Abstract
Oral squamous cell carcinomas are characterized by complex, often near-triploid karyotypes with structural and numerical variations superimposed on the initial clonal chromosomal alterations. We used immunohistochemistry combined with classical cytogenetic analysis and spectral karyotyping to investigate the chromosomal segregation defects in cultured oral squamous cell carcinoma cells. During division, these cells frequently exhibit lagging chromosomes at both metaphase and anaphase, suggesting defects in the mitotic apparatus or kinetochore. Dicentric anaphase chromatin bridges and structurally altered chromosomes with consistent long arms and variable short arms, as well as the presence of gene amplification, suggested the occurrence of breakage–fusion–bridge cycles. Some anaphase bridges were observed to persist into telophase, resulting in chromosomal exclusion from the reforming nucleus and micronucleus formation. Multipolar spindles were found to various degrees in the oral squamous cell carcinoma lines. In the multipolar spindles, the poles demonstrated different levels of chromosomal capture and alignment, indicating functional differences between the poles. Some spindle poles showed premature splitting of centrosomal material, a precursor to full separation of the microtubule organizing centers. These results indicate that some of the chromosomal instability observed within these cancer cells might be the result of cytoskeletal defects and breakage–fusion–bridge cycles.
Genetic instability or changes in chromosome structure and numbers is an important facet of oncogenesis. The consequence of genetic instability can be an alteration in copy number of one or more genes, a change in gene expression, or a change in gene structure such that the protein sequence is altered (1). These genetic changes can lead to either increased or diminished protein activity or can create a new gain-of-function activity for the altered protein. Genetic instability can result from changes in chromosome structure, through errors in DNA metabolism, repair, recombination, or other rearrangements of the DNA sequence (2), or from misregulation of the cell cycle, for example, uncoupling DNA replication from cell division (3) or centrosomal duplication from division (4). Abnormalities in the chromosomal segregational apparatus are also likely to play an important role in genetic instability. These include centrosomal defects, defects in kinetochore–microtubule attachment, and movement of chromosomes relative to the poles. While it is likely that all of these changes contribute to carcinogenesis, the extent to which each plays a part is still largely uncharacterized.
One of the most striking manifestations of genetic instability in cancer cells is the variation observed in the karyotypes of different cells, even within the same tumor. A well-characterized example is squamous cell carcinomas of the oral cavity. The karyotypes of oral squamous cell carcinomas (OSCC) are complex, often near triploid, and are composed of multiple numerical and structural abnormalities, including deletions, balanced and unbalanced translocations, isochromosomes, dicentric chromosomes, and homogeneously staining regions (5). Despite the numerous and diverse abnormalities, classical cytogenetic analyses of OSCC cells have revealed several consistent chromosomal changes, such as breakpoints at bands 1p11–1p13, 3p14, and 11q13 (6) and amplification of band 11q13 (7, 8). Chromosomal rearrangements may be driven and/or fixed by activation of oncogenes or loss of tumor suppressor genes. For example, the 11q13 amplicon includes CCND1, the cyclin D1 gene, which plays a role in promoting cell division (9).
We have begun to investigate the source of chromosomal instability in cultured cells derived from human OSCC. We have found high levels of both numerical and structural chromosome variations, including marker chromosomes and dicentrics. During mitotic division, we observed multiple segregational defects, including multipolar spindles, lagging chromosomes, and anaphase bridges, leading to chromosomal breakage at telophase into centric and acentric micronuclei. In some cases, the anaphase bridges contained DNA from chromosome 11. An additional observed defect was the untimely and unnatural division of the nuclear mitotic apparatus protein NuMA at the centrosomes, apparently an early step in the splitting of the spindle poles. These observations support the conclusion that many of the cell-to-cell variations in chromosome structure and numbers in cancer cells result from (i) dicentric chromosomes forming anaphase bridges, which break and reform similar to that observed originally in maize by Barbara McClintock (10) and (ii) specific structural defects in the spindle and chromosome segregational machinery.
Materials and Methods
Cell Culture.
In this study, we examined three of our OSCC cell lines, UPCI:SCC003, UPCI:SCC131, and UPCI:SCC172, uncloned outgrowths developed from primary tumors from consenting patients who had not been treated previously with chemotherapy or radiation therapy (S.M.G., J. K. Reddy, S. Comsa, K. M. Rossie, C. M. Lese, M. S., B. N. Appel, R. Wagner, E. N. Myers, and J. T. Johnson, unpublished work). The cells were cultured in MEM with Earle's salts supplemented with nonessential amino acids (Life Technologies, Grand Island, NY), l-glutamine, gentamicin, and 10% FBS (Irvine Scientific).
