Abstract
Drosophila olfactory (ORs) and gustatory (GRs) receptors are evolutionarily unrelated to vertebrate ORs or nematode chemosensory receptors. Insect ORs display a reverse membrane topology compared with conventional G-protein-coupled receptors, suggesting that the mammalian scheme of chemosensory signal transduction cannot directly apply to insects. Experimental studies of GR membrane topology are lacking. We analysed the distribution of amino acid sites in GRs and ORs that show evidence for divergence under either positive selection or relaxed purifying constraints, in the genomes of 12 Drosophila species and found significant differences between these two receptor types. This suggests that insect ORs and GRs have distinct molecular properties and mechanisms of ligand recognition and/or signal transduction.
Keywords: gustatory receptors, olfactory receptors, Drosophila, divergent selection
1. Introduction
Chemosensory processes are vital to the relationship between an organism and its environment. The evolutionary divergence of chemoperception underlies processes including mate choice, ecological specialization and speciation (Smadja & Butlin 2009). The peripheral receptors involved in olfaction, identified in mammals, belong to a family of membrane-bound G-protein-coupled receptors (GPCRs), which conduct signal transduction through G-protein activation (Buck & Axel 1991). In vertebrates, these have seven hydrophobic transmembrane (TM) domains that are linked by three extracellular (ELs) and three intracellular (ILs) loops. The NH2-tail is located extracellularly, while the COOH-tail is cytoplasmic. GPCRs vary in the location of their ligand recognition region, but olfactory receptors (ORs) are thought to bind ligands in ligand-binding pockets of the TM regions (Schwartz et al. 2006).
ORs and gustatory receptors (GRs) of insects were originally classified as GPCRs (Clyne et al. 1999, 2000). However, in Drosophila, it has recently been found that the vertebrate model of protein structure does not apply for ORs. These form dimers with a co-expressed receptor, OR83b, and the membrane orientation of the ORs and OR83b is reversed with the NH2-tail intracellular and the COOH-tail extracellular (Benton et al. 2006; Wistrand et al. 2006; Lundin et al. 2007). Dimerization of ORs occurs through coupling of the COOH-terminal region with OR83b, and interactions between the third cytoplasmic loop of ORs and OR83b imply that this is a conserved mechanism of the Drosophila OR family (Benton et al. 2006).
The insect GRs are closely related to the ORs (Robertson et al. 2003), with some GRs also expressed in the antenna (Jones et al. 2007). However, GR neurons do not express OR83b and responses to ligands are independent of OR83b expression (Benton et al. 2006). Recent studies of signal transduction in Drosophila ORs have shown that they serve as ion channels and as a GPCR, suggesting that the signal transduction consists of a fast and transient ion conductance and a long-lasting non-selective cation conductance (Sato et al. 2008; Smart et al. 2008; Wicher et al. 2008).
The mechanism of ligand recognition by insect ORs and GRs is unknown. By analogy with mammals, it was hypothesized that odorants are recognized at the sensillum pore and transported to underlying ORs by small soluble odorant-binding proteins, present in the lymph surrounding receptor neurons (Hekmat-Scafe et al. 2002).
Twelve Drosophila genome sequences are available (Consortium 2007), allowing analyses of the evolution of chemosensory genes across species that differ in chemosensory-driven behaviours such as oviposition and mate choice. Gene duplication and pseudogenization show patterns possibly related to the degree of ecological specialization (McBride & Arguello 2007) or population biology (Gardiner et al. 2008). Which regions of the chemosensory receptors show evidence of evolutionary constraint or divergence? Functionally important regions may be inferred from sequence conservation. Divergence will be minimal in constrained regions, but relatively high functional divergence rates can occur through either relaxed constraints or positive diversifying selection (Nielsen 2005). Here, we investigate the distribution of amino acids that show accelerated rates of evolutionary divergence of ORs and GRs in the 12 Drosophila species.
