comparison of the annotation-supported model and the ARTS TSS prediction program. Chromosome 16 is divided into 50-nt chunks, and the prediction having the largest value within each chunk is computed for each program. Positive chunks contain RefSeq or CAGE starts, respectively. Negative chunks comprise downstream gene portions for RefSeq, and all non-positive chunks for CAGE. The area under the curve (auROC) provides a performance measure given these chunk definitions.
Using the limited data available, we investigated annotation-supported model output for noncoding genes by scanning over a hand-curated set of 20 putative human and mouse miRNA primary transcript start sites having some degree of experimental support (see Methods). We applied the annotation-supported model to scan 8-kb regions surrounding the supported start sites. We observed that 70% of these scans are qualitatively and quantitatively similar to those of the annotation-supported test set (see Supplemental material), exhibiting a high-probability region containing the TSS and other nearby probability peaks delineating annotated starts for many of the surrounding mRNAs and ESTs. The remaining 30% of scans generally have very few probable single-peak start site regions in the vicinity, suggesting that the start sites for these particular transcripts are not likely to be high-propensity start locations. This is consistent with the view that Pol II non-protein coding genes are also transcribed by a variety of promoter types that broadly tend to correlate with specificity of expression. Supplemental Figure 3 illustrates a typical example of a miRNA scan that is consistent with a single-peak TSS, while Supplemental Figure 4 shows an example that has no indicated single-peak TSS near the annotated start of the putative primary transcript.
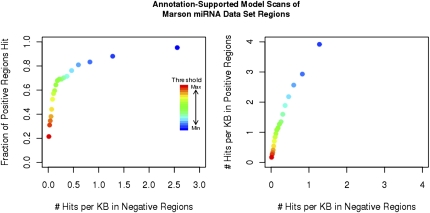
We additionally investigated annotation-supported model output on a set of predicted miRNA promoter regions based on histone H3 trimethylation data in human and mouse embryonic stem cells (Marson et al. 2008). In contrast to the hand-curated set of 20 transcripts above, this set is significantly larger but provides putative miRNA primary transcript start regions on the order of several kilobases in length as opposed to specific experimentally supported TSS. Regions in this set may overlap the actual miRNA precursor foldback, or be as far as 250 kb away from it. Starting from the 268 nongenic mouse miRNA start regions in the set, we retained 84 unique regions after selecting the upstream-most miRNA from each cluster, and requiring that there was some distance between the start region and the miRNA, but that no annotated UCSC Known Gene start site was contained in this intervening sequence (see Methods). We then defined a set of positive regions as the 84 putative miRNA start regions, and a set of negative regions composed of all sequence between the miRNA start regions and the miRNA locations themselves. We scanned both positive regions (161 kb) and negative regions (2833 kb) and compared the density of probability peak hits in each type of sequence (Fig. 8). We observed a dramatically lower density of hits in the negative regions—6.2-fold less than in positive regions at a probability threshold of 0.5, a level where 68% of positive regions contain one or more hits. This increases to approximately a 10-fold difference at higher probability thresholds. Our predictions therefore correlate well with the Marson data set predictions, and given the high precision of our predictor, can be used to locate specific TSS within the larger regions from Marson et al. (2008). When we compared the percentage of positive regions hit with the negative region hit density (Fig. 8), we observed that this percentage declines with decreasing probability threshold at a distinctly more rapid rate after ∼70% of positive regions are hit. This agrees well with our finding on the hand-curated miRNA TSS set that suggested that ∼70% of miRNA primary transcripts have strong single-peak start site predictions.
Figure 8.
Output of the annotation-supported model on the Marson putative miRNA TSS region data set. Each positive region is predicted by the Marson data set to contain one or more miRNA TSS, whereas negative regions are not predicted to contain any miRNA TSS. (Left) The curve compares the percentage of positive regions at each probability threshold (color) hit by an annotation-supported model probability peak to the number of hits per kilobase (hit density) within the negative regions. (Right) The curve compares hit density within the positive regions to hit density within the negative regions.
Single-peak CAGE tag clusters do not constitute one homogeneous set
Our use of logistic regression allowed us to investigate the importance of model features directly by examining their logistic regression coefficient values. Logistic regression coefficients describe the partial contribution of each feature to a predictive model; because our model features approximate regional binding affinities, these values provide insight into the predictive roles of each binding element. In the annotation-supported model, three prominent canonical sequence elements have very large coefficients (TATA-box, Initiator, and GC-content), while a second tier of prominent coefficients tunes performance. This model structure, with precisely the same elements in the top and second tiers of coefficients, is consistently repeated across all cross-validation partitions as well as the final annotation-supported model. In particular, 17 out of 20 binding factors appear in all 10 cross-validation sets with values >0.1, while the remaining three factors do so in at least eight out of 10 cross-validation sets. Figure 9 provides a graphical breakdown of logistic regression coefficients for the annotation-supported model, according to binding element. A complete set of enriched elements for all data sets is provided in the Supplemental material.
Figure 9.
