Abstract
Viralload (VL) assessment of cytomegalovirus (CMV) by real-time PCR is an important tool for diagnosing and monitoring CMV viremia in patients with compromised immune systems. We report results from a sample exchange organized by members of the Association for Molecular Pathology that compared PCR results from 23 laboratories; 22 such laboratories used a laboratory-developed real-time PCR assay and one laboratory used a competitive PCR assay. The samples sent to each laboratory were comprised of a dilution panel of CMV virion-derived reference materials that ranged from 0 to 500,000 copies/ml. Accuracy, linearity, and intralaboratory precision were established for the different laboratory-developed assays. Overall, PCR results were linear for each laboratory (R2 > 0.97 in all but two). While 13 laboratories showed no significant quantitative assay bias, 10 laboratories reported VLs that were significantly different compared with expected values (bias range, −0.82 to 1.4 logs). The intralaboratory precision [mean coefficient of variance of 2% to 5% (log-scale)] suggested that changes in VLs of less than 3- to fivefold may not be significantly different. There was no significant association between laboratory-specific technical variables (PCR platform, calibrator, extraction method) and assay linearity or accuracy. These data suggested that, within each laboratory, relative VL values were linear, but additional method standardization and a CMV DNA reference standard are needed to allow laboratories to achieve comparable numeric results.
Cytomegalovirus (CMV) is a double-stranded DNA virus that is a member of the Herpesviridae family. Establishment of persistent and latent infections with CMV in the human host is a common occurrence. With seropositivity rates at 80% and CMV viral reactivation being associated with a compromised immune system, CMV is considered one of the most common opportunistic pathogens leading to life-threatening illnesses in the immunocompromised patient. Quantitative CMV viral load (VL) testing has been shown to be useful in diagnosing active disease, screening for preemptive therapy, and monitoring therapeutic response.1,2,3,4,5,6,7,8,9,10,11 Therefore, accurate, sensitive, and precise diagnostic tests that can detect and quantitate CMV viral load are essential.
Despite the need for accurate quantitative CMV viral load measurement, no U. S. Food and Drug Administration-approved assay is currently available. Typically, CMV is quantitated using the pp65 antigenemia assay or using real-time PCR. The pp65 antigenemia assay is a useful surrogate marker for CMV viral load; however, it is only useful in patients with sufficient leukocyte counts, is labor-intensive, and has strict sample requirements.12,13 The use of a quantitative PCR assessment for CMV viral load provides significantly better sensitivity and precision.12,14,15 Within individual laboratories, real-time PCR assays are able to quantify CMV DNA accurately in a closed system and over a wide dynamic range, allowing for quick turnaround-time and less possibility for contamination.2,5,12,14,15,16 However, given the heterogeneity of assay systems, the comparison of VL values between different laboratories may not be as accurate or precise as intralaboratory VLs. Toward that goal, we report results from a multilaboratory sample-exchange study designed to assess the performance characteristics of laboratory-developed real-time PCR assays using common CMV reference materials.
Materials and Methods
Purpose of the Study
The purpose of this study was to compare different laboratory-developed CMV DNA real-time PCR assay performance characteristics using common CMV reference materials. Laboratories enrolled in the study by subscribing through the Association for Molecular Pathology (AMP) CMV Working Group Listserv. Participation was limited to the first 25 laboratories that enrolled.
Materials Supplied
Four sets of AcroMetrix OptiQuant CMV Viral DNA Panels (Benicia, CA), each comprised of a 5-sample serial dilution of intact CMV viral particles, were sent to participating laboratories by overnight courier on dry ice. Laboratories were instructed to store the panel members at −70°C until use. The CMV Viral DNA panel was prepared by diluting and adjusting a concentrated CMV virion solution from a CMV-producing cell line in normal human plasma non-reactive for antibodies to other infectious agents. The normal human plasma also contained 0.05% sodium azide, and 0.05% gentamicin sulfate as preservatives. The highest member of the dilution series (determined to have a CMV viral load of 500,000 copies/ml using the Roche COBAS Amplicor CMV Monitor test; Indianapolis, IN) was serially diluted to obtain the other four members of the panel. Panel members had CMV DNA concentrations of 0, 500, 5000, 50,000, and 500,000 copies/ml.
