Abstract
Aims
Alcoholism is a chronic relapsing disorder with an enormous societal impact. Understanding the genetic basis of alcoholism is crucial to characterize individuals' risk and to develop efficacious prevention and treatment strategies.
Methods
We examined the available scientific literature to provide an overview of different approaches that are being integrated increasingly to advance our knowledge of the genetic bases of alcoholism. Examples of genes that have been shown to influence vulnerability to alcoholism and related phenotypes are also discussed.
Results
Genetic factors account for more than 50% of the variance in alcoholism liability. Susceptibility loci for alcoholism include both alcohol-specific genes acting either at the pharmacokinetic or pharmacodynamic levels, as well as loci moderating neuronal pathways such as reward, behavioral control and stress resiliency, that are involved in several psychiatric diseases. In recent years, major progress in gene identification has occurred using intermediate phenotypes such as task-related brain activation that confer the advantage of increased power and the opportunity of exploring the neuronal mechanisms through which genetic variation is translated into behavior. Fundamental to the detection of gene effects is also the understanding of the interplay between genes as well as genes/environment interactions. Whole Genome Association studies represent a unique opportunity to identify alcohol-related loci in hypothesis-free fashion. Finally, genome-wide analyses of transcripts and chromatin remodeling promise an increase in our understanding of the genome function and of the mechanisms through which gene and environment cause diseases.
Conclusions
Although the genetic bases of alcoholism remain largely unknown, there are reasons to think that more genes will be discovered in the future. Multiple and complementary approaches will be required to piece together the mosaic of causation.
Keywords: Alcoholism, GABA, HTT, intermediate phenotypes, MAOA, review, WGA
INTRODUCTION
Alcohol use disorders (AUD), including dependence and abuse, are common and etiologically complex diseases with a 12-month prevalence of 8.6% in the United States [1]. Alcohol is the most widely used addictive drug world-wide. According to the World Health Organization (WHO), there are 2 billion alcohol users (http://www.who.int/substance_abuse/facts/global_burden/en/). The impact on public health in terms of mortality and morbidity [2], and in terms of societal cost, is enormous (http://www.drugabuse.gov/about/welcome/aboutdrugabuse/magnitude). Understanding the genetic basic of alcoholism is a crucial step for the development of efficient prevention strategies and personalized treatments. For this purpose, it is important to identify genes predisposing individuals to alcoholism, genes moderating consequences of alcohol exposure, clinical course and treatment response, mechanisms through which genes exert their effects on behavior and interactions of genes with other genes and with environmental factors.
Although it is clearly known that genetic factors play a role in alcoholism, identification of the specific genes involved has proved challenging. Alcoholism is genetically complex, without a clear pattern of Mendelian inheritance. Major determinants of complexity are likely to include genetic heterogeneity (see Glossary at the end of this paper), heterogeneity at the level of neurobiological vulnerability, polygenicity, phenocopies, gene × environment interaction and incomplete penetrance. In recent years, the collection of large family-based and case–control samples informative for linkage and association, the more refined characterization of phenotypes, the use of statistical methods that take into account the multi-factorial etiology of alcoholism and advances in molecular genetic technologies have overcome many of the limits of early genetic studies. Consequently, several genetic loci that moderate vulnerability to alcoholism have been identified. At the phenotype level, major progress has been made through the use of intermediate phenotypes. In this regard, neuroimaging techniques, such as magnetic resonance, allow for a direct access to neuronal structure and activity in living humans and have been crucial to increase power to identify the effects of genetic variants in risk as well as to provide information on the neurobiological mechanisms through which genes exert their effects on behavior. As mentioned, alcoholism is a multi-factorial disease and several genes, each of small effect, as well as environmental variables, are likely to be involved. As will be discussed, in some instances common functional alleles of small effect have been identified, and in other cases uncommon alleles of strong effect are also known. From what is known, and from the large part of the genetic variance in risk that is still unexplained by genes identified so far, it is clear that methods are needed to identify additional loci whose individual influence is small at the population level. Our ability to detect gene effects is dependent upon the context in which their effects are measured, and it is becoming clear that we cannot ignore that genes act within a complex network that includes other genes, environmental variables and developmental timing. Finally, impressive advances have occurred in molecular genetic technologies. Although it is yet not technically feasible to re-sequence every base or genotype every known polymorphic site, technologies now available enable genome-wide searches for disease-causing genes with the linkage disequilibrium approach and genome-wide studies of gene expression and chromatin modifications that reflect the epigenetic response of the genome to environmental exposures, including the interactions of gene variations with those exposures.
This paper provides an overview of different approaches that are being integrated increasingly to advance our knowledge of the genetic bases of alcoholism. Examples of genes that alter risk for alcoholism and related phenotypes and treatment response are also discussed.
BEHAVIOUR GENETIC STUDIES
Heritability of alcoholism
Family, adoption and twin studies have clearly demonstrated that genetic factors are important in moderating vulnerability to alcoholism. Based on analyses of large, well-characterized cohorts of twins including nearly 10 000 twin pairs, alcoholism is a moderately to highly heritable psychiatric disease, with a heritability of more than 0.5 [3]. For comparison, heritability of generalized anxiety disorder has been estimated to be 0.32 [4] and that of autism has been estimated to be 0.9 [5]. Although early studies suggested that alcohol dependence was more heritable in men compared to women, more recent twin studies, based on larger sample sizes, have shown that genes contribute to vulnerability to alcohol dependence about equally in both sexes [6].
Shared versus unshared etiology
Alcoholism coexists frequently with other addictions, including illicit substance abuse and nicotine dependence more often than would be expected by chance [7]. Such comorbidity between disorders can indicate the existence of etiological factors that are shared (co-causation), but can also reflect inter-causation. Behavior genetic studies can establish the origins of comorbidity and evaluate the extent to which liability to different diseases is shared or unshared. This is conducted by looking at the cross-inheritance of different diseases within families and in twin pairs. Twin and family studies on alcoholism have revealed that genetic factors acting on alcoholism include both alcohol-specific genetic factors as well as genetic factors that are shared with other addictions (for review see Goldman & Bergen) [8]. For example, results from several twin studies [9-13] have detected consistently a considerable overlap in the genetic liability to alcoholism and nicotine dependence, particularly in individuals who drink or smoke heavily. Rates of smoking are declining; however, studies reported during the past 20 years have indicated that as many as 80% of alcohol-dependent individuals are heavy smokers [14,15]. Approximately 50% of the genetic vulnerability to nicotine dependence is shared with alcoholism, whereas 15% of the genetic vulnerability to alcoholism is shared with nicotine dependence [9].
Besides other addictions, alcoholism also coexists frequently with other psychiatric diseases, including both internalizing disorders (e.g. depression and anxiety) as well as externalizing disorders [e.g. antisocial personality disorder (ASPD), conduct disorder (CD) and attention deficit hyperactivity disorder (ADHD)] [1,7,16]. Twin studies reveal consistently the existence of shared genetic influences between alcoholism and externalizing disorders [17-20]. Longitudinal studies have shown that externalizing disorders of childhood such as CD and ADHD are important risk factors for the subsequent development of alcoholism [21]. Evidence from twin studies for shared genetic influences between alcoholism and internalizing disorders are more controversial [18,22,23]. However, longitudinal studies have shown that anxiety disorders such as panic disorder and social phobia predict subsequent alcohol problems in adolescents and young adults [24].
Identification of the specific etiological factors reflecting both the shared and unshared liability to alcoholism requires a redefinition of this disease and a process of simplification and deconstruction of etiology that may be achievable through the use of intermediate phenotypes.