Logistic regression coefficients above 0.05 for the annotation-supported model. TATA-box, GC content within a 100-nt region, and Initiator elements are dominant, but highly accurate performance relies heavily on many other TFs. A complete listing of all factors and binding element abbreviations is provided in the Supplemental material.
While annotation-supported TSS comprise the majority of single-peak promoters, a considerable fraction of CAGE tags does not coincide with annotated 5′-ends; rather, these tags are found in gene-poor areas or in the interior of annotated transcripts. We refer to these clusters as the CAGE-only-supported set, that is, CAGE single-peak TSS that did not fall into the annotation-supported category, and split them into training and test sets just as for the annotation-supported tag clusters (see Methods). We note that many CAGE-only-supported TSS (51%) lie in the introns and exons of annotated gene transcripts, and an overwhelming portion of the remainder (∼80%) fall within mapped expressed sequence tags (ESTs). Overall, only 14% of tag clusters fall within CpG islands, a drastically lower fraction than for annotation-supported tags. When we applied the annotation-supported model to the CAGE-only-supported test set, performance dropped to a significantly lower auROC of 0.71. At a threshold of 0.5, only 109 out of 1240 samples (∼9%) are predicted to be a TSS. In this subset, 65% overlap with CpG islands, and 67% of successful predictions fall outside annotated genes, a strong deviation from the overall pattern (14% CpG islands, 49% outside of genes).
This striking difference in performance could result from two different scenarios. One possibility is that while these clusters do, indeed, represent capped transcripts, they do not, in fact, correspond to transcription start sites. The alternative is that this set constitutes a different set of Pol II promoters, for which different sequence features are discriminative. To investigate this, we applied the same positional enrichment and TF selection method (see Methods) to determine the binding elements that are positionally enriched in the CAGE-only-supported training set. We observed that CAGE-only-supported TSS as a group are not only enriched for a different set of binding elements, but even when certain elements are enriched in both sets, they may have different regions of positional enrichment (Supplemental Fig. 5). Furthermore, by using the CAGE-only-supported training set to retrain the model, we observed an improvement to an auROC value of 0.80 in cross-validation (Supplemental Fig. 6). The final CAGE-only-supported model achieved an auROC of 0.83 on the CAGE-only-supported test set. The CAGE-only-supported model distributes smaller but approximately equal coefficient weights across a larger top tier of coefficients (Supplemental Fig. 7). GC content plays a very small role compared to other coefficients in the model. Furthermore, there is much more variation among the individual cross-validation partitions in regard to which coefficients are chosen among the top-tier coefficients. About 10% of TSS in this class are identified with high resolution, and the remaining TSS are only identified at low classifier thresholds where many additional probability peaks are called.
Given the success of our approach on annotation-supported promoters, one could expect our strategy to work well on other strongly peaked CAGE tag clusters. The results demonstrate that even with a retrained model, known transcription factors cannot describe the CAGE-only set nearly as accurately as the annotation-supported set. While there is the possibility that our set of PWMs is not adequate, and that other as-yet-unknown TFs are responsible for positioning Pol II for this subset, it certainly leaves open the possibility that these clusters are, in fact, not representing Pol II TSS.
Discussion
In this study, we determined a set of features, based solely on DNA affinity for known binding elements, that are sufficient to define single-peak TSS at near-perfect-accuracy levels. The available high-quality CAGE data on the precise patterns of initiation events allow us to define several subclasses and study separate models for TSS close to annotated protein-coding genes, as well as for TSS only supported by CAGE tags but not close to TSS of annotated genes.
The success of the TSS model trained on currently annotated genes derives from this highly informative data set, along with biologically motivated feature definition, feature selection, and interpretability. Using a background-corrected signal that accounts for local dinucleotide sequence composition was also a key factor in observing positional specificity of factor enrichment, and therefore in creating a high-resolution classifier. The built-in feature selection property of L1-regularized logistic regression penalizes features that are not predictive of outcome. The model can therefore not only determine which TFs tend to be predictive of TSS location within their regions of enrichment, but also precisely which windows within that region are most predictive. The contrasting negative data set was chosen very stringently in order to force our model to distinguish single-peak sites with high spatial resolution. We found that choosing a negative example set very near to the single-peak TSS sites themselves came at a small cost of reducing apparent cross-validation performance, with a substantial benefit of reducing noise, particularly in GC-rich areas.
The set of CAGE tag clusters not falling near annotated gene starts warrants further investigation. The vast majority of these clusters do not show the same set of features as annotation-supported TSS. While training a specific model improved prediction performance on this set, it emerged that the features we use do not accurately represent these clusters. At this point, it is open as to whether these clusters correspond to TSS that could be as reliably modeled using other yet-to-be identified features, or if these clusters, in fact, do not represent initiation events. A small fraction of this set, on the order of 10%, are similar to annotation-supported TSS and can be predicted by our model; we expect these clusters to correspond to alternative start sites of known genes, or start sites of as-yet-unannotated, possibly noncoding genes.