Test Procedure
Participating laboratories used the DNA extraction procedure that was appropriate to their laboratory-developed CMV test system to extract CMV DNA from the CMV panel member samples. Four replicate samples of each panel member were extracted and amplified separately. Each participant laboratory was instructed to test the samples as “unknowns” using the calibrators particular to their assay. The results from each laboratory's assay had to meet validity criteria for their laboratory-developed CMV DNA test system. Results were reported in the units of measure appropriate to the assay (eg, copies/ml), along with the calculated mean, SD, and percent coefficient of variance (% CV) for each panel member replicate. Individual sample results that did not meet the laboratory's validity criteria were excluded. Panel member results that fell outside each laboratory's dynamic range were reported as such (eg, <50 copies/ml or >500,000 copies/ml). Each laboratory participant forwarded copies of all raw data to the study center for comparative analysis and statistical evaluation. CMV VL results were compared between individual laboratory values (observed) and the reference panel expected values, by linear regression of the log10 transformed data. For each laboratory, bias, if significant, was defined as the mean difference between observed and expected log-VL values. Bias was considered significant when a paired t-test (observed versus expected VL) yielded a P value below 0.05 (two-sided).
Results and Discussion
Twenty-five laboratories were enrolled in this study; however, data were only provided by 23 laboratories. Twenty-two of these laboratories used an in-house developed real-time PCR assay while the COBAS Amplicor Monitor PCR platform was used by one laboratory. Table 1 lists the test system parameters for each laboratory including the instrument platform, the extraction method, and the calibrator. Twelve laboratories reported results using Roche's LightCycler Real-Time PCR System instrument. Six laboratories used one of the Applied Biosystems (ABI; Foster City, CA) Real-Time PCR System instruments, and the remaining laboratories used either the iCycler (BioRad Laboratories, Hercules, CA), SmartCycler (Cepheid, Sunnyvale, CA), or COBAS Amplicor Monitor PCR platform. TIB MolBiol (Berlin, Germany), Advanced Biotechnologies Inc (ABI, Columbia MD), and in-house produced plasmids were the most commonly used calibrators, and the majority of laboratories used a Qiagen (Valencia, CA) product for DNA extraction (n = 12). The region of CMV targeted by the PCR primers was reported by 16 laboratories. Ten laboratories used glycoprotein B (UL55) and two used DNA polymerase (UL54) as the PCR target. One laboratory, each, reported using primers for pp65 (UL83), HindIII X region, UL117, and the immediate early gene (UL123).
Table 1.
PCR Platforms, Extraction Methods, and Calibrators Used by Participating Laboratories
| Lab# | Instrument / platform | Extraction method | Calibrator |
|---|---|---|---|
| 1 | LightCycler | QIAamp ultraSens | TIB plasmid* |
| 2 | LightCycler | QIAamp DNA mini kit | TIB plasmid |
| 3 | LightCycler | QIAamp minElute | TIB plasmid |
| 4 | LightCycler | Qiagen blood mini Kit | TIB plasmid |
| 5 | LightCycler | QIAamp DNA mini kit | TIB plasmid |
| 6 | LightCycler | Gentra pureGene | CMVAD169 quantitated viral DNA† |
| 7 | LightCycler | Roche highPure | TIB plasmid |
| 8 | LightCycler | Roche magnaPure | TIB plasmid |
| 9 | LightCycler | Roche magnaPure | In-house plasmid |
| 10 | LightCycler | Roche magnaPure | In-house culture |
| 11 | LightCycler | Roche magnaPure | In-house plasmid |
| 12 | LightCycler | Roche highPure | CMVAD169 quantitated viral DNA |
| 13 | ABI 7000 | bioMerieux nucliSens | CMVAD169 quantitated viral DNA |
| 14 | ABI 7500 | Qiagen M48 automated | In-house plasmid |
| 15 | ABI 7700 | QIAamp DNA mini kit | CMVAD169 quantitated viral DNA |
| 16 | ABI 7700 | Qiagen biorobot | In-house plasmid |
| 17 | ABI 7900HT | Gentra capture column | ATCC CMVAD-169 |
| 18 | ABI Prism 7900HT | Roche magnaPure | In-house purified DNA |
| 19 | SmartCycler | Qiagen blood mini kit | CMVAD169 quantitated viral DNA |
| 20 | SmartCycler | QIAamp minElute | OptiQuant CMV viral DNA‡ |
| 21 | iCycler | Qiagen viral kit | CMVAD169 quantitated viral DNA |
| 22 | iCycler | Qiagen blood mini kit | CMVAD169 quantitated viral DNA |
| 23 | COBAS AmplicorCMVmonitor | Amplicor CMV monitor | Kit quantitation standard§ |
TIB MolBiol.