Classical Cytogenetic Analysis.
The cell cultures were harvested by using standard cytogenetic techniques (11). Metaphase cells from all three cell lines were digitally imaged and karyotyped after trypsin-Giemsa banding (7) with a CytoVision Ultra System (Applied Imaging, Santa Clara, CA). Karyotypes have been or will be published elsewhere (ref. 7, and C. M. Lese, T. Ried, W. Gottberg, J. W. Wilson, S. C. Reshmi, J. K. Reddy, J. T. Johnson, E. N. Myers, and S.M.G., unpublished work). To examine the number of centromeres per chromosome, the metaphase cells were stained with C-banding using barium hydroxide and Giemsa stain (12).
Chromosome 11 Painting.
For chromosome 11 painting of cells growing in situ on Lab-Tek chamber slides (Nalge Nunc, Naperville, IL), the cell culture medium was removed by aspiration, and the cells were fixed in 3:1 methanol:acetic acid for 100 min, rinsed several times with 70% acetic acid solution, and air dried. The slides were aged for 30 min in 2× SCC at 37°C, pretreated with RNase for 1 hr at 37°C, then with pepsin for 10 min at 37°C, followed by postfixation in 1% formaldehyde for 7 min. Slides were denatured for 2 min at 70°C in 70% formamide/2× SSC, then dehydrated and air-dried prior to hybridization. The Spectrum Green directly labeled WCP11 probe (Vysis, Downers Grove, IL) was hybridized according to the manufacturer's directions. Slides were counterstained with the DNA-specific fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI; Sigma) in antifade solution (Oncor), stored at −20°C prior to viewing, analyzed with an Olympus (New Hyde Park, NY) BHS fluorescence microscope, and images were captured with a CytoVision Ultra.
Spectral Karyotyping (SKY).
Prior to SKY, the metaphase cells were G-banded as above. Within 24 hr, the slides were destained and hybridized using the SkyPaint 24 color fluorescence in situ hybridization (FISH) painting kit (Applied Spectral Imaging, Carlsbad, CA) as described in (13). Briefly, the G-banded slides were washed twice in xylene for 5 min each, rinsed in methanol for 5 min, and dehydrated through a graded ethanol series. Slides were postfixed immediately (10 min in 1% formaldehyde/1× PBS/50 mM MgCl2), washed for 5 min in PBS, dehydrated through a graded ethanol series, and air-dried. Slides were denatured, hybridized, and washed according to the manufacturer's instructions. Hybridized metaphase cells were captured and analyzed on the SkyVision I System with SkyView software (Applied Spectral Imaging).
Immunofluorescence.
Cells at passage 4–35 were grown on glass coverslips (Corning) for 24–72 hr, fixed in methanol for 30 min at −20°C, and air-dried. Samples were either processed within 1 hr or stored at −20°C with desiccant. The coverslips were hydrated in PBS containing 1% BSA and subject to indirect immunofluorescence with the following primary Abs: rabbit Abs to NuMA at 1:10,000, human autoimmune anti-centromere from a patient with the (CREST) variant of scleroderma (calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasias) at 1:80 or undiluted mouse monoclonal anti-tubulin. The coverslip with primary Ab was placed in a humid chamber at 37°C for 1 hr and then washed several times with PBS/1% BSA. The secondary Abs were Cy3-conjugated sheep anti-mouse IgG (Sigma) used at 1:1,000, FITC-conjugated goat anti-mouse (Sigma) at 1:100, or FITC-conjugated goat anti-human IgG at 1:100 (Roche Molecular Biochemicals). In all cases, the coverslips were counterstained with DAPI at 1–3 μg/ml.
Results
For these analyses, we selected three of our OSCC cell lines, designated UPCI:SCC003, UPCI:SCC131, and UPCI:SCC172. The TP53 gene (exons 5–8) in all three tumors and derived cell lines was sequenced and was found to be wild type. The tumor tissue was negative for human papillomavirus by the PCR. To examine the variation in chromosome number and structure between different cells within a single culture, G-banding and SKY were performed. The latter technique utilizes a DNA-hybridization probe mixture in which individual chromosomes are labeled with one to five fluorophores. Chromatin from each chromosome can be discriminated by spectral imaging as having a distinct dye spectrum, enabling characterization of the origin of segments of rearranged chromosomes (Fig. 1A). The karyotype of the UPCI:SCC172 cell line was observed to be highly aneuploid, containing gains, losses, and structural abnormalities of numerous chromosomes, including 1, 3, 7, 9, and 11. No two karyotypes were identical, varying in chromosome copy number and/or structure. This is illustrated in the supplemental material (Table 1) on the PNAS web site (www.pnas.org) for five well-characterized UPCI:SCC172 cells examined by G-banding and SKY. Karyotypic variability is observed for most of the more than 30 OSCC cell lines we have karyotyped by G-banding (C. M. Lese, T. Ried, W. Gottberg, J. W. Wilson, S. C. Reshmi, J. K. Reddy, J. T. Johnson, E. N. Myers, and S.M.G., unpublished work).