INTERMEDIATE PHENOTYPES
Alcohol abuse and dependence as defined by the Diagnostic and Statistical Manual of Mental Disorders (DSM, American Psychiatric Association) are highly heterogeneous entities. DSM classification is based upon symptom clusters and as such is, to a large extent, an outcome-based classification, not reflecting multiple etiologies of vulnerability. One strategy to discover gene effects in etiologically complex diseases is through deconstruction of complexity into parts that are likely to be less etiologically heterogeneous. Intermediate phenotypes access mediating mechanisms of gene and environmental effects. Heritable intermediate phenotypes that are disease-associated have been termed endophenotypes, and these include biochemical, endocrinological, neuroanatomical, cognitive and neuropsychological parameters [25].
Several intermediate phenotypes have been linked specifically to alcoholism, perhaps reflecting alcohol-specific genetic liability. These include alcohol-induced flushing, which is a protective intermediate phenotype, and low response to the effects of alcohol, which is an endophenotype predictive of risk. The genetic origins of alcohol-induced flushing are discussed later. The level of response to alcohol is believed to reflect mainly pharmacodynamic variation in response [26]. A low response to alcohol predicts increased risk of developing alcohol use disorders [27-29] and has been associated with genetic variation in the serotonin transporter gene and in the gene encoding the subunit α6 of the γ-aminobutyric acid receptor A (GABRA6) [30].
Other intermediate phenotypes predict diatheses that include alcoholism as well as other psychiatric diseases. Relevant phenotypes in this regard include electrophysiological, psychological, neuroendocrinological and, more recently, neuroimaging phenotypes. Neuroimaging provides access to the neuronal mechanisms underlying emotion, reward and craving and therefore represents an extraordinary tool to link genes to the neuronal pathways that produce behaviors (for review see Meyer-Lindenberg & Weinberger [31]; Xu et al. [32]). For example, amygdala activation after exposure to stressful stimuli predicts anxiety and captures inter-individual differences in emotional response and stress resiliency [33]. On the other hand, activation of the pre-frontal cortex during working memory performance is used to evaluate pre-frontal cognitive function which is impaired in several psychiatric diseases.
Several of the most relevant intermediate phenotypes for alcoholism are summarized in Table 1, which also includes the functional loci that have been linked consistently to such intermediate phenotypes.
Table 1.
Gene effects on intermediate phenotypes for alcoholism.
| Domain | Mode of assessment | Locus |
|---|---|---|
| Attention/dyscontrol | Low amplitude P300 | COMT Val158Met |
| Frontal cognitive tasks | MAOA–LPR | |
| Task-specific MRI | ||
| Questionnaires | ||
| Pharmacokinetics | Flushing questionnaires | ALDH2 Glu487Lys |
| Metabolite levels | ADH1B His47Arg | |
| Level of response | Alcohol challenge/clamp | |
| Questionnaire | ||
| Treatment response | Response in clinical trials | OPRM1 Asn40Asp |
| Anxiety/dyscontrol | α, β resting EEG power | |
| Mesolimbic reward | Task-specific MRI, PET | |
| Stress/resiliency | Task-related MRI, PET | HTTLPR (La, S, Lg) |
| Endocrine responses | COMT Val158Met | |
| Questionnaires | NPY | |
| Brain volume/structure | Structural MRI | BDNF Met66Val |
| MAOA–LPR | ||
| HTTLPR | ||
| COMT Val158Met |
FINDING GENES FOR ALCOHOLISM
Two main strategies have been used and are integrated increasingly to identify genetic variations influencing alcoholism vulnerability and other alcoholism-related phenomena: the candidate gene approach and the genome-wide linkage approach. In the former, genes known to influence processes involved in the pathogenesis or treatment of alcoholism are selected. In the latter the whole genome is interrogated simultaneously in a hypothesis-free fashion. A point of integration between the methods is the study of candidate genes located in chromosome regions implicated by genome-wide scans.
Candidate genes
Alcoholism is a disease of complex etiology. The process of addiction involves cellular molecular networks in which hundreds of genes are involved. As a result, many candidate genes for alcoholism have been postulated. So far, only a few functional loci moderating vulnerability to alcoholism have been identified. Some of these genes (e.g. catechol-O-methyltransferase: COMT) act on neurobiological mechanisms such as reward, behavioral control and stress resiliency that are involved in alcoholism as well as in other psychiatric diseases. In contrast, other genes (e.g. alcohol-metabolizing genes) are likely to reflect alcohol-specific liability. The use of intermediate phenotypes and studies taking into account the interaction between genes and the environment have been critical to identify and understand these loci, some of which are discussed below (see also Table 1).
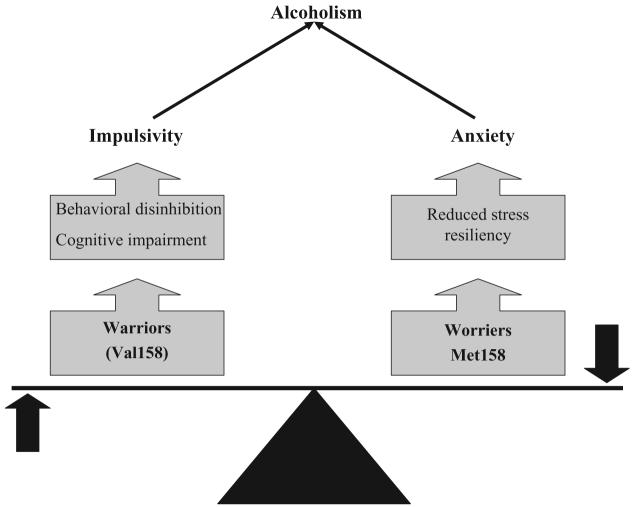
COMT: warriors versus worriers
Different pathways can lead to the same outcome in different individuals. For this reason, alternative alleles at the same locus may increase vulnerability to alcoholism through different effects on brain function and behavior, and these risk effects may even be opposing. COMT is an enzyme that metabolizes dopamine (DA), norepinephrine (NE) and other catecholamines. COMT plays an important role in the regulation of dopamine levels in the pre-frontal cortex because of the paucity of dopamine transporter in this region [34,35]. COMT knock-out mice have increased levels of dopamine dramatically in the pre-frontal cortex [36,37]. Val158Met is a common functional polymorphism located in the coding sequence of the human COMT gene in which the amino acid at codon 158 can be either valine (Val) or methionine (Met). This missense variant alters enzyme activity by affecting enzyme stability [38,39]. The Val158 allele is three- to fourfold more active than Met158 [40] and the alleles act co-dominantly [41]. Therefore, Val158 is predicted to lower dopamine level in the frontal cortex. In line with this idea, the Val158 allele has been linked to inefficient frontal lobe function evaluated with different methodologies [42-44]. On the other hand, the Met158 allele, although associated with better cognitive performance, is associated with decreased stress resiliency and increased anxiety. This allele has been associated with increased anxiety among women in two independent populations [45], with an increased pain–stress response and a lower pain threshold [46,47], and increased amygdala reactivity to unpleasant stimuli [48]. From an evolutionary perspective, balanced selection both against and for the Met and Val alleles has probably maintained these alleles across populations and generations. A heuristic for the phenomenon of maintenance on the basis of counterbalancing selection for stress resiliency (Val) and cognition (Met) is the ‘warrior’ (Val158) versus ‘worrier’ (Met158) model (Fig. 1) [49]. In certain addicted populations, for example polysubstance abusers [50], Val158 is associated with addiction, probably through the externalizing domain. On the other hand, in addicted populations with high frequencies of internalizing disorders such as late-onset alcoholics in Finland [51] and Finnish social drinkers [52], increased risk appears to be conferred by the Met158 allele.
Figure 1.