2. Material and methods
Homologues of the ORs and GRs of Drosophila melanogaster were identified and aligned as previously reported (Gardiner et al. 2008). Orthologues were tested for selection using the M1–M2 and M7–M8 models of the codeml program in PAML (Yang 1997). M1–M2 comparisons are very stringent and can lack power to detect signatures of diversifying selection; these were not significant for any locus. M7 and M8 impose less constraint on the distribution of ω, with M8 having the most power to detect signatures of diversifying selection on a fraction of sites (Anisimova et al. 2002). When M7–M8 comparisons were significant (p<0.05), we used M8a (ω=1) and M8 (ω free) models to investigate the potential roles of reduced purifying selection versus positive selection (Swanson et al. 2003). The Bayes empirical Bayes (BEB) method was used to calculate the posterior probability (PP) that each codon is from a class of sites under diversifying selection in the M8 model (Yang et al. 2005).
Sites identified with PP>0.50 were analysed; we refer to these as ‘candidate sites’ that show accelerated divergence, either under positive selection or relaxed purifying constraints (in cases when the evidence for selection was not significant in the M8a–M8 tests). We also compared the distribution of sites with a higher PP (above 0.80), increasing the stringency. Corrections for multiple tests have not been applied, so that these should be viewed as a class of candidate sites for which there is some evidence of divergence.
We used information on domain structure of Drosophila GRs and ORs at the ExPASy proteomics server (http://ca.expasy.org/) and predicted the secondary structure and membrane topology using TMHMM (Krogh et al. 2001) and Tmpred (Hofmann & Stoffel 1993).
3. Results
Previously (Gardiner et al. 2008), we detected that there is evidence (p<0.05) of positive selection overall for 10 Gr and 10 Or genes (based on significant M7–M8 comparisons; see also table 2 in the electronic supplementary material), containing 8141 amino acids. Within these, BEB identified 67 GR and 74 OR amino acids (see table 1 in the electronic supplementary material). Approximately 55 per cent of these sites have PP ranging from 0.50 to 0.70, and the remainder have PP higher than 0.70, approximately 9 per cent of sites have PP>0.90.
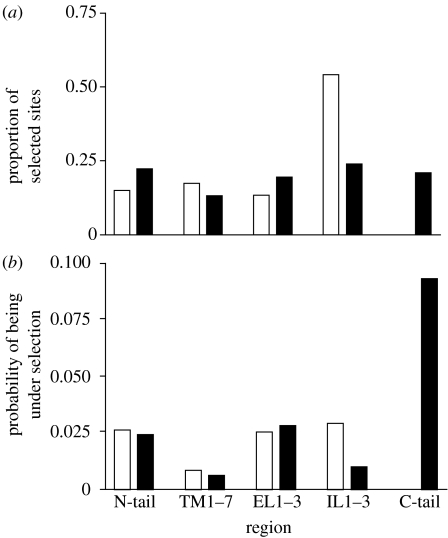
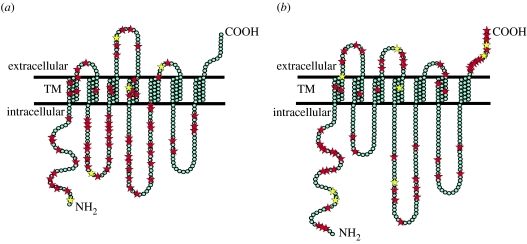
Seven TM domains were identified in all ORs and GRs. The TMHMM and Tmpred algorithms predicted an intracellular orientation of the NH2-tails and an extracellular orientation of the COOH-tails, supporting the ‘reverse’ membrane topology of Drosophila receptors. The majority of candidate sites in ORs are found in loop regions (approx. 68%), approximately 16 per cent of sites are located in TM regions (mainly in TM1 and TM4), and 15 per cent of sites are located on the cytoplasmic NH2-termini (figures 1a and 2), in general agreement with Guo & Kim (2007). Sites mapped to the loops have a surprising distribution; approximately 13 per cent of candidate sites were found in the ELs and 55 per cent mapped to the ILs, where we found them mostly in the IL1 (17 sites) and IL2 (18 sites).
Figure 1.
(a) Distribution of the sites showing evidence of divergence by protein regions (NH2-tail, TM domains 1–7 (TM1–7), extracellular loops 1–3 (EL1–3), intracellular loops 1–3 (IL1–3), COOH-tail). (b) The overall probability of an amino acid showing evidence of divergent selection. OR, olfactory receptor (white bars); GR, gustatory receptor (black bars).