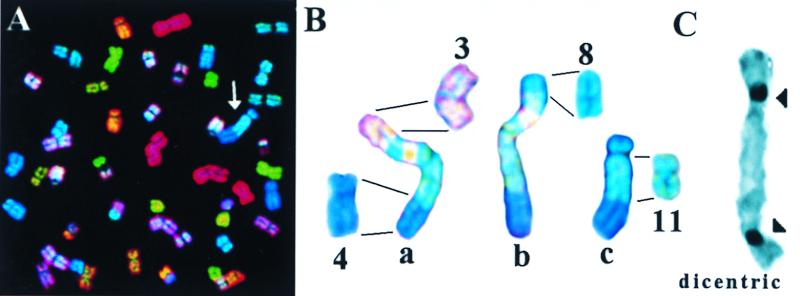
Figure 1.
(A) Display image of metaphase chromosomes from a Colcemid-arrested UPCI:SCC172 cell after SKY analysis. Arrowhead identifies the marker chromosome “c” shown in B. (B) Normal chromosomes 3, 4, 8, and 11 from this culture are shown. Marker chromosomes, also from UPCI:SCC172, are indicated as a, b, and c. (C) A representative dicentric chromosome is shown from a Colcemid-arrested UPCI:SCC131 cell C-banded to highlight the centromeres.
Of particular note was the observation that many cells within UPCI:SCC172 displayed what appeared to be related chromosomal structural variations, examples of which are shown in Fig. 1B. The SKY analysis shows these exceptional chromosomes to include segments of chromosomes 3, 4, 8, and 11. The marker chromosomes, labeled a–c, appear to have much of their DNA in common, but have different short-arm sequences. All three marker chromosomes share portions of chromosomes 4 and 11, but have different short-arm terminal segments derived from chromosomes 3 and 8. Unfortunately, cytogenetic nomenclature ignores this sort of variation, focusing only on “clonal” events in common between cells. Marker chromosomes have also been observed in the UPCI:SCC003 cell line, but the UPCI:SCC131 line has not produced sufficiently high-quality metaphase spreads to hybridize optimally with the SKY technique. These results demonstrate the remarkable evolutionary variation in chromosome structure between different OSCC cells within the same culture. Because the cell lines were uncloned outgrowths from tumor biopsies, it is possible that the variant marker chromosomes may have evolved in the primary tumors prior to culture. To determine whether the chromosomal structural variation in the cultured cells included changes in the number of centromeres per chromosome, we also performed C-banding of metaphase chromosome spreads, which revealed the presence of varying numbers of dicentric chromosomes (Fig. 1C).
To determine the source of this chromosomal variation, we examined mitotic segregation in the OSCC cell lines by indirect immunofluorescence with Abs to NuMA to mark the centrosomes (provided by Duane Compton, Dartmouth Medical School, Hanover, NH), CREST scleroderma autoimmune serum to mark the kinetochores (provided by Carol Feghali and Thomas Medsger, University of Pittsburgh, Pittsburgh, PA), and anti-tubulin Abs to mark the spindle (provided by Charles Walsh, University of Pittsburgh). During the metaphase stage of mitosis, chromosomes attach to both spindle poles by means of kinetochore microtubules and align equidistantly between the two poles to form the chromosomal metaphase plate (14). In the OSCC cell lines, most of the metaphase spindles appeared structurally and functionally normal (Fig. 2A, and see Fig. 3). The most striking visible difference from the norm was the appearance of multipolar spindles. In these aberrant structures, the chromosomes appeared to attempt to achieve alignment between the multiple poles, forming bizarre Y or cruciform shapes (Fig. 2B).