Our TSS model can be used as a high-resolution predictor to identify TSS when scanning genomic sequences. While many other learning algorithms may be used for promoter prediction, we selected our classifier not simply to optimize performance, but also to offer specific insight on which TFs commonly play a significant role in the determination of single-peak promoter location. While other discriminative algorithms such as SVMs may perform comparably well on our feature set, they are often more difficult to interpret (Sonnenburg et al. 2008), whereas logistic regression is a method exactly suited to provide a probabilistic classification outcome from continuous features that illuminates how that outcome was derived. In the annotation-supported and CpG-island models, TATA, Initiator, and GC content are the single most dominant signals as expected. However, in sharp contrast to previous approaches whose automatically derived feature sets mostly centered on TATA and GC content (Down and Hubbard 2002), a numerous second layer of features collectively provides a large contribution to predictive value. Among the second tier of elements, several factors have been suggested to be over-represented at specific regions in the vicinity of TSS, for example, YY1 and CREB (Xi et al. 2007), but none have been previously used to predict TSS location.
We observe that within all models, the regions of enrichment for some TFs are broadly defined despite the local background correction, and effectively act as “GC sponges,” whereas elements with narrowly defined regions of enrichment provide locational specificity on top of this GC enrichment information. Inclusion of a specific GC content variable simplifies the model by readily explaining broad increases in GC content near a TSS, thereby reducing GC sponges. It has recently been observed that GC content is anticorrelated with nucleosome occupancy (Lee et al. 2007), lending a sensible biological explanation for its prominence in the model. Together, these observations suggest a possible DNA code in which broadly defined affinity for GC-rich binding elements such as SP1 can serve to recruit these factors to nucleosome-free regions, while TFs with narrowly defined regions of enrichment are likely to interact directly with core Pol II machinery to help refine the location of transcription initiation. This biological model is consistent with other recent studies that observed that many TFs in higher eukaryotes have strong biases for binding sites to be highly position-specific if they are close to the start of a gene (Tabach et al. 2007).
When examining the regression coefficients in more detail, it is apparent that for many elements including the canonical TATA-box, coefficients are highest within the central part of the region of enrichment as expected. Negative coefficients, however, are equally as important as positive coefficients; the model learns not only where the factor should be, but also where it should not be observed in relation to a TSS location. Regions of enrichment for canonical elements agree with literature-supported models of spacing with respect to the initiation site. Intriguingly, some TFs are enriched in certain single-peak CAGE data sets within regions that agree with their literature-described positions, while others show a different narrowly defined region of enrichment. One particularly striking case of this is the DPE element, a binding element that has experimental support in Drosophila but largely theoretical support via conservation evidence in vertebrates (Burke et al. 1998). In the annotation-supported data set, the strongest peak of enrichment for this element is, in fact, located upstream of the TSS. However, we need to interpret each of these cases with some caution, as it may also happen that binding element motifs in some cases serve as surrogates for other factors, that is, that the enrichment of one factor actually reflects the preference of a different, possibly unknown factor with a somewhat similar binding preference.
There is a strong indication that most miRNA genes are also transcribed by Pol II; however, the majority of their primary transcripts (pri-miRNAs) remain uncharacterized because of the experimental difficulty of isolating these rapidly degraded transcripts (Kim and Nam 2006). Current experimental evidence suggests that pri-miRNAs may be very long, with examples ranging from ∼4 kb (Cai et al. 2004) to >50 kb in length (Fukuda et al. 2007). It further suggests that mature miRNAs are not necessarily located near the start of these transcripts. High-resolution genomic scans are therefore of particular utility for investigating promoter architecture in this situation. Our application of the annotation-supported model to a set of 20 putative miRNA primary transcripts with some degree of experimental support conservatively suggests that ∼70% of miRNAs may have one or more single-peak promoters. Our investigation on the Marson data set (Marson et al. 2008) indicates that annotation-supported model predictions correlate well with miRNA start regions predicted using histone H3 trimethylation data, and supports the idea that up to ∼70% of miRNA transcripts are likely to be associated with a strong single-peak TSS. Our work strongly suggests that single-peak promoters of non-protein coding genes can be distinguished at high resolution on the genome.
The annotation-supported model provides an alternative way to describe TSS location based solely on DNA affinity for known binding elements. Unlike other methods designed for high-resolution scanning such as the CoreBoost program, it does not require separate treatment of CpG-rich regions, regions defined by ChiP-chip data, or any other prior knowledge about the nature of a sequence to be scanned. A priori, we did not know what performance to expect when our model was applied to promoters with broad initiation patterns rather than single peaks; these broad TSS are currently estimated to outnumber single-peak promoters in the genome. When applied in a chromosome-wide scan, model performance over RefSeq genes as well as other CAGE start sites was competitive with ARTS, an SVM-based TSS predictor using a modular kernel and thousands of features that has demonstrated superior performance in genome-wide scans over an array of other methods. This is particularly remarkable considering that the peak types of TSS in these data sets are unknown, and that such a course chunked-genome comparison yields the advantage to ARTS as a predictor with broader regional identification as opposed to a high-spatial-resolution signal. Our analyses thus strongly suggest that the annotation-supported model is suitable for high-resolution de novo TSS prediction on the genome in the absence of experimental data. While the model predicts the probability that a given location on the genome is a narrowly localized high-propensity start location, it is not designed to predict the relative number of CAGE tags present or to operate with single-nucleotide resolution. Our investigation therefore implies a complementary role for this classifier in conjunction with the first-order Markov model described in Frith et al. (2008). The annotation-supported model may first be applied to search for probable single-peak TSS locations, suggesting suitable regions for experimental scrutiny, followed by an analysis of relative start site propensity at the single-nucleotide level.