Catechol-O-methyltransferase (COMT): the warrior (Val158) versus worrier (Met158) model
Alcohol metabolizing enzymes: ALDH2 and ADH1B
In contrast to the complex and not completely understood effects of alcohol on the central nervous system, the hepatic metabolism of alcoholism is well understood and the most convincing examples of alcoholism-related genes are the alcohol dehydrogenase IB (ADH1B) and aldehyde dehydrogenase 2 (ALDH2) genes that encode for two enzymes catalyzing consecutive steps in alcohol degradation. ADH metabolizes ethanol to acetaldehyde, a toxic intermediate, which is then metabolized to acetate by ALDH. The most important functional loci at these genes are the His47Arg polymorphism in the ADH1B gene, where Arg47 is a superactive variant acting in co-dominant fashion, and the ALDH2 Glu487Lys, in which the Lys487 allele inactivates ALDH2 dominantly. Either higher activity of ADH1B or lower activity of ALDH2 lead to accumulation of acetaldehyde following alcohol assumption which, in turn, causes an unpleasant and aversive response, termed the flushing reaction. The flushing reaction is characterized by facial flashing, headache, hypotension, palpitations, tachycardia, nausea and vomiting. Interestingly, the genotype-determined flushing reaction is equivalent to the effects of disulfiram (a drug used to prevent relapse), and certain antiprotozoal drugs, including metronidazole, that inhibit ALDH. In Chinese and Japanese populations where both His47 and Lys487 are highly abundant, and in Jewish populations where His47 is abundant, many individuals carry genotypes that are protective against the development of alcoholism. Both polymorphisms have been associated with alcohol dependence [53] as well as with risk of developing cancers of the oropharynx and esophagus [54]. The protective effect seems to vary across environments [55] and shows genotype–genotype additivity [56].
Predicting treatment response: OPRM1
Currently available medications for the treatment of alcoholism have limited efficacy. Therefore, the identification of genetic variation predicting treatment response would be useful for improving the clinical management of patients by personalizing treatment. Naltrexone, a μ-opioid receptor antagonist, is one of three pharmacotherapies currently approved for the maintenance of abstinence in alcoholism, the others being acamprosate and disulfiram. Results from clinical trials have shown moderate efficacy of naltrexone and efforts to identify genetic variants that moderate this effect have focused upon the gene encoding the μ-opioid receptor 1 (OPRM1). A functional polymorphism within the coding sequence of this gene (Asn40Asp) has been described [57]. In cell culture, μ-opioid receptors encoded by the Asp40 variant bind β-endorphin and activate G-protein-coupled protein potassium ion channels with three times greater potency than receptors encoded by the Asn40 variant [57].This polymorphism has been linked inconsistently to alcoholism and other addictions [57-61]. Recent evidence supports that this locus predicts response to naltrexone in clinical trials [62,63] with Asp40 being associated with better response, although another study was negative [64].
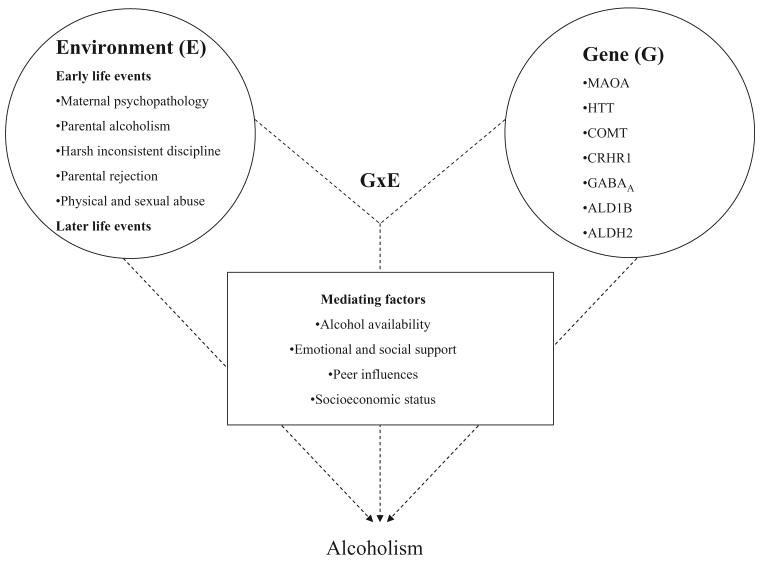
Gene × environment interaction: MAOA and HTT
Alcoholism occurs due to individual choice, environmental and genetic determinants and interactions between factors within these three domains of causation (Fig. 2). Environmental factors include alcohol availability, parental attitudes, peer pressure, underage drinking and childhood maltreatment. From this perspective the ability to detect gene effects is dependent upon context and timing [65]. Severe childhood stress and neglect increase vulnerability to alcoholism but also several alcoholism-related psychiatric diseases including ASPD, CD, anxiety and depression, with the risks of these common diseases being elevated several-fold in the stress-exposed [66,67]. However, not all subjects exposed to environmental stressors develop alcoholism or other psychiatric diseases, indicating that people differ widely in stress resiliency. Genetic variation is likely to account partially for much of the differential vulnerability. Gene × environment interaction (G × E) occurs when the effect of exposure to an environmental factor on a person's health is conditional upon his or her genotype (for review see Caspi & Moffitt [68]). The knowledge that differences in DNA sequence moderate individuals in their resiliency or vulnerability to environmental pathogens is well known for several complex diseases, including psychiatric diseases as well as cancer, diabetes and cardiovascular, infective and immune diseases, etc. G × E effects within the context of psychiatric diseases have been described for several genes including monoamine oxidase A (MAOA) [69], the serotonin transporter (HTT) [70], COMT [71], the corticotropin releasing hormones receptor 1 gene [72] and the dopamine transporter [73]. So far, results appear to be particularly robust because of validation from different perspectives for MAOA and HTT.
Figure 2.
Main and interactive effects of genetic and environmental risk factors for alcoholism
MAOA is an X-linked gene encoding monoamine oxidase A, a mitochondrial enzyme that metabolizes monoamine neurotransmitters including NE, DA and serotonin (5-HT). MAOA knock-out mice have higher levels of DA, 5-HT and NE, and manifest increased aggressive behavior and stress reactivity [74]. In the human, different MAOA genetic variants impair MAOA activity to different degrees and the reduction in enzyme activity appears to parallel the effect on behavior. In 1993, Brunner et al. [75] reported a Dutch family in which eight males were affected by a syndrome characterized by borderline mental retardation and impulsive behavior including impulsive aggression, arson, attempted rape, fighting and exhibitionism. The syndrome is due to a stop-codon variant in the eighth exon of MAOA leading to complete and selective deficiency of MAOA activity. More recently a common MAOA polymorphism influencing MAOA transcription was discovered [76]. This locus, termed the MAOA-linked polymorphic region (MAOA–LPR), is a variable number tandem repeat (VNTR) located approximately 1.2 kb upstream from the MAOA start codon and within the gene's transcriptional control region [76,77]. Alleles at this VNTR have a different number of copies of a 30-base pairs (bp) repeated sequence with the three and four repeats alleles being by far the most common. Alleles with four repeats are transcribed more efficiently than alleles with three copies of the repeat and are therefore associated with higher MAOA activity [76]. In a large longitudinal cohort of boys, Caspi et al. [69] found that MAOA–LPR moderates the effect of childhood maltreatment on vulnerability to develop antisocial behavior. In this study maltreated boys with the low activity genotype were more likely to develop antisocial problems later in life than boys with the high activity genotype. Several studies have tried to replicate this finding, and a recent meta-analysis revealed a significant pooled effect [78]. More recently it has been shown that the MAOA × environment interaction described for males is also valid for women. In a sample of Native American women with high rates of childhood sexual abuse (CSA), it was shown that the effect of CSA on risk of developing alcoholism and ASPD was contingent upon the MAOA–LPR genotype [79]. Sexually abused women who were homozygous for the low-activity MAOA–LPR allele had higher rates of alcoholism, particularly antisocial alcoholism, compared with women who were homozygous for the high-activity allele. Heterozygous women displayed an intermediate risk pattern. In contrast, there was no relationship between alcoholism/antisocial behavior and MAOA–LPR genotype in women who had not been sexually abused.