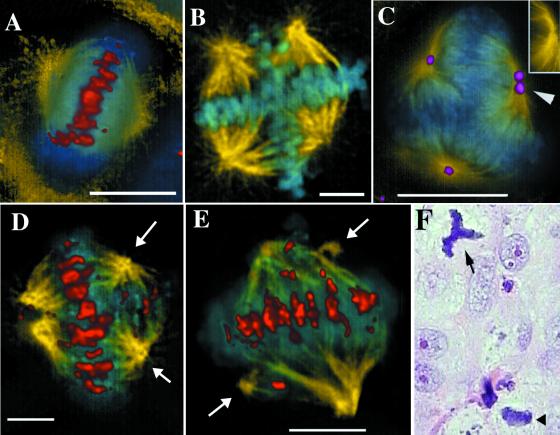
Figure 2.
UPCI:SCC131 cells were immunolabeled with Abs to tubulin (yellow), and kinetochores (red in A, D, and E) or NuMA (purple in C) and counterstained with the DNA dye, DAPI (blue). A normal metaphase from this culture is shown in A, and multipolar metaphase spindles in B–E. (C, Inset) The anti-tubulin image alone of the pole marked with an arrowhead. (D and E) Arrows mark minor spindle poles. (Bars, 5 μm.) (F) A tissue section from a formalin-fixed, paraffin-embedded, laryngeal carcinoma was stained with hematoxylin and eosin. An atypical tripolar mitosis (arrow) and a normal bipolar mitotic figure (arrowhead) are shown.
Figure 3.
UPCI lines SCC131, SCC172, and SCC003 were treated with anti-tubulin immunolabeling and DAPI staining and examined for five segregational abnormalities. The percentages of metaphase cells with multipolar spindles, interphase cells with micronuclei, metaphase, and anaphase cells with lagging chromosomes, and anaphase cells with chromatin bridges are given, as well as the percentage of metaphase cells with normal spindles. The mean and SEM from three coverslips are shown. A total for each cell line of 25–81 anaphase cells was counted for anaphase bridges and lagging chromosomes, 86–164 metaphase cells for multipolar spindles and lagging chromosomes, 1264–1593 interphase cells for micronuclei, and 110–370 metaphase cells for normal spindles.
To identify the position of the spindle poles unambiguously, we immunolabeled them with Abs to NuMA (15). NuMA is a nuclear matrix protein that relocates to the spindle poles during mitosis. NuMA staining was seen at all spindle poles, but, in some, the NuMA labeling was seen to have split into two parts, even though only a single microtubule organizing center (MTOC) was present (Fig. 2C, Inset shows the marked pole without NuMA staining). This division of the NuMA immunolabeling is likely to be a precursor to full spindle pole separation found in multipolar spindles. When we stained the cells with anti-kinetochore Abs, we observed that the multiple spindle poles did not necessarily have an equal influence on the chromosomes migrating to the metaphase plate. In some cases, the chromosomes were evenly divided between the poles (Fig. 2B), but, in others, there was a marked asymmetry in metaphase chromosomal alignment. In Fig. 2D, nearly normal metaphase alignment is achieved, with the chromosomes equidistant between one major pole and two minor poles (arrows). In addition, a small subset of the chromosomes is aligned between the two minor poles. In Fig. 2E, two minor sites of microtubule organization (arrows) have separated from the major poles. Each minor pole draws one or two chromosomes away from the metaphase alignment between the major poles. Thus, the different poles, presumably depending on their microtubule-nucleation ability, exerted different influences on the final alignment of the chromosomes. These examples are not artifacts of cell culture. Fig. 2F shows an example of a tripolar spindle from a tissue sample obtained from a laryngeal carcinoma biopsy.
We also observed segregational errors associated with bipolar spindles in the OSCC cells, including lagging metaphase and anaphase chromosomes and anaphase bridges. The segregational defects were counted and are compared between cell lines in Fig. 3. Anaphase chromatin forming continuous bridges connecting the two sets of separating chromosomes is shown in Fig. 4. Anaphase bridges can be formed by dicentric chromosomes being pulled simultaneously toward both spindle poles and are consistent with our observation of multicentric chromosomes in these cultures. To demonstrate that the bridges contained dicentric chromosomes, we immunostained with anti-kinetochore Abs. Anaphase bridges usually contained centromeres, typically seen as two or more well-separated fluorescent signals (Fig. 4A), although centromereless bridges were also observed. Thus, both the presence of multicentric chromosomes and the kinetochore staining of bridges suggested that at least some of the bridges were caused by chromosomes being pulled simultaneously toward both poles in anaphase.