Figure 2.
Membrane topology of receptors and distribution of selected sites: (a) OR and (b) GR. Red stars, sites with PP>0.5; yellow stars, sites with PP>0.9.
Candidate sites in GRs show a different pattern with fewer found in the loop regions (43%) and more on the NH2 (22%) and COOH (21%) tails of the protein (figures 1a and 2). The sites mapped to the loops are nearly equally distributed between the extracellular (19% of sites) and intracellular surfaces (24% of sites); most sites were found in the EL2 (7 sites) and IL2 (10 sites). Approximately 13 per cent of sites mapped to TMs, mostly in TM1. The distribution of candidate sites among regions differed between the receptor types (G4=31.43, p<0.0001).
Because the number of amino acids differs between regions and loci, we also analysed the proportion of sites per locus showing evidence of selection, using a binomial logistic regression, effectively controlling for the number of amino acids within a region. The incidence of potential selection varied between regions (deviance ratio=9.51, Χ4,902<0.001), and there was a significant interaction between region and receptor type (deviance ratio=10.08, Χ4,902<0.001). The main differences were that amino acids were more likely to be identified in the COOH-tail of GRs and in the IL1–3 regions of ORs (figure 1b).
We analysed the distribution of sites with PP>0.80 (19 OR and 16 GR sites). Again, the majority of sites (approx. 53%) mapped to the ILs in the ORs, but only approximately 13 per cent of sites mapped to the GRs. We did not find candidate sites on the COOH-tail in the ORs, but approximately 31 per cent of GR candidate sites mapped here.
After testing the strength of selection with the stringent M8a model, we find that only six genes (four Ors and two Grs) show significant evidence of diversifying selection (see table 2 in the electronic supplementary material). More than half of the candidate sites in these OR loci mapped to the ILs.
4. Discussion
We detected evidence of increased functional divergence at 20 loci, but only six showed a signal of diversifying selection. Thus, a proportion of the candidate sites identified for these six loci have diverged under positive selection, but, for the rest of the genes, rapid divergence was probably due to relaxed purifying constraints. In Drosophila ORs, the majority of candidate sites were in the ILs. This is unexpected because these regions are likely to interact with secondary messengers and other proteins involved in signal transduction. We predicted these to be under constraint rather than diversifying or relaxed selection. The COOH-tail of ORs is essential for coupling with OR83b, and highly divergent OR proteins share the strongest similarity within the COOH-termini (Benton et al. 2006). None of the candidate sites mapped to the COOH-tail. Only a few mapped to IL3, which interacts with IL3 of OR83b during dimerization (Benton et al. 2006), and therefore is expected to be under constraint. The majority of candidate sites mapped to IL1 and IL2, which do not interact with OR83b, and their role in odour recognition or signal transduction are unknown. The prevalence of divergent sites here may mean that relaxed constraints act on these regions; however, Or genes that showed the strongest signal of positive selection also had most sites within IL1 and IL2.
Surprisingly, the distribution of candidate sites in GRs differed from ORs, despite similar secondary structures. More sites mapped on the COOH-tail and fewer in the ILs in GRs. Evidence of positive selection was weakest for Gr genes (M8a and M8 comparisons were significant only for two Gr genes). Thus, more rapid divergence was probably due to relaxed purifying constraints. In this case, the prevalence of the candidate sites on the COOH-tail in GRs indicates that this domain is subject to less constraint than in ORs. Despite Drosophila GRs being evolutionarily related to the ORs, there are significant functional differences between these receptors. The olfactory system recognizes volatile ligands through the expression of ORs in the antenna and maxillary palps. GRs mediate response to soluble compounds and pheromones (Thorne et al. 2004), CO2 (Jones et al. 2007), and can be expressed in the central nervous system, possibly indicating non-gustatory roles (Thorne & Amrein 2008). The Drosophila olfactory system functions through OR/OR83b complexes, while GRs function independently of OR83b (Benton et al. 2006), suggesting that ORs and GRs have independent molecular properties as well as different functions.