Genes can also influence the probability of stress exposure, a phenomenon known as G × E correlation. G × E correlation results in a contamination of genetic and environmental effects leading to difficulties in interpretating G × E interaction. In this regard, animal models are extremely useful because they offer the possibility of controlling environmental influences enhancing gene effects and eliminating confounds that might underlie correlations. Macaques with histories of stress exposure, such as maternal separation early after birth, show increased alcohol consumption, higher impulsive aggression and incompetent social behavior and serotonin dysfunction, and consume more alcohol and have increased behavioral and endocrine responsivity to stress compared to mother-reared animals (for review see Barr et al. [80]). Consistent with studies on humans, a polymorphism of MAOA in the rhesus macaque that is orthologous to the human MAOA–LPR has been associated with aggression, and this association depended on whether or not the monkey had been separated from the mother. The low-activity genotype was associated with higher aggression only in mother-reared male monkeys [81].
The effect of MAOA on the hippocampus, a brain region which is involved in the processing of emotional experience, may underlie the interaction between MAOA and childhood trauma. Carriers of the low activity variant of MAOA–LPR display hyperactivation of the hippocampus and amygdala during the retrieval of negatively valenced emotional material but not during the retrieval of neutral material [82]. Therefore, the increased sensitivity to adverse experiences of carriers of the low activity MAOA genotype might be due to their impaired ability to extinguish adverse memories and conditioned fears. Other important mechanisms that are likely to underlie G × E interactions include hormonal effects. Steroid hormones including estrogens, progesterones, androgens and glucocorticoids modulate MAOA expression [83,84]. Both glucorticoids and androgens increase MAOA expression through response elements that are located within the MAOA promoter [83]. Recently, in a sample of criminal alcoholics, it has been shown that the effect of testosterone on aggression and alcoholism is contingent upon MAOA–LPR genotype [85]. Indeed, a positive correlation between testosterone level and antisocial behavior was found only among carriers of the low-activity allele, suggesting that this VNTR might influence the effect of testosterone on the MAOA promoter.
HTT is responsible for serotonin re-uptake and is a key regulator of serotonin availability in the synaptic cleft. In two unrelated families, an uncommon coding region mutation, Ile425Val was found in subjects with treatment-resistant obsessive compulsive disorder and carriers also had other diseases including alcoholism, Asperger's syndrome, social phobia, anorexia nervosa and tic disorder [86]. Additional rare HTT missense variants have been described in recently autism [87]. A common polymorphism of the HTT promoter region (5-HTTLPR) affects expression, with the major alleles involving 16 (L) or 14 (S) copies of a 20–23 bp imperfect repeated sequence [87]. Recently, it has been shown that 5-HTTLPR is actually a functionally tri-allelic locus due to a relatively common, functional A > G substitution within the L allele [88]. The low transcribing s allele has been associated inconsistently with anxiety and alcoholism. However, the effect of this allele on behavior appears stronger if stress exposure is taken into account. 5-HTTLPR moderates the impact of stressful life events on risk of depression and suicide [70,89,90]. Carriers of the low transcribing s allele exhibit more depression and suicidality following stressful life events compared to individuals with two copies of the L allele [70]. Furthermore, 5-HTTLPR has been shown to moderate the functions of brain regions, such as the amygdala, that are critical in emotional regulation and response to environmental changes. Carriers of the low-activity allele display increased amygdala reactivity to fearful stimuli [33], reduced amygdala volume [91] and enhanced functional coupling between the amygdala and the ventromedial pre-frontal cortex [92]. Closer to the function of the gene, an effect of 5-HTTLPR genotype on transporter expression in brain in vivo has been reported in some studies [93] but not in others [94]. Consistent with findings in humans, the macaque orthologous rs-5HTTLPR polymorphism was shown to influence alcohol consumption and stress response, depending on rearing conditions. Carriers of the low expressed genotype who were separated from their mother at an early age displayed higher stress reactivity and ethanol preference [95]. The combined effect of rh-HTTLPR and environment on stress reactivity suggests that the influence of HTTLPR on behavior might be traced to altered regulation of the hypothalamic–pituitary–adrenal (HPA) axis.
Genome-wide scans
Genome-wide scans, including whole genome linkage and whole genome association (WGA), allow the hypothesis-free mapping of disease-causing loci within the genome. Chromosome regions and genes implicated by these studies, as well as potential strengths and limitations of WGA methodologies, are discussed below.
Whole genome linkage
In whole genome linkage studies a panel of polymorphisms is tested for meiotic linkage to a disease (marker-disease coupling or repulsion) in family-based samples, identifying chromosome regions that are shared more often among phenotypically concordant relatives compared to phenotypically discordant family members. The implicated chromosomal regions are usually broad, for example greater than 10 megabases. Therefore, a more refined search for candidate genes within the region of interest is subsequently conducted. To perform whole-genome linkage analysis for alcoholism, several large family-based data sets have been collected. These include the Collaborative Study on the Genetics of Alcoholism (COGA) [96], the Roscommon study of Irish families [97]; a sample of multiplex families collected in the Pittsburg area [98]; and samples collected from relatively isolated populations including Native Americans [99-100] and Finns [101, Enoch et al. unpublished]. Such isolated populations, and large families within them, are likely to confer the advantage of reduced genetic heterogeneity. A non-exhaustive list of convergent findings across studies includes a region on chromosome 4q, that contains the alcohol dehydrogenase (ADH) gene cluster [96,97,99,100], and a chromosome 4p region near the centromere containing a γ-aminobutyric acid receptor (GABAA) gene cluster [96,99]. In the COGA sample there was also evidence for linkage to chromosomes 1 and 7, and to chromosome 2 at the location of an opioid receptor gene [96]. A region on chromosome 1 was linked to alcoholism and affective disorder in the COGA data set [102], supporting further the existence of a genetic overlap between alcoholism and internalizing disorders. Linkage analyses have also been conducted with intermediate phenotypes for alcoholism including low response to alcohol [29], neurophysiological endophenotypes such as P300 [103] and reduced alpha power (Enoch et al., unpublished), and chromosome regions identified by these studies overlap partially with those reported for alcoholism.
The GABAA receptor: γ-aminobutyric acid (GABA) is the primary inhibitory neurotransmitter in the central nervous system. GABAA receptor-mediated chloride currents into neurons are facilitated by various drugs including ethanol, benzodiazepines and barbiturates. Several lines of evidence suggest that GABA is involved in many effects of alcohol including tolerance, dependence and cross-tolerance to benzodiazepines and barbiturates. A series of mouse ethanol-related behaviors, including preference, withdrawal severity and sedation sensitivity, map quantitative trait loci (QTL) regions where GABAA receptor-gene clusters are located [104,105]. In the rat, a Arg100Gln missense variant located in the GABAA α6 subunit gene (GABRA6) was associated with variation in ethanol and benzodiazepine sensitivity [103]. In humans, AUD has been linked to both the chromosome 4 [99,106] and chromosome 5 [107] GABA clusters. Linkage signals appear to derive from GABRA6 on chromosome 5 [107] and the GABAA α2 (GABRA2) [106,108] on chromosome 4. A human variant of GABRA6 (Pro385Ser), which is located in the chromosome 5 cluster, was associated with sensitivity to alcohol [30] and benzodiazepine [109].