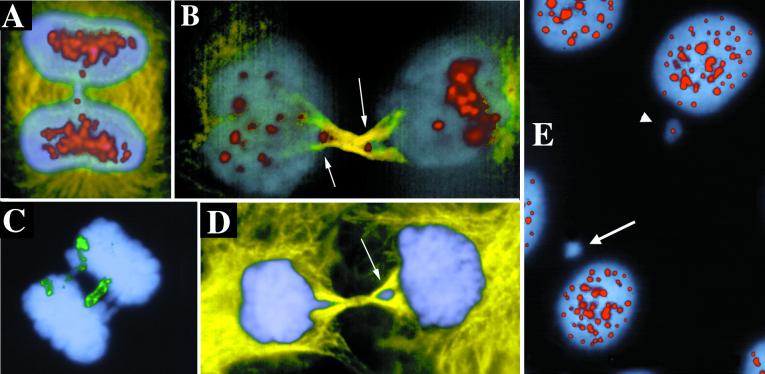
Figure 4.
Anaphase bridges containing centromeres and chromosome 11. Immunolabeling with Abs to tubulin (yellow), centromeres (red), and with DAPI (blue) and FISH with a chromosome 11 paint probe (green). (B) Arrows point to centromeres trapped in the forming midbody as these late telophase cells divide. (D) Arrow points to the trapped lagging chromosome excluded from the reforming nucleus of the cell on the right. (E) Some micronuclei are immunonegative for anti-centromere Abs. Arrow points to negative micronucleus, and arrowhead points to positive. Examples are from UPCI:SCC131.
Anaphase bridging has been proposed to be a mechanism for gene amplification (16), and the UPCI:SCC003, -131, and -172 cells lines express amplification of chromosomal band 11q13 in the form of a homogeneously staining region (ref. 7, and X.H., W.S.S., M.S., C. M. Lese, and S.M.G., unpublished work). To determine whether chromosome 11 could be found in the anaphase bridges, we used chromosomal painting with chromosome 11 sequences as a FISH probe. Chromosome 11 was found in 16% of anaphase bridges, suggesting that anaphase bridges may be involved in amplification of chromosome 11 in these OSCC cells (Fig. 4C).
In some cells, the anaphase bridge chromatin was excluded from the reforming nucleus. Fig. 4B illustrates a cell in telophase, as demonstrated by formation of the anti-tubulin-positive midbody region between the dividing daughter cells. At this stage, the nuclear envelope should be reforming, but the cells have not yet resolved their anaphase bridge, with centromere staining persisting in the midbody region (arrows in Fig. 4B). At later stages of division, it appears that these lagging chromosomes are excluded from the reforming nucleus and incorporated into micronuclei separate from the main genome and from each other (Fig. 4D). This exclusion of bridged chromatin from the cell nucleus most likely explains the high frequency of micronuclei observed previously in oral cancer cells (17). When the micronuclei were stained with Abs to kinetochores, both centric and acentric micronuclei were observed (Fig. 4E). Thirty percent (n = 120) of the micronuclei contained kinetochores as judged by immunostaining with CREST autoimmune serum. Apparently, chromosome fragmentation occurred as the chromosomes were pulled simultaneously in both directions, producing both acentric fragments and centric chromosomes.
Discussion
Chromosomal loss and rearrangement are known to be important signs of genetic instability in cancer cells, but the mechanisms behind these changes are unclear. We have begun an investigation of the contribution of segregational errors to chromosomal instability using oral carcinoma cells as our model system. In these cultures, we found frequent variations in chromosome numbers and structure between different cells from the same tumor cell culture. We believe that many of these abnormalities can be explained by chromosomal segregational defects.
The OSCC cells clearly had difficulty achieving normal metaphase alignment and anaphase chromosome separation. One simple model is that the kinetochores are defective for movement, and that this single defect causes chromosomes to lag at both metaphase and anaphase. Consistent with this model, approximately equal numbers of lagging chromosomes were observed in both metaphase and anaphase cells. One way to test this directly would be a real-time analysis of chromosome movement to determine whether the same chromosomes express a lagging phenotype in both metaphase and anaphase. Alternatively, lagging anaphase chromosomes, like bridged chromosomes, could also be the result of dicentric chromosomes, if one centromere is only partially active, as originally suggested by Fraccaro et al. (18).
One specific source of delay in anaphase progression was the formation of anaphase chromatin bridges. Anaphase bridges are rare in normal mammalian cells, but are common in virally infected and tumor cells (19) or in patients with chromosome breakage syndromes, such as Bloom syndrome (20). Anaphase bridges can be formed as a result of defects in telomere structure or length (21), defects in DNA replication (22), recombination (23), or translocations that introduce a second centromere in the chromosome (24).