Our study suggests several worthwhile future directions of investigation. An exhaustive analysis of all current TRANSFAC and Jaspar binding elements with positional weight matrices may yield an even larger number of elements that display regions of positional enrichment with respect to single-peak TSS. In particular, this may help to increase performance on the CAGE-only supported TSS set—the lower level of success of this model is less surprising if one considers that the TF binding models currently included in our study were selected because of previously reported enrichment upstream of coding genes. Furthermore, including higher-order interactions in the logistic regression model may reveal specific combinations of the enriched subregions that are predictive of single-peak TSS location, suggesting modules that are active in single-peak promoters. A breakdown of CAGE tags by tissue type may enable single-peak TSS prediction with some degree of tissue specificity, particularly for those tissues in which a large number of tags become available for training. By expanding the scope to include broad-peak and multi-modal CAGE tag distributions, one can also investigate the extent to which other promoters with other TSS distribution types are accurately identified using the current local DNA binding affinity model, or if not, whether it can be adapted to these initiation distributions. Finally, many current studies suggest that the incorporation of epigenetic information such as histone modification and nucleosome location data can prove fruitful in predicting TSS location.
Methods
Data sets
A “CAGE tag” is a 20–21-nt 5′ cDNA end that has been mapped to the genome. A “CAGE tag cluster” (TC) is composed of tags that overlap on the same strand by one or more nucleotide positions. Our analysis uses two groups of mouse single-peak CAGE tag clusters, defined by the authors of the original high-throughput experimental study in a subsequent analysis of TATA-initiation site spacing (Ponjavic et al. 2006). A single-peak TC contains at least 50 tags and has a distance of <4 nt between the 25 and 75 tag density percentiles. Each TC in the twin-TSS subgroup of single-peak TCs has a neighboring TSS within 4 nt of the highest TSS peak that contains at least 25% of the tags in the highest peak, and together these two positions contain >75% of tags within the cluster (461 TCs in total). The single-TSS subgroup consists of single-peak TCs that are not in the twin-TSS subgroup (2399 TCs in total). In brief, both subgroups have a very tiny region within the cluster that contains the vast majority of tags. As detailed below, one subgroup is used for training and the other for independent testing. In all cases, the highest peak is considered the representative TSS within the cluster. According to estimates from the RIKEN authors, peaked TSS comprise ∼45% of all tag clusters.
The single-TSS and twin-TSS groups are each further subdivided for analysis. Each group is split into annotation-supported and CAGE-only-supported subgroups. The annotation-supported subgroup contains only TCs that fall within 500 nt of an annotated UCSC Known Gene or RefSeq gene start. The CAGE-only-supported subgroup contains all remaining TCs. For comparative analyses, the annotation-supported subgroup is additionally split in an alternative way into the CpG-island and non-CpG-island subgroups. The CpG-island subgroup contains only annotation-supported TCs where the representative TSS lies within a CpG-island, and the non-CpG-island subgroup contains all remaining annotation-supported TCs. All CpG islands are defined using EMBOSS newcpgreport, the application used in the production of CpG island database CPGISLE (Larsen et al. 1992). Supplemental Table 1 provides a chart of the TC counts in each subset of the single-TSS and twin-TSS groups.
We use each of the four data subsets of the single-TSS group (annotation-supported/CAGE-only-supported, CpG-island/non-CpG-island) for model training and cross-validation, and the respective subsets of the twin-TSS group for completely independent testing. A training set is produced from each single-TSS data subset in the following way: Each TC contains a representative TSS, and together the set of genomic locations of these TSS comprises the positive examples. For each TSS in the positive set, a group of 20 intergenic locations is drawn at random from the region between 100 nt and 4 kb upstream of the TSS. Additionally, one location is drawn at random from the annotated CDS of mouse UCSC Known Genes. Intergenic and CDS locations comprise the negative examples. Therefore, each training set is composed of positive, negative intergenic, and negative CDS examples in a 1:20:1 ratio. Figure 3B provides a visual summary of how positive and negative examples in a training set are derived. An independent test set is produced from each twin-TSS data subset by taking all twin-TSS locations as positive examples, while negative examples are composed of 100,000 randomly selected locations from the most recent mouse genome build (mm9).
All CAGE tags were mapped to the mm5 mouse genome build in their definition (Carninci et al. 2006), and therefore positive and intergenic samples must be taken from this build. CDS examples in each training set are drawn from the latest mouse genome build, mm9. Data set composition of ∼5% CDS was chosen to broadly reflect the low fraction of coding sequence in the mouse and human genomes. All data sets are made available in the Supplemental material.