Whole-genome association
Large-scale genotyping techniques have recently been made available and genome-wide analyses for several complex diseases including amyotrophic lateral sclerosis [110], Type 2 diabetes [111], neuroticism [112], obesity [113] and human immunodeficiency virus 1 (HIV-1) [114] have been run within large data sets of unrelated individuals using dense panels including more than 500 000 polymorphisms. These WGA studies have the advantage of increased power for detecting effects of relatively common alleles (>0.05) and more refined localization of signals to smaller chromosome regions compared to family-based linkage analyses, which have a reciprocal advantage of being powerful for detecting effects of rare and uncommon alleles that are present in only a small proportion of probands and their families.
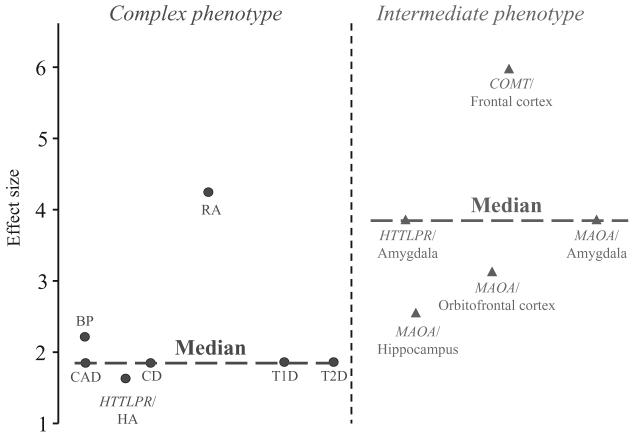
A WGA scan has been run recently in a sample of unrelated alcohol-dependent (n = 120) and control (n = 160) individuals sampled from the COGA pedigrees [115]. This study identified several candidate genes that might moderate vulnerability to alcoholism and whose products are involved in cellular signaling, gene regulation, development, cell adhesion and Mendelian disorders. However, these findings are weakened by the small sample size and the consequent lack of power to identify exhaustively common alcoholism-causing alleles. Indeed, the typical effect size of genetic variations acting on complex diseases is small [116] and the median sample size required for a WGA study to have 90% power even at an alpha level of 0.05 has been estimated to be as high as 15 000 participants [117]. A WGA study searching for genes influencing seven complex disorders, including bipolar disorder, have been completed recently in a sample of 14 000 cases (2000 cases per disease) and 3000 controls and can be considered as illustrative of the general potential of WGA (Welcome Trust study [118]). For the seven diseases studied in the Wellcome Trust study, a total of 24 genome-wide significant signals were found. The maximum genotype-specific odds ratios for significant disease loci ranged from 1.5 to 18.5, with a median of 1.9 (see Fig. 3). Furthermore, the disease loci found accounted for only a small portion of the genetic variance of these diseases (consistent with the common allele/moderate effect model). For example, the 16p12 locus found for bipolar disorder with an odds ratio of approximately 2.1 accounts, perhaps, for 4% of the genetic risk for bipolar disorder at the general population level, although its impact might be higher within certain families. WGA in large case–controls data sets has not yet been reported for alcoholism. However, this approach, even when large case–control samples will be available for alcoholism, will not enable identification of the complete list of genetic loci involved in alcoholism. Another important strategy that can be used to increase power of WGA studies is through the use of intermediate phenotypes. Indeed, effect sizes of genetic variations acting on intermediate phenotypes more proximal to gene action and including measures such as task-related brain activation appear to be larger than effects on complex disease phenotypes [116]. In Fig. 3 effect sizes reported for well-characterized functional loci including MAOA–LPR [82], 5-HTTLPR [33] and COMTVal158Met [42] are compared to odds ratios reported for the complex diseases studied by the Welcome Trust and to the odds ratio reported by a large meta-analysis [119] evaluating the effect of 5-HTTLPR on harm avoidance, which can also be considered a complex phenotype (Fig. 3, see also Goldman & Ducci [116]). A whole-genome scan on intermediate phenotypes relevant for alcoholism is an alternative and potentially very powerful approach in the search for the alcoholism-causing genes.
Figure 3.
Genotype-specific effect size (odds ratios) for complex versus intermediate phenotypes. CAD = coronary artery disease; T1D = Type 1 diabetes; T2D = Type 2 diabetes; CD = Crohn's disease; BP = bipolar disorder; RA = rheumatoid arthritis; HA = harm avoidance; MAOA = monoamine oxidase A; HTT = serotonin transporter; LPR = linked polymorphic region; COMT = catechol-O-methyltransferase
As well as power and the false-negative issue, another important concern of WGA analyses is false positive findings due to the extremely high number of markers evaluated and statistical tests performed. In this regard, particular attention has been placed on the threshold used to defined statistical significance and on replication in independent populations. Furthermore, current marker panels used in WGA are composed of single nucleotide polymorphisms (SNPs) and do not include other types of genetic variation such as copy number variations (CNVs), whose importance in disease susceptibility is being recognized increasingly. In this regard, recent whole-genome analysis has revealed that both CNVs and SNPs can alter patterns of mRNA expression, but the SNP variation does not track most of the effects of CNVs, indicating that both types of polymorphisms should be interrogated to perform a comprehensive genome-wide evaluation of the effects of genomic variation on disease vulnerability [120]. Finally, the ultimate validation of WGA results will be represented by the identification of the specific causing variants within the disease-associated genomic regions.
UNDERSTANDING GENE FUNCTION
Our understanding of the protein-coding function of the genome is still limited, and we have even less understanding of non-protein coding transcripts and genome elements regulating gene expression. However, new technologies including gene expression profiling using microarrays and methods to evaluate chromatin remodeling are available, enabling a detailed exploration of the genome function and promising an increase of our knowledge of mechanisms through which genetic variation influence protein level/function and environment interacts with the genome.
DNA microarray
DNA microarrays are used for the simultaneous measurement of the expression of thousands of genes and represent a tool that can be used to identify genes that are expressed differentially within QTL linked previously to specific alcohol-related phenotypes using inbred or selected lines of animals. Indeed, several strains of animals have been generated that display differences in alcohol-related behaviors, such as animals with high or low sensitivity to various effect of alcohol or with high or low preference for alcohol [104]. Kerns et al. [121] studied differences in basal or ethanol-responsive gene expression in the brains of two strains of mice that differ markedly in a number of behavioral responses to ethanol. Several genes that were expressed differentially in the two strains as well as several ethanol-regulated genes were found within brain regions that are involved in reward, including the nucleus accumbens, pre-frontal cortex and ventral tegmental area. Multiplex evaluation of gene expression by microrrays enables the exploration of gene networks. Tabakoff et al. [122] compared the mRNA expression profiles of mouse strains displaying marked differences in acute tolerance to alcohol and results from this study indicate the importance of a signal transduction cascade that involves the glutamatergic pathway.
Epigenetics
Complex epigenetic mechanisms that regulate gene activity without altering DNA code have been shown to produce long-lasting changes in gene expression essential to development and cellular differentiation and to adaptation to environmental changes. These mechanisms, that include DNA methylation, post-translational covalent modifications of histones, nucleosome sliding and nucleosome and histone substitution, cause modifications of chromatin conformation which, in turn, regulate gene expression (for review see Tsankova et al. [123]). For example, the amount of DNA methylation in promoter regions correlates with gene inactivation. Chromatin remodeling can be studied at the single locus level; however, methods such as the chromatin immunoprecipitation in combination with DNA sequencing (ChIP-seq) allow for high-resolution genome-wide analysis. Alcohol exposure has been shown to cause changes in chromatin structure in rat brain [124,125]. Alpha synuclein (SNCA) is a gene that maps to a QTL for alcohol preference, and expression of alpha synuclein is increased in different brain areas in rats displaying alcohol preference [126]. Recently, an increase in alpha synuclein promoter DNA methylation has been found in patients with alcoholism [127], and genetic variation within the human SNCA gene has been linked to alcohol craving [128]. DNA methylation also appears to regulate expression of alcohol dehydrogenase [129] and HTT [130] genes.