Anaphase bridges were originally observed by Barbara McClintock (10), who found that chromosomes in maize could break, exposing a novel end, which after replication fused with the sister chromatid to form a dicentric bridge in anaphase. These bridges then can go through a cycle of breakage, fusion, and bridging (BFB). An alternative mechanism involving mitotic recombination and quadriradial formation has also been proposed (20). BFB cycles have been proposed to be an important mechanism of gene amplification in the acquisition of drug resistance (25–27). Recent work has shown a role for fragile sites in BFB-induced gene amplification (16). Telomeric fusions and BFB cycles have also been reported in other cancer cells and are thought to lead to specific types of chromosomal rearrangements, including deletions and ring chromosomes (28–30). We believe that many of the chromosomal differences we are seeing between OSCC cells in culture are because of BFB cycles. BFB cycles could explain the high variation between cells within a culture, because dicentric chromosomes are inherently unstable and may change in virtually every generation (31). As an example, BFB cycles most likely produce the amplification of chromosome 11q13 with concomitant deletion of distal sequences (8). This is in contrast to recombinational models for gene amplification (23), which would not produce the chromatin bridges we have observed in OSCC cells unless the recombinational intermediates persisted into anaphase.
Micronuclei are commonly observed in oral cancer cells and are an important biomarker for this disease (17, 32). Micronuclei could be formed either from lagging anaphase chromosomes or anaphase bridges, both of which occur in our OSCC cultures. In human lymphocytes from healthy volunteers, micronuclei are rare and nearly all contain intact chromosomes, and were interpreted to result from lagging anaphase chromosomes (33). In contrast, more than two-thirds of the micronuclei from the OSCC cells studied here were negative for centromere immunostaining, suggesting that many of the micronuclei in oral cancer cells result from chromosome-fragmentation events, such as could occur with anaphase bridging. The appearance of acentric micronuclei is consistent with the expected DNA damage resulting from BFB cycles. Pharmacological agents that lead to DNA damage usually result in acentric micronuclei, whereas agents that produce aneuploidy, by interference with microtubule dynamics, usually yield centromere-positive micronuclei (33, 34). These observations, combined with our data showing dicentric chromosomes being excluded from the reforming nucleus, indicate that many of the micronuclei in oral cancer cells result from anaphase bridges.
The frequency of 20–30% micronuclei in our OSCC samples is much higher than the 0.21–0.7% of exfoliated oral epithelial cells from healthy volunteers (35). However, the frequency of 30% centric micronuclei in this study is roughly similar to the 50% centromere-positive micronuclei observed in these cells (35). This comparison suggests that the defect in OSCC cells that results in micronucleus formation, compared with normal oral epithelium, is not in the resolution of dicentric chromosomes, but rather the frequency with which they form. In patients with Bloom syndrome, one centromere of dicentric chromosomes is usually inactivated (20). We propose that this mechanism for centromere inactivation may be lost in our OSCC cells, contributing to the high levels of BFB events and the resulting genomic instability of these cultures.
Multipolar spindles are frequently observed in cancer cells (19), but the origin is unclear. One model proposes that multipolar spindles are the result of excess centrioles, for example, from centriolar replication in the absence of division. Neoplastic progression in transgenic mice with an elastase-SV40 large T antigen gene was associated with increased numbers of centrioles, up to five at a single pole (36). However, whereas pericentriolar material is required for microtubule nucleation at spindle poles (37), centrioles are not. When multipolar spindles were induced by Colcemid treatment in Chinese hamster ovary cells, 60% of the MTOCs of tripolar cells lacked any centriole (38). It seems clear that multipolar spindles can be the result of separation of the pericentriolar material without centriolar duplication. Our results indicate that the NuMA protein is able to form duplicated foci prior to spindle pole body separation. At the spindle pole, NuMA binds to the microtubule motor dynein, and the dynein-activating complex dynactin (39, 40) and can polymerize and has been proposed to tether and stabilize microtubules at the spindle poles (39, 41, 42). We suggest that accumulation of otherwise normally functioning NuMA at the spindle poles of these cancer cells may recruit enough microtubule crosslinking activity, for example, the dynein motor, to induce splitting of the MTOC, leading to multipolar spindles. Consistent with this interpretation, microtubule asters can be formed in vitro in the absence of centrosomes if dynein is added (43). The splitting of the poles can produce major or minor MTOCs, as defined by the number of microtubules and the extent of influence on chromosome alignment. This variation in activity could simply be the result of the total amount of pericentriolar material that each pole receives when they separate or whether specific unidentified factors or modifications are present. We favor the interpretation that the amount of pericentriolar material that each nascent pole receives from the original determines its relative activity, but additional studies are required to address this issue.