We constructed a miRNA putative primary transcript data set by identifying 20 miRBase miRNAs (Griffiths-Jones et al. 2006) located within transcripts that have some degree of experimental support. To date, mammalian miRNA TSS data have been difficult to obtain on a large scale because miRNA primary transcripts are rapidly cleaved and degraded in the cell nucleus (Kim and Nam 2006). Transcripts containing five miRNAs have explicit literature support as miRNA primary transcripts: hsa-mir-23a (Lee et al. 2004), hsa-mir-21 (Cai et al. 2004), hsa-mir-155 (Tam 2001; Tam and Dahlberg 2006), mmu-mir-223 (Fukao et al. 2007), and mmu-mir-199a-2 (Fukuda et al. 2007). An additional 15 transcripts are curated from several UCSC data sources. UCSC gene sets contain transcripts from cDNA libraries or other clone sources that are annotated as entirely noncoding or with a few atypically small exons. These transcripts are often identified either explicitly as noncoding or as producing an unknown protein product. Additionally, the ENCODE regions provide several sources of experimental evidence for UCSC annotated start sites, including the Stanford Promoter set (Trinklein et al. 2003).
We also constructed a set of “positive regions” predicted to contain miRNA primary transcript start sites from a recent study by Marson et al. (2008), along with a corresponding set of “negative regions” deemed less likely to contain these start sites according to the same data. We collectively refer to these regions as the “Marson data set.” In order to select positive regions from Marson et al. (2008), we started from the 268 miRNA TSS regions not specifically labeled as Genic in the Supplemental material provided by the authors. Many of these were labeled as putative start regions for more than one miRNA (since mature miRNAs can be transcribed together in a cluster on a single primary transcript); among such regions, we selected the upstream-most miRNA as the unique cluster representative. From this set of TSS regions associated with a unique miRNA, we selected those regions with a non-zero distance between the TSS region and the location of its associated mature miRNA. Some of these cases contained an annotated UCSC Known Gene start site between the TSS region and the miRNA; we removed these cases. The 84 remaining TSS regions comprise the set of positive regions. These regions are associated with 81 unique miRNA cluster representatives. Each positive region has a corresponding negative region, defined as the sequence between the positive region and the miRNA location. In the case in which a miRNA is associated with more than one positive region, the negative regions associated with the more upstream positive regions are defined as the sequence between the end of the positive region and the start of the next positive region downstream. Thus, each positive region is predicted to contain one or more miRNA TSS, and each Negative region, in contrast, is not predicted to contain a miRNA TSS. Negative regions also do not contain any UCSC Known Gene start by definition.
Calculation of background-corrected TF binding site scores
Features were designed to approximate the DNA binding affinities of TFs to a particular genomic region, and these approximate affinities were computed using the method of log-likelihood scoring for positional weight matrices (PWMs) (Stormo 2000). Each TF is represented by a PWM, in our case, a matrix of frequencies with which this TF is expected to bind certain DNA motifs. We used the standard method of adding pseudocounts to eliminate zero-valued matrix entries (we add 0.25 pseudocounts). The standard scoring method can be viewed as sliding this PWM along a DNA sequence, and at each nucleotide position computing the likelihood that the DNA motif at this particular location was generated by the PWM description versus the likelihood that the motif was generated by a background frequency model. The log of this ratio of likelihoods defines the score at a particular position, and a high positive score implies that a DNA location is a probable binding site for the TF.
The background model is usually defined as the set of single-nucleotide frequencies within a large set of promoters in a particular genome. However, in mammalian genomes, the dinucleotide base composition can change dramatically within the local vicinity of a TSS. As an obvious example, a TSS within a CpG island contains a much higher number of CG dinucleotides than the surrounding sequence. As a result, the standard background model can make TF scores in the region of a TSS “look big” for a slightly GC-rich PWM or “look small” for a slightly AT-rich PWM. Figure 1 shows this concept. In order to examine whether a TF is enriched at a particular location within the vicinity of the TSS, we wanted to use a scoring method that discounts enrichment arising solely from the relationship between PWM composition and background composition.
To this end, we used a local dinucleotide background model, where frequencies are calculated within a 500-nt window of the position at which the log-likelihood score is computed. This type of model is known in the literature as a local first-order Markov background model (Blanchette et al. 2006). For simplicity, we call this method the local background correction. It is used for all scoring computations in this study. Figure 1 illustrates how a locally background-corrected binding affinity signal elucidates the specific region of positional enrichment for TFs. In theory, one may use background models of increasingly high order to discount for local sequence content, at risk of fitting the background signal to an undesirable degree. The overall idea of choosing a first-order background model is to use the simplest possible method that accounts for fluctuations in GC content that are not related to the presence of any specific binding element.