CONCLUSION
Although most of the genetic determinants of alcoholism remain to be discovered there are reasons for optimism. In recent years a technological revolution has occurred producing a shift from single-locus studies to genome-wide searches. The genomes trascriptome, epigenome and, to some extent, proteome, can now be assessed at a level of detail that was previously inconceivable. Innovations are required at the analytical level to integrate and validate the massive amounts of data produced by these new technologies and different approaches. However, these tools promise to increase our understanding of the mechanisms by which genetic variation alters molecular function and predisposes individuals to alcoholism and other diseases.
GLOSSARY
- Genetic heterogeneity
a model of genetic determinism in which different alleles lead to the same phenotype in different individuals, but an individual allele can suffice to produce the phenotype.
- Polygenicity
a model of genetic determinism in which many alleles function in combination to produce a phenotype.
- Phenocopies
a phenotype of environmental origin that mimics a phenotype of genetic origin.
- Penetrance
the probability of expressing a phenotype that is determined by a genotype.
- Intermediate phenotype
mechanism-related manifestation of a more etiologically complex phenotype.
- Allele
alternative form of a genetic locus.
- Epigenetics
biological mechanisms that regulate gene activity without altering DNA sequence, and that can result in long-lasting changes in gene expression.
- Genetic locus
a particular location on a chromosome.
- Microarray
a genetic analysis tool containing a large number of features such that many different DNAs, RNAs or proteins can be measured simultaneously.
- Inbred lines
strains by repeated matings of genetically related individuals such that most or all genetic variation has been eliminated.
- Chromatin remodeling
dynamic changes in DNA/protein structure altering the packaging and function of DNA.
- Quantitative trait locus (QTL)
region of a chromosome that contains one or more genetic loci that contribute to a phenotypic difference.
References
- 1.Grant BF, Stinson FS, Dawson DA, Chou SP, Dufour MC, Compton W, et al. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2004;61:807–16. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- 2.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 3.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 4.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158:1568–78. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 5.Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:472–86. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- 6.Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–96. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 7.Kessler RC, Crum RM, Warner LA, Nelson CB, Schulenberg J, Anthony JC. Lifetime co-occurrence of DSM-III-R alcohol abuse and dependence with other psychiatric disorders in the National Comorbidity Survey. Arch Gen Psychiatry. 1997;54:313–21. doi: 10.1001/archpsyc.1997.01830160031005. [DOI] [PubMed] [Google Scholar]
- 8.Goldman D, Bergen A. General and specific inheritance of substance abuse and alcoholism. Arch Gen Psychiatry. 1998;55:964–5. doi: 10.1001/archpsyc.55.11.964. [DOI] [PubMed] [Google Scholar]
- 9.Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–90. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- 10.Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 11.Hopfer CJ, Stallings MC, Hewitt JK. Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. J Stud Alcohol. 2001;62:717–23. doi: 10.15288/jsa.2001.62.717. [DOI] [PubMed] [Google Scholar]
- 12.True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–61. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- 13.Koopmans JR, van Doornen LJ, Boomsma DI. Association between alcohol use and smoking in adolescent and young adult twins: a bivariate genetic analysis. Alcohol Clin Exp Res. 1997;21:537–46. [PubMed] [Google Scholar]
- 14.Burling TA, Ziff DC. Tobacco smoking: a comparison between alcohol and drug abuse inpatients. Addict Behav. 1988;13:185–90. doi: 10.1016/0306-4603(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 15.DiFranza JR, Guerrera MP. Alcoholism and smoking. J Stud Alcohol. 1990;51:130–5. doi: 10.15288/jsa.1990.51.130. [DOI] [PubMed] [Google Scholar]
- 16.Grant BF, Stinson FS, Dawson DA, Chou SP, Ruan WJ, Pickering RP. Co-occurrence of 12-month alcohol and drug use disorders and personality disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2004;61:361–8. doi: 10.1001/archpsyc.61.4.361. [DOI] [PubMed] [Google Scholar]
- 17.Fu Q, Heath AC, Bucholz KK, Nelson E, Goldberg J, Lyons MJ, et al. Shared genetic risk of major depression, alcohol dependence, and marijuana dependence: contribution of antisocial personality disorder in men. Arch Gen Psychiatry. 2002;59:1125–32. doi: 10.1001/archpsyc.59.12.1125. [DOI] [PubMed] [Google Scholar]
- 18.Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–37. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 19.Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–24. [PubMed] [Google Scholar]
- 20.Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96:684–95. [PubMed] [Google Scholar]
- 21.Sher KJ, Bartholow BD, Wood MD. Personality and substance use disorders: a prospective study. J Consult Clin Psychol. 2000;68:818–29. [PubMed] [Google Scholar]
- 22.Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. Alcoholism and major depression in women. A twin study of the causes of comorbidity. Arch Gen Psychiatry. 1993;50:690–8. doi: 10.1001/archpsyc.1993.01820210024003. [DOI] [PubMed] [Google Scholar]
- 23.Prescott CA, Aggen SH, Kendler KS. Sex-specific genetic influences on the comorbidity of alcoholism and major depression in a population-based sample of US twins. Arch Gen Psychiatry. 2000;57:803–11. doi: 10.1001/archpsyc.57.8.803. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann P, Wittchen HU, Hofler M, Pfister H, Kessler RC, Lieb R. Primary anxiety disorders and the development of subsequent alcohol use disorders: a 4-year community study of adolescents and young adults. Psychol Med. 2003;33:1211–22. doi: 10.1017/s0033291703008158. [DOI] [PubMed] [Google Scholar]
- 25.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 26.Schuckit MA. Biological, psychological and environmental predictors of the alcoholism risk: a longitudinal study. J Stud Alcohol. 1998;59:485–94. doi: 10.15288/jsa.1998.59.485. [DOI] [PubMed] [Google Scholar]
- 27.Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–81. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez LA, Wilson JR, Nagoshi CT. Does psychomotor sensitivity to alcohol predict subsequent alcohol use? Alcohol Clin Exp Res. 1993;17:155–61. doi: 10.1111/j.1530-0277.1993.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuckit MA, Smith TL, Kalmijn J, Tsuang J, Hesselbrock V, Bucholz K. Response to alcohol in daughters of alcoholics: a pilot study and a comparison with sons of alcoholics. Alcohol Alcohol. 2000;35:242–8. doi: 10.1093/alcalc/35.3.242. [DOI] [PubMed] [Google Scholar]
- 30.Hu X, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An expanded evaluation of the relationship of four alleles to the level of response to alcohol and the alcoholism risk. Alcohol Clin Exp Res. 2005;29:8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- 31.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–27. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 32.Xu K, Ernst M, Goldman D. Imaging genomics applied to anxiety, stress response, and resiliency. Neuroinformatics. 2006;4:51–64. doi: 10.1385/NI:4:1:51. [DOI] [PubMed] [Google Scholar]
- 33.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–3. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 34.Lewis DA, Melchitzky DS, Sesack SR, Whitehead RE, Auh S, Sampson A. Dopamine transporter immunore-activity in monkey cerebral cortex: regional, laminar, and ultrastructural localization. J Comp Neurol. 2001;432:119–36. doi: 10.1002/cne.1092. [DOI] [PubMed] [Google Scholar]
- 35.Mazei MS, Pluto CP, Kirkbride B, Pehek EA. Effects of catecholamine uptake blockers in the caudate-putamen and subregions of the medial prefrontal cortex of the rat. Brain Res. 2002;936:58–67. doi: 10.1016/s0006-8993(02)02542-8. [DOI] [PubMed] [Google Scholar]