Our findings provide strong support for the model that a substantial proportion of the genetic instability in cancer cells is because of specific structural defects in chromosome segregation. These include BFB cycles leading to gene amplification, chromosome fragmentation and loss, and segregational errors from separation of microtubule-organizing material. In fact, it appears that all of the karyotypic changes we see in these cultures may be explained by the observed segregational defects. The formation of supernumerary spindle poles appears to be preceded by the separation of the centrosomal protein NuMA into distinct foci. The extra poles could have only a minor influence on the metaphase alignment, pulling one or a few chromosomes from the main body of chromosomes or major effects, splitting the chromosomes equally in four or more directions simultaneously. It will be of interest to determine whether the relative amounts of NuMA that partition to these minor poles determine their ability to capture chromosomes and perturb the bipolar metaphase alignment.
Supplementary Material
Acknowledgments
The cell lines were initiated and cared for initially by Dr. Christa M. Lese, Ms. Jaya Reddy, or Ms. Seia Comsa, the latter two under the guidance of Ms. Robin Wagner. We thank Dr. Chandramohan S. Ishwad and Ms. Diana Kerestan for their high quality DNA sequence analyses and human papilloma virus testing; Drs. Charles Walsh, Duane Compton, Carol Feghali, and Thomas Medsger for Abs; and Dr. Thomas Ried of the National Cancer Institute and Mr. Dirk Soenksen, Ms. Cathy Jaenish, and Ms. Elizabeth Weeks of Applied Spectral Imaging for training and assistance with the spectral karyotyping. These studies were supported by an Oral Cancer Prevention, Research, and Treatment Center Planning Grant (P20DE12378) (to Dr. Eugene Myers), Grants CB-171 and JFRA-624 from the American Cancer Society (to W.S.), National Institutes of Health Grants F32DE05707 (to M.S.), R01DE10513, R01DE12008, P30CA47904-11S39019, and S10RR11879 (to S.M.G.), Smokeless Tobacco Research Council Grant 0660 (to S.M.G.), and the Project Grant 1P60DE13059-01 of the Oral Cancer Center at the University of Pittsburgh (to W.S. and S.M.G.). Cytogenetic analyses were carried out in the University of Pittsburgh Cancer Institute Cytogenetics Facility.
Abbreviations
- OSCC
oral squamous cell carcinomas
- DAPI
4′,6-diamidino-2-phenylindole
- SKY
spectral karyotyping
- MTOC
microtubule organizing center
- BFB
breakage–fusion–bridge
- FISH
fluorescence in situ hybridization
- NuMA
nuclear mitotic apparatus protein
- CREST
calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasias
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Miyagawa K. Int J Hematol. 1998;67:3–14. doi: 10.1016/s0925-5710(97)00113-8. [DOI] [PubMed] [Google Scholar]
- 3.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 4.Winey M. Curr Biol. 1996;6:962–964. doi: 10.1016/s0960-9822(02)00639-5. [DOI] [PubMed] [Google Scholar]
- 5.Cowan J M. Otolaryngol Clin North Am. 1992;25:1073–1087. [PubMed] [Google Scholar]
- 6.Jin Y, Jin C, Wennerberg J, Mertens F, Hoglund M. Cancer Res. 1998;58:5859–5865. [PubMed] [Google Scholar]
- 7.Lese C M, Rossie K M, Appel B N, Reddy J K, Johnson J T, Myers E N, Gollin S M. Genes Chromosomes Cancer. 1995;12:288–295. doi: 10.1002/gcc.2870120409. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Hoglund M, Jin C, Martins C, Wennerberg J, Akervall J, Mandahl N, Mitelman F, Mertens F. Genes Chromosomes Cancer. 1998;22:312–320. [PubMed] [Google Scholar]
- 9.Akervall J A, Michalides R J, Mineta H, Balm A, Borg A, Dictor M R, Jin Y, Loftus B, Mertens F, Wennerberg J P. Cancer. 1997;79:380–389. [PubMed] [Google Scholar]
- 10.McClintock B. Cold Spring Harbor Symp Quant Biol. 1941;9:72–81. [Google Scholar]
- 11.Heo D S, Snyderman C, Gollin S M, Pan S, Walker E, Deka R, Barnes E L, Johnson J T, Herberman R B, Whiteside T L. Cancer Res. 1989;49:5167–5175. [PubMed] [Google Scholar]