Positional enrichment and TF selection
In order to understand which TFs may be enriched with respect to TSS location in a particular data set, we began with a list of 39 known TFs and canonical binding elements from two sources. The first source comes from an analysis that uses a context-free grammar-based TF scanning model to find elements that show some degree of positional enrichment within mouse and human promoters (Schug 2005). Subsequent analyses support the enrichment findings for elements in this original study (Stepanova et al. 2005; Xi et al. 2007). All TRANSFAC elements are represented by PWMs in the TRANSFAC 9.4 and Jaspar databases. We then added several elements to this list from the recent literature that are contained in Jaspar Pol-II 2008 (Bryne et al. 2008). The complete list of elements (provided in the Supplemental material) ranges from canonical binding elements such as TATA and Initiator to less well-known factors such as VBP (von Hippel-Lindau binding protein) and RREB (Ras-responsive element binding protein).
Using this list of elements and the local background correction method described above, we scanned each PWM over a 1-kb region on either side of each TSS in the training data set under consideration. At each nucleotide position with respect to the TSS, we summed all positive scores over the examples in the TSS data set. This procedure was performed on both the sense and anti-sense DNA strands. In some cases, an element is represented by more than one TRANSFAC or Jaspar PWM, and in these cases we select the PWM that displays the greatest enrichment on either strand according to our scoring method. The result is a histogram of cumulative positive scores for this data set within 1 kb of the TSS position, for each binding element and each strand. (Results are shown for each training set in the Supplemental material.)
A region of positional enrichment is then computed for each element and strand as follows: First, the location of the maximum cumulative score is determined; if this score peak location is not within 100 nt of the TSS, this element–strand combination is discarded. Next, the average of all cumulative scores >1 kb from the TSS is computed and stored as the background average. We then step upstream from the score peak, one nucleotide position at a time, until the cumulative score falls below the background average at least five times. We perform the same procedure stepping downstream from the score peak, and the difference between upstream and downstream stopping locations determines the width of the region of positional enrichment (this width is not allowed to exceed 500 nt). Thus, each region of enrichment is described by score peak location and score peak width.
Building the feature set
We use the binding elements and their regions of positional enrichment to build a set of features describing each training example. For both positive and negative examples, we compute all scores and feature values by treating each location as a putative TSS. For each binding element and strand, we scan the element's PWM over the region of positional enrichment using the local background correction. As illustrated in Figure 3A, we subdivide the region into five center-overlapping windows of equal width, with two flanking windows on either side of the region. For very wide score peaks (score peak width >200 nt), flanking window size is equal to that of the other windows; in all other cases, flanking window size is equal to score peak width. Within each window, positive scores are summed to produce the feature value. Additionally, GC percentage is computed within 100 nt on either side of the putative TSS location. Therefore the total number of features is seven times the number of binding element–strand combinations with defined regions of positional enrichment, plus one for GC content.
Training and classification
We use L1-regularized logistic regression for training and classification of the data in each training set. This method effectively performs automatic feature selection by penalizing the use of more variables; it eliminates the least significant features to the model. Our software pipeline uses the l1_logreg package, an efficient C implementation of the interior-point method for L1-regularized logistic regression (Koh et al. 2007). The L1 penalty parameter defines the degree to which a large number of features is tolerated. We select the L1 parameter as part of the cross-validation process. L1 is chosen to optimize classification performance as measured by auROC (area under the ROC curve), and provides information on variable selection stability. We divide the data set into 10 parts, where each part contains an equal number of positive, negative intergenic, and negative CDS examples. Each cross-validation set contains eight parts for training, one part for validation (selection of the optimal L1 parameter), and one part for independent testing. For each of the 10 cross-validation sets, a logistic regression model is trained for each value of L1 in (0.0001, 0.0002,…, 0.01) and tested on the validation part. l1_logreg is always applied using the feature data standardization option. In all data sets and partitions, we observe that plotting validation auROC for each L1 value results in a smooth curve with a global optimum. We then apply the optimal L1 model to the test part for an independent estimate of performance as measured by auROC.
As a baseline performance comparison for each data set, we also train a näive Bayes classifier using precisely the same features and cross-validation partitions. The vast majority of features are not normally distributed over the positive or negative example sets; their distributions are heavily right-skewed because the region of enrichment for a particular factor will have near-zero scores for many examples, while fewer examples attain a variety of very high scores. We therefore use an empirical näive Bayes implementation (BioMaLL version 0.83, http://www.geneprediction.org/biomall/), where the number of feature discretization bins is optimized over the validation set. Finally, we also retrain the generalized HMM defined in the McPromoter classifier. We perform cross-validation on each data set using the same training, validation, and test partitions for performance comparison. All performance comparison curves and auROC values here and throughout this work are computed using the ROCR package (Sing et al. 2005) for the R statistical computing language. Detailed summaries of all results for each method over all training sets are provided in the Supplemental material.