- 36.Gogos JA, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, et al. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci USA. 1998;95:9991–6. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yavich L, Forsberg MM, Karayiorgou M, Gogos JA, Mannisto PT. Site-specific role of catechol-O-methyltransferase in dopamine overflow within prefrontal cortex and dorsal striatum. J Neurosci. 2007;27:10196–209. doi: 10.1523/JNEUROSCI.0665-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scanlon PD, Raymond FA, Weinshilboum RM. Catechol-O-methyltransferase: thermolabile enzyme in erythrocytes of subjects homozygous for allele for low activity. Science. 1979;203:63–5. doi: 10.1126/science.758679. [DOI] [PubMed] [Google Scholar]
- 39.Weinshilboum R, Dunnette J. Thermal stability and the biochemical genetics of erythrocyte catechol-O-methyltransferase and plasma dopamine-beta-hydroxylase. Clin Genet. 1981;19:426–37. doi: 10.1111/j.1399-0004.1981.tb00740.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–21. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 42.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–22. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malhotra AK, Kestler LJ, Mazzanti C, Bates JA, Goldberg T, Goldman D. A functional polymorphism in the COMT gene and performance on a test of prefrontal cognition. Am J Psychiatry. 2002;159:652–4. doi: 10.1176/appi.ajp.159.4.652. [DOI] [PubMed] [Google Scholar]
- 44.Goldberg TE, Egan MF, Gscheidle T, Coppola R, Weickert T, Kolachana BS, et al. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889–96. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- 45.Enoch MA, Xu K, Ferro E, Harris CR, Goldman D. Genetic origins of anxiety in women: a role for a functional catechol-O-methyltransferase polymorphism. Psychiatr Genet. 2003;13:33–41. doi: 10.1097/00041444-200303000-00006. [DOI] [PubMed] [Google Scholar]
- 46.Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003;299:1240–3. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]
- 47.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14:135–43. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 48.Smolka MN, Schumann G, Wrase J, Grusser SM, Flor H, Mann K, et al. Catechol-O-methyltransferase val158met genotype affects processing of emotional stimuli in the amygdala and prefrontal cortex. J Neurosci. 2005;25:836–42. doi: 10.1523/JNEUROSCI.1792-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu G, Lipsky RH, Xu K, Ali S, Hyde T, Kleinman J, et al. Differential expression of human COMT alleles in brain and lymphoblasts detected by RT-coupled 5′ nuclease assay. Psychopharmacology (Berl) 2004;177:178–84. doi: 10.1007/s00213-004-1938-z. [DOI] [PubMed] [Google Scholar]
- 50.Vandenbergh DJ, Rodriguez LA, Miller IT, Uhl GR, Lachman HM. High-activity catechol-O-methyltransferase allele is more prevalent in polysubstance abusers. Am J Med Genet. 1997;74:439–42. [PubMed] [Google Scholar]
- 51.Tiihonen J, Hallikainen T, Lachman H, Saito T, Volavka J, Kauhanen J, et al. Association between the functional variant of the catechol-O-methyltransferase (COMT) gene and type 1 alcoholism. Mol Psychiatry. 1999;4:286–9. doi: 10.1038/sj.mp.4000509. [DOI] [PubMed] [Google Scholar]
- 52.Kauhanen J, Hallikainen T, Tuomainen TP, Koulu M, Karvonen MK, Salonen JT, et al. Association between the functional polymorphism of catechol-O-methyltransferase gene and alcohol consumption among social drinkers. Alcohol Clin Exp Res. 2000;24:135–9. [PubMed] [Google Scholar]
- 53.Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132:607–21. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- 54.Yokoyama A, Omori T, Yokoyama T, Sato Y, Mizukami T, Matsushita S, et al. Risk of squamous cell carcinoma of the upper aerodigestive tract in cancer-free alcoholic Japanese men: an endoscopic follow-up study. Cancer Epidemiol Biomarkers Prev. 2006;15:2209–15. doi: 10.1158/1055-9965.EPI-06-0435. [DOI] [PubMed] [Google Scholar]
- 55.Tu GC, Israel Y. Alcohol consumption by orientals in North America is predicted largely by a single gene. Behav Genet. 1995;25:59–65. doi: 10.1007/BF02197242. [DOI] [PubMed] [Google Scholar]
- 56.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–81. [PMC free article] [PubMed] [Google Scholar]
- 57.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergen AW, Kokoszka J, Peterson R, Long JC, Virkkunen M, Linnoila M, et al. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2:490–4. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 59.Kranzler HR, Gelernter J, O'Malley S, Hernandez-Avila CA, Kaufman D. Association of alcohol or other drug dependence with alleles of the mu opioid receptor gene (OPRM1) Alcohol Clin Exp Res. 1998;22:1359–62. [PubMed] [Google Scholar]
- 60.Sander T, Gscheidel N, Wendel B, Samochowiec J, Smolka M, Rommelspacher H, et al. Human mu-opioid receptor variation and alcohol dependence. Alcohol Clin Exp Res. 1998;22:2108–10. [PubMed] [Google Scholar]
- 61.Gelernter J, Kranzler H, Cubells J. Genetics of two mu opioid receptor gene (OPRM1) exon I polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol Psychiatry. 1999;4:476–83. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- 62.Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28:1546–52. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 63.Anton RF, Oroszi G, O'Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRMI) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65:135–44. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31:555–63. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 65.Bergen SE, Gardner CO, Kendler KS. Age-related changes in heritability of behavioral phenotypes over adolescence and young adulthood: a meta-analysis. Twin Res Hum Genet. 2007;10:423–33. doi: 10.1375/twin.10.3.423. [DOI] [PubMed] [Google Scholar]
- 66.Robin RW, Chester B, Rasmussen JK, Jaranson JM, Goldman D. Prevalence, characteristics, and impact of childhood sexual abuse in a Southwestern American Indian tribe. Child Abuse Negl. 1997;21:769–87. doi: 10.1016/s0145-2134(97)00038-0. [DOI] [PubMed] [Google Scholar]
- 67.Walker EA, Gelfand AN, Gelfand MD, Koss MP, Katon WJ. Medical and psychiatric symptoms in female gastroenterology clinic patients with histories of sexual victimization. Gen Hosp Psychiatry. 1995;17:85–92. doi: 10.1016/0163-8343(94)00058-l. [DOI] [PubMed] [Google Scholar]
- 68.Caspi A, Moffitt TE. Gene–environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–90. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 69.Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–4. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 70.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–9. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 71.Caspi A, Moffitt TE, Cannon M, McClay J, Murray R, Harrington H, et al. Moderation of the effect of adolescent-onset cannabis use on adult psychosis by afunctional polymorphism in the catechol-O-methyltransferase gene: longitudinal evidence of a gene–environment interaction. Biol Psychiatry. 2005;57:1117–27. doi: 10.1016/j.biopsych.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 72.Blomeyer D, Treutlein J, Esser G, Schmidt MH, Schumann G, Laucht M. Interaction between CRHR1 gene and stressful life events predicts adolescent heavy alcohol use. Biol Psychiatry. 2007;63:146–51. doi: 10.1016/j.biopsych.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 73.Brookes KJ, Mill J, Guindalini C, Curran S, Xu X, Knight J, et al. A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use ofalcohol during pregnancy. Arch Gen Psychiatry. 2006;63:74–81. doi: 10.1001/archpsyc.63.1.74. [DOI] [PubMed] [Google Scholar]
- 74.Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–6. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–80. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 76.Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–9. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- 77.Deckert J, Catalano M, Syagailo YV, Bosi M, Okladnova O, Di Bella D, et al. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet. 1999;8:621–4. doi: 10.1093/hmg/8.4.621. [DOI] [PubMed] [Google Scholar]
- 78.Taylor A, Kim-Cohen J. Meta-analysis of gene–environment interactions in developmental psychopathology. Dev Psychopathol. 2007;19:1029–37. doi: 10.1017/S095457940700051X. [DOI] [PubMed] [Google Scholar]
- 79.Ducci F, Enoch MA, Hodgkinson C, Xu K, Catena M, Robin RW, et al. Interaction between a functional MAOA locus and childhood sexual abuse predicts alcoholism and antisocial personality disorder in adult women. Mol Psychiatry. 2007;13:334–47. doi: 10.1038/sj.mp.4002034. [DOI] [PubMed] [Google Scholar]