- 12.Sumner A T. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- 13.Veldman T, Vignon C, Schrock E, Rowley J D, Ried T. Nat Genet. 1997;15:406–410. doi: 10.1038/ng0497-406. [DOI] [PubMed] [Google Scholar]
- 14.Nicklas R B. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 15.Compton D A, Szilak I, Cleveland D W. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 17.Benner S E, Lippman S M, Wargovich M J, Lee J J, Velasco M, Martin J W, Toth B B, Hong W K. Int J Cancer. 1994;59:457–459. doi: 10.1002/ijc.2910590403. [DOI] [PubMed] [Google Scholar]
- 18.Fraccaro M, Lo Curto F, Vogel W. Prog Clin Biol Res. 1978;26:181–202. [PubMed] [Google Scholar]
- 19.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. New York: Elsevier; 1990. pp. 96–99. [Google Scholar]
- 20.Therman E, Trunca C, Kuhn E M, Sarto G E. Hum Genet. 1986;72:191–195. doi: 10.1007/BF00291876. [DOI] [PubMed] [Google Scholar]
- 21.Harley C B, Villeponteau B. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 22.Poupon M F, Smith K A, Chernova O B, Gilbert C, Stark G R. Mol Biol Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith K A, Gorman P A, Stark M B, Groves R P, Stark G R. Cell. 1990;63:1219–1227. doi: 10.1016/0092-8674(90)90417-d. [DOI] [PubMed] [Google Scholar]
- 24.Hastie N D, Allshire R C. Trends Genet. 1989;5:326–331. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 25.Smith K A, Stark M B, Gorman P A, Stark G R. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Martin S, Trask B, Hamlin J L. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 27.Ishizaka Y, Chernov M V, Burns C M, Stark G R. Proc Natl Acad Sci USA. 1995;92:3224–3228. doi: 10.1073/pnas.92.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondello C, Casati A, Riboni R, Nuzzo F. Cancer Genet Cytogenet. 1995;79:41–48. doi: 10.1016/0165-4608(94)00085-p. [DOI] [PubMed] [Google Scholar]
- 29.Sawyer J R, Sammartino G, Husain M, Lewis J M, Anderson B, Boop F A. Genes Chromosomes Cancer. 1993;8:69–73. doi: 10.1002/gcc.2870080202. [DOI] [PubMed] [Google Scholar]
- 30.Sawyer J R, Thomas E L, Roloson G J, Chadduck W M, Boop F A. Cancer Genet Cytogenet. 1992;60:152–157. doi: 10.1016/0165-4608(92)90008-v. [DOI] [PubMed] [Google Scholar]
- 31.Riboni R, Casati A, Nardo T, Zaccaro E, Ferretti L, Nuzzo F, Mondello C. Cancer Genet Cytogenet. 1997;95:130–136. doi: 10.1016/s0165-4608(96)00248-8. [DOI] [PubMed] [Google Scholar]
- 32.Stich H F, Parida B B, Brunnemann K D. Int J Cancer. 1992;50:172–176. doi: 10.1002/ijc.2910500203. [DOI] [PubMed] [Google Scholar]
- 33.Ford J H, Schultz C J, Correll A T. Am J Hum Genet. 1988;43:733–740. [PMC free article] [PubMed] [Google Scholar]
- 34.Degrassi F, Tanzarella C. Mutat Res. 1988;203:339–345. doi: 10.1016/0165-1161(88)90030-1. [DOI] [PubMed] [Google Scholar]
- 35.Moore L E, Titenko-Holland N, Quintana P J, Smith M T. J Toxicol Environ Health. 1993;40:349–357. doi: 10.1080/15287399309531800. [DOI] [PubMed] [Google Scholar]
- 36.Levine D S, Sanchez C A, Rabinovitch P S, Reid B J. Proc Natl Acad Sci USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gould R R, Borisy G G. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keryer G, Ris H, Borisy G G. J Cell Biol. 1984;98:2222–2229. doi: 10.1083/jcb.98.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merdes A, Ramyar K, Vechio J D, Cleveland D W. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 40.Mattagajasingh S N, Huang S C, Hartenstein J S, Snyder M, Marchesi V T, Benz E J. J Cell Biol. 1999;145:29–43. doi: 10.1083/jcb.145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C H, Snyder M. Mol Biol Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dionne M A, Howard L, Compton D A. Cell Motil Cytoskeleton. 1999;42:189–203. doi: 10.1002/(SICI)1097-0169(1999)42:3<189::AID-CM3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 43.Verde F, Berrez J M, Antony C, Karsenti E. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.