Evaluation on an independent set of TSS
Tenfold cross-validation on a particular data set using L1-regularized logistic regression results in a set of 10 models, each with its own optimal L1 parameter and performance estimate on a separate test partition. A final model is created by taking an average of these L1 values and training on the entire training data set (all 10 parts) using this consensus value of L1. This results in one model for the annotation-supported and CAGE-only-supported training sets, which we will call the “annotation-supported model” and the “CAGE-only-supported model,” respectively. For each training set, the final model is tested on the independent test set of similarly annotated single-peak TSS. This provides a performance evaluation on data that has not previously been seen by the classifier in cross-validation.
Scanning over genomic sequence: Coding genes
The annotation-supported model is used to classify each position in the region from 4 kb upstream to 4 kb downstream of each TSS in the annotation-supported test set. At each position, the model predicts the probability that this position is a single-peak TSS. Because the subwindows covering each region of enrichment are not a single nucleotide in width, this signal is conservatively smoothed using a median filter with window width equal to that of the smallest feature subwindow (5 nt). A probability peak is called a “hit” for a given threshold if the signal exceeds the threshold value. Moving from upstream to downstream over the scanned region, the signal is considered to enter a peak when it exceeds the threshold, and to exit a peak when it falls below the threshold and remains below for at least 10 nt. We consider a location as a TSS hit if the probability peak contains a TSS. By computing average distance from a TSS hit center to the TSS itself, we assess how well the TSS hits approximate TSS location when scanning the genome.
Figure 5 shows the outcome of this assessment for the annotation-supported model, along with ARTS, CoreBoost, and retrained McPromoter outcomes for comparison. Output from ARTS and CoreBoost was obtained directly from the authors of these programs for 10 kb surrounding each of the 266 regions in the annotation-supported test set. Because CoreBoost requires 1.3 kb of flanking sequence for its predictions, there are 300-nt regions on either end of the 8-kb test regions for which no TSS predictions are made. This technically confers a slight advantage to CoreBoost in Figure 5 comparisons; however, we consider this negligible for practical purposes. Additionally, the feature generation methods of different programs may be affected to varying degrees by large portions of uncalled bases (appearing as Ns) in a sequence; this could potentially be an issue for older assemblies such as mm5 where the mapped CAGE tag data are available. For purposes of equitable comparison, care was taken that no sequence in the test set contained more than 25% Ns within a 1500-nt window on either side of a TSS.
We applied the annotation-supported model to mouse chr 16, and also obtained SVM score predictions at single-nucleotide resolution for the entire chromosome from the authors of the ARTS program. Both programs used the mm5 assembly so that outcomes on CAGE data could be compared. Two data sets were selected for comparison: (1) RefSeq genes as defined by the UCSC mm5 refGene track; and (2) CAGE starts, defined as locations on chr 16 having 10 or more CAGE tags by the UCSC mm5 rikenCageCtssPlus and rikenCageCtssMinus tracks. We then implemented the chunking method for genome-wide performance comparison on the RefSeq gene set exactly as described for ARTS in Sonnenburg et al. (2006). Each annotated start is given a buffer of ±20 nt, which we will call the start region. Chromosome 16 is divided into chunks of size 50 nt (Fig. 7) and 500 nt (Supplemental Fig. 2). Positive chunks are defined as those overlapping a start region, and negative chunks are defined as non-positive chunks overlapping any part of a RefSeq gene. In our case, any chunk (positive or negative) associated with a RefSeq gene whose annotated start was within 500 nt of an annotation-supported training set CAGE TSS is removed from consideration. For a particular chunk size and program, the largest predicted value within a chunk is then taken to represent the chunk. Finally, performance comparison curves and auROC values are computed using these definitions. For the CAGE set, an identical procedure is used (including removal of any chunk that includes a training TSS), except that negative chunks are defined as any chunk of sequence on chr 16 that has not been labeled as positive or removed from consideration.
Scanning over genomic sequence: Noncoding genes
For an additional perspective on noncoding RNAs, we scanned [TSS − 4 kb, TSS + 4 kb] regions from a set of 20 putative human and mouse miRNA primary transcripts. The miRNA putative primary transcript data set used is detailed above in the “Data sets” section. Scans were performed with the annotation-supported model exactly as described for the 8-kb genomic scans over the annotation-supported test set. Using the annotation-supported model, we also scanned the positive and negative regions of the Marson data set (described above in the “Data sets” section). We call a probability peak a “hit” using exactly the same criteria described above for scanning over coding regions. We examine the degree to which our predictions correlate with the Marson data set predictions by computing two types of “performance curves” shown in Figure 8. At a series of probability thresholds ranging from 0.05 to 0.95 in increments of 0.05, we compute the number of hits per kilobase (hit density) separately for the positive regions and for the negative regions. We plot these two hit densities in Figure 8 on the right. For each threshold, we also compute the number of positive regions in the data set that contain at least one hit, and compare the percentage of positive regions hit to the hit density within the negative regions in Figure 8 on the left.
Availability
The annotation-supported classifier is publicly available as an open source command-line tool at http://tools.igsp.duke.edu/generegulation/S-Peaker.