- 80.Barr CS, Newman TK, Becker ML, Parker CC, Champoux M, Lesch KP, et al. The utility of the non-human primate; model for studying gene by environment interactions in behavioral research. Genes Brain Behav. 2003;2:336–40. doi: 10.1046/j.1601-1848.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 81.Newman TK, Syagailo YV, Barr CS, Wendland JR, Champoux M, Graessle M, et al. Monoamine oxidase A gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 2005;57:167–72. doi: 10.1016/j.biopsych.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Meyer-Lindenberg A, Buckholtz JW, Kolachana B, Hariri AR, Pezawas L, Blasi G, et al. Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc Natl Acad Sci USA. 2006;103:6269–74. doi: 10.1073/pnas.0511311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ou XM, Chen K, Shih JC. Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J Biol Chem. 2006;281:21512–25. doi: 10.1074/jbc.M600250200. [DOI] [PubMed] [Google Scholar]
- 84.Youdim MB, Banerjee DK, Kelner K, Offutt L, Pollard HB. Steroid regulation of monoamine oxidase activity in the adrenal medulla. FASEB J. 1989;3:1753–9. doi: 10.1096/fasebj.3.6.2495232. [DOI] [PubMed] [Google Scholar]
- 85.Sjöberg RL, Ducci F, Barr CS, Newman TK, Dell'osso L, Virkkunen M, et al. A non-additive interaction of a functional MAO-A VNTR and testosterone predicts antisocial behavior. Neuropsychopharmacology. 2008;33:425–30. doi: 10.1038/sj.npp.1301417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, et al. Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry. 2003;8:933–6. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- 87.Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L, et al. Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet. 2005;77:265–79. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–31. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 89.Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–26. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy A, Hu XZ, Janal MN, Goldman D. Interaction between childhood trauma and serotonin transporter gene variation in suicide. Neuropsychopharmacology. 2007;32:2046–52. doi: 10.1038/sj.npp.1301331. [DOI] [PubMed] [Google Scholar]
- 91.Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, et al. 5-HTTLPR polymorphism impacts human cingulate–amygdala interactions: a genetic susceptibility mechanism for depression. Nat Neurosci. 2005;8:828–34. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- 92.Heinz A, Braus DF, Smolka MN, Wrase J, Puls I, Hermann D, et al. Amygdala–prefrontal coupling depends on a genetic variation of the serotonintransporter. Nat Neurosci. 2005;8:20–1. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- 93.Heinz A, Jones DW, Mazzanti C, Goldman D, Ragan P, Hommer D, et al. A relationship between serotonin transporter genotype and in vivo protein expression and alcohol neurotoxicity. Biol Psychiatry. 2000;47:643–9. doi: 10.1016/s0006-3223(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 94.Shioe K, Ichimiya T, Suhara T, Takano A, Sudo Y, Yasuno F, et al. No association between genotype of the promoter region of serotonin transporter gene and serotonin transporter binding in human brain measured by PET. Synapse. 2003;48:184–8. doi: 10.1002/syn.10204. [DOI] [PubMed] [Google Scholar]
- 95.Barr CS, Newman TK, Lindell S, Shannon C, Champoux M, Lesch KP, et al. Interaction between serotonin transporter gene variation and rearing condition in alcohol preference and consumption in female primates. Arch Gen Psychiatry. 2004;61:1146–52. doi: 10.1001/archpsyc.61.11.1146. [DOI] [PubMed] [Google Scholar]
- 96.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–15. [PubMed] [Google Scholar]
- 97.Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–11. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- 98.Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128:102–13. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–21. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 100.Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129:110–15. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- 101.Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL, et al. Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry. 1998;55:989–94. doi: 10.1001/archpsyc.55.11.989. [DOI] [PubMed] [Google Scholar]
- 102.Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–24. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- 103.Porjesz B, Begleiter H, Wang K, Almasy L, Chorlian DB, Stimus AT, et al. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002;61:229–48. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 104.Crabbe JC, Phillips TJ, Buck KJ, Cunningham CL, Belknap JK. Identifying genes for alcohol and drug sensitivity: recent progress and future directions. Trends Neurosci. 1999;22:173–9. doi: 10.1016/s0166-2236(99)01393-4. [DOI] [PubMed] [Google Scholar]
- 105.Korpi ER, Kleingoor C, Kettenmann H, Seeburg PH. Benzodiazepine-induced motor impairment linked to point mutation in cerebellar GABAA receptor. Nature. 1993;361:356–9. doi: 10.1038/361356a0. [DOI] [PubMed] [Google Scholar]
- 106.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, et al. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- 108.Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141:599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwata N, Cowley DS, Radel M, Roy-Byrne PP, Goldman D. Relationship between a GABAA alpha 6 Pro385Ser substitution and benzodiazepine sensitivity. Am J Psychiatry. 1999;156:1447–9. doi: 10.1176/ajp.156.9.1447. [DOI] [PubMed] [Google Scholar]
- 110.Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–88. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 111.Salonen JT, Uimari P, Aalto JM, Pirskanen M, Kaikkonen J, Todorova B, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81:338–45. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2007;13:302–12. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome–wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole–genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, et al. Pooled association genome scanning for alcohol dependence using 104 268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141:844–53. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ducci F, Goldman D. Deconstruction of vulnerability to complex diseases: enhanced effect sizes and power of intermediate phenotypes. ScientificWorld J. 2007;7:124–30. doi: 10.1100/tsw.2007.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64:203–13. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- 118.Wellcome Trust Case Control Consortium Genome–wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet. 2004;127:85–9. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- 120.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–66. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tabakoff B, Bhave SV, Hoffman PL. Selective breeding, quantitative trait locus analysis, and gene arrays identify candidate genes for complex drug-related behaviors. J Neurosci. 2003;23:4491–8. doi: 10.1523/JNEUROSCI.23-11-04491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 124.Mahadev K, Vemuri MC. Effect of ethanol on chromatin and nonhistone nuclear proteins in rat brain. Neurochem Res. 1998;23:1179–84. doi: 10.1023/a:1020778018149. [DOI] [PubMed] [Google Scholar]
- 125.Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126–32. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- 126.Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, et al. Alpha-synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci USA. 2003;100:4690–5. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–70. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- 128.Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, et al. Association of alcohol craving with alpha-synuclein (SNCA) Alcohol Clin Exp Res. 2007;31:537–45. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 129.Dannenberg LO, Chen HJ, Tian H, Edenberg HJ. Differential regulation of the alcohol dehydrogenase 1B (ADH1B) and ADH1C genes by DNA methylation and histone deacetylation. Alcohol Clin Exp Res. 2006;30:928–37. doi: 10.1111/j.1530-0277.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 130.Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144:101–5. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]