Acknowledgments
We are grateful to the NERC (grant NE/C003187/1), and to Daniel Barker, Shiela Unkles and anonymous reviewers for their advice.
Supplementary Material
Analysed amino acids with order number of D. melanogaster proteins and posterior probability calculated by BEB.
The results of the M1–M2, M7–M8, M8a–M8 comparisons for 20 loci that show strongest signature of selection.
References
- Anisimova M., Bielawski J.P., Yang Z. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S.W., Vosshall L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. doi:10.1371/journal.pbio.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. doi:10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Clyne P.J., Warr C.G., Freeman M.R., Lessing D., Kim J., Carlson J.R. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. doi:10.1016/S0896-6273(00)81093-4 [DOI] [PubMed] [Google Scholar]
- Clyne P.J., Warr C.G., Carlson J.R. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. doi:10.1126/science.287.5459.1830 [DOI] [PubMed] [Google Scholar]
- Consortium (12 Drosophila Genomes Consortium) Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. doi:10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Gardiner A., Barker D., Butlin R.K., Jordan W.C., Ritchie M.G. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol. Ecol. 2008;17:1648–1657. doi: 10.1111/j.1365-294X.2008.03713.x. doi:10.1111/j.1365-294X.2008.03713.x [DOI] [PubMed] [Google Scholar]
- Guo S., Kim J. Molecular evolution of Drosophila odorant receptor genes. Mol. Biol. Evol. 2007;24:1198–1207. doi: 10.1093/molbev/msm038. doi:10.1093/molbev/msm038 [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe D.S., Scafe C.R., McKinney A.J., Tanouye M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. doi:10.1101/gr.239402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMbase—a database of membrane spanning proteins segments. J. Biol. Chem. 1993;374:166. [Google Scholar]
- Jones W.D., Cayirlioglu P., Kadow G.I., Vosshall L.B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. doi:10.1038/nature05466 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. doi:10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Lundin C., Käll L., Kreher S.A., Kapp K., Sonnhammer E.L., Carlson J.R., von Heijne G., Nilsson I. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581:5601–5604. doi: 10.1016/j.febslet.2007.11.007. doi:10.1016/j.febslet.2007.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.S., Arguello R.J. Five Drosophila genomes reveal non-neutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. doi:10.1534/genetics.107.078683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. doi:10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Warr C.G., Carlson J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. doi:10.1073/pnas.2335847100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Pellegrino M., Nakagawa T., Nakagawa T., Vosshall L.B., Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. doi:10.1038/nature06850 [DOI] [PubMed] [Google Scholar]
- Schwartz T.W., Frimurer T.M., Holst B., Rosenkilde M.M., Elling C.E. Molecular mechanism of 7TM receptor activation—a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. doi:10.1146/annurev.pharmtox.46.120604.141218 [DOI] [PubMed] [Google Scholar]
- Smadja C., Butlin R.K. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity. 2009;102:77. doi: 10.1038/hdy.2008.55. doi:10.1038/hdy.2008.55 [DOI] [PubMed] [Google Scholar]
- Smart R., et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. doi:10.1016/j.ibmb.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Nielsen R., Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Thorne N., Amrein H. Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J. Comp. Neurol. 2008;506:548–568. doi: 10.1002/cne.21547. doi:10.1002/cne.21547 [DOI] [PubMed] [Google Scholar]
- Thorne N., Chromey C., Bray S., Amrein H. Taste perception and coding in Drosophila. Curr. Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. doi:10.1016/j.cub.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Wicher D., Schafer R., Bauernfeind R., Stensmyr M.C., Heller R., Heinemann S.H., Hansson B.S. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. doi:10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- Wistrand M., Kall L., Sonnhammer E.L.L. A general model of G protein-coupled receptor sequences and its application to detect remote homologs. Protein Sci. 2006;15:509–521. doi: 10.1110/ps.051745906. doi:10.1110/ps.051745906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comp. Appl. BioSci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. doi:10.1093/bioinformatics/13.5.555 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W.S.W., Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. doi:10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysed amino acids with order number of D. melanogaster proteins and posterior probability calculated by BEB.
The results of the M1–M2, M7–M8, M8a–M8 comparisons for 20 loci that show strongest signature of selection.