Acknowledgments
We thank Jasmina Ponjavic and Albin Sandelin for providing the single-TSS and twin-TSS CAGE tag clusters, which they defined for use in their analysis of TATA-initiation site spacing. Many thanks to Sören Sonnenburg, Gunnar Rätsch, Xiaoyue Zhao, and Michael Zhang for graciously taking the time to provide us with large-scale output from the ARTS and CoreBoost programs. We are also grateful to Jonathan Schug for many insightful discussions at the beginning stages of this project and for the contributions of his dissertation work, which pointed us toward a large set of TFs with regions of positional enrichment. U.O. and M.M. are funded by NIH grants R01 HG 004065 and P50 GM 081883. A.G.H. and M.M. were supported by a National Science Foundation career award Grant (DBI-0238295). A.G.H. is also supported by a grant from the General Secretary of Research and Technology of Greece (Grant Nr. 340: Program 397).
Footnotes
[Supplemental material is available online at www.genome.org. The annotation-supported classifier is publicly available as an Open Source command-line tool at http://tools.igsp.duke.edu/generegulation/S-Peaker.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.085449.108.
Note added in proof
A study concurrent with ours also examined CAGE-only supported tag clusters, i.e., gene-internal clusters that fall >500 nt from the annotated gene start (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). The authors provided initial evidence for the intriguing possibility that these clusters arise from processed and recapped mRNA transcripts, in agreement with our observation that the sequence properties of this group is distinct from bona fide transcription start sites.
References
- Abeel T., Saeys Y., Rouze P., Van de Peer Y. ProSOM: Core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics. 2008;24:i24–i31. doi: 10.1093/bioinformatics/btn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic V.B., Seah S.H. Dragon Gene Start Finder: An advanced system for finding approximate locations of the start of gene transcriptional units. Genome Res. 2003;13:1923–1929. doi: 10.1101/gr.869803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic V.B., Seah S.H., Chong A., Zhang G., Koh J.L., Brusic V. Dragon Promoter Finder: Recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Bucher P., Trifonov E.N. Compilation and analysis of eukaryotic Pol II promoter sequences. Nucleic Acids Res. 1986;14:10009–10026. doi: 10.1093/nar/14.24.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T.W., Willy P.J., Kutach A.K., Butler J.E., Kadonaga J.T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harb. Symp. Quant. Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cavin Perier R., Junier T., Bucher P. The Eukaryotic Promoter Database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Grosse I., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P.C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Fukuda Y., Kiga K., Sharif J., Hino K., Enomoto Y., Kawamura A., Nakamura K., Takeuchi T., Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Goni J.R., Perez A., Torrents D., Orozco M. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 2007;8:R263. doi: 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Koh K., Kim S.-J., Boyd S. An interior-point method for large-scale l1-regularized logistic regression. J. Mach. Learn. Res. 2007;8:1519–1555. [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Stemmer G., Harbeck S., Niemann H. Stochastic segment models of eukaryotic promoter regions. Pac. Symp. Biocomput. 2000;2000:380–391. doi: 10.1142/9789814447331_0036. [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G.C., Niemann H., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J., Lenhard B., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Schug J. University of Pennsylvania Press; Philadelphia: 2005. “Integrating gene expression signals with bounded collection grammars”. Ph.D. Thesis, [Google Scholar]
- Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Sonnenburg S., Zien A., Ratsch G. ARTS: Accurate recognition of transcription starts in human. Bioinformatics. 2006;22:e472–e480. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- Sonnenburg S., Zien A., Philips P., Ratsch G. POIMs: Positional oligomer importance matrices—understanding support vector machine-based signal detectors. Bioinformatics. 2008;24:i6–i14. doi: 10.1093/bioinformatics/btn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova M., Tiazhelova T., Skoblov M., Baranova A. A comparative analysis of relative occurrence of transcription factor binding sites in vertebrate genomes and gene promoter areas. Bioinformatics. 2005;21:1789–1796. doi: 10.1093/bioinformatics/bti307. [DOI] [PubMed] [Google Scholar]
- Stormo G.D. DNA binding sites: Representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yamashita R., Nakai K., Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y., Brosh R., Buganim Y., Reiner A., Zuk O., Yitzhaky A., Koudritsky M., Rotter V., Domany E. Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site. PLoS One. 2007;2:e807. doi: 10.1371/journal.pone.0000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- Tam W., Dahlberg J.E. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.J., Saldanha A.J., Myers R.M. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hannenhalli S. A mammalian promoter model links cis elements to genetic networks. Biochem. Biophys. Res. Commun. 2006;347:166–177. doi: 10.1016/j.bbrc.2006.06.062. [DOI] [PubMed] [Google Scholar]
- Wang J., Ungar L.H., Tseng H., Hannenhalli S. MetaProm: A neural network based meta-predictor for alternative human promoter prediction. BMC Genomics. 2007;8:374. doi: 10.1186/1471-2164-8-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Yu Y., Fu Y., Foley J., Halees A., Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 2007;17:798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Xuan Z., Zhang M.Q. Boosting with stumps for predicting transcription start sites. Genome Biol. 2007;8:R17. doi: 10.1186/gb-2007-8-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Ruan J., Wang G., Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput. Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]