Advanced Biotechnologies Inc.
AcroMetrix.
Roche Diagnostics.
The quantitative CMV VL test results from each of the 23 participating laboratories are shown in Table 2. The log10 transformed viral load values expected for the dilution series were 2.70, 3.70, 4.70, and 5.70 log copies/ml, respectively. The mean (log) measured VL for all 23 laboratories was 2.96 (n = 75), 3.81 (n = 85), 4.77 (n = 91), and 5.66 (n = 92) for the panel members respectively, and measured VLs correlated linearly with expected VLs (r = 0.9998; P < 0.0001; Figure 1). The mean observed VL of the 500 copies/ml sample (2.96) was slightly (but significantly) higher than the expected value (2.70) (P = 0.0005), while the observed VLs of the other three dilutions were not significantly different from expected values. The absence of a significant “observed” versus “expected” viral load difference in 3 of the 4 CMV-positive panel members suggests that the viral load value of the panel members was accurately assigned. Panel member 0 was negative for CMV DNA, and all laboratories appropriately reported VL values less than their lower limit of detection. With the exception of three laboratories (2, 17 and 19) that failed to detect CMV in the sample with an expected 500 copies/ml [one of which (2) also failed to detect CMV at 5000 copies/ml], all other CMV-positive samples were reported as “CMV detectable” by all laboratories.
Table 2.
CMV DNA Viral Load Values Obtained from Participating Laboratories
| Mean copies/ml (log) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Member 1 |
Member 2 |
Member 3 |
Member 4 |
|||||
| Laboratory | 2.70 copies/ml | 3.70 copies/ml | 4.70 copies/ml | 5.70 copies/ml | R2 | Slope | Bias | Bias P |
| 1 | 4.12 | 4.37 | 5.08 | 5.97 | 0.94755 | 0.63125 | NS | 0.08 |
| 2 | * | * | 4.00 | 4.78 | NS | 0.09 | ||
| 3 | 3.27 | 4.04 | 5.18 | 5.74 | 0.98133 | 0.85886 | NS | 0.07 |
| 4 | 2.92 | 3.89 | 5.01 | 5.75 | 0.98767 | 0.94608 | NS | 0.15 |
| 5 | 2.54 | 3.17 | 4.47 | 5.02 | 0.97411 | 0.85588 | −0.44 | 0.04 |
| 6 | 3.02 | 3.64 | 4.79 | 5.94 | 0.98114 | 1.00206 | NS | 0.28 |
| 7 | 3.53 | 4.72 | 5.77 | 6.58 | 0.99112 | 1.02726 | 0.93 | 0.0007 |
| 8 | 3.06 | 3.98 | 4.97 | 6.02 | 0.99937 | 0.98805 | 0.30 | 0.0005 |
| 9 | 2.79 | 3.59 | 4.76 | 5.70 | 0.99473 | 0.99357 | NS | 0.93 |
| 10 | 3.36 | 4.31 | 5.27 | 6.25 | 0.99998 | 0.96493 | 0.59 | 0.0001 |
| 11 | 2.77 | 3.84 | 4.92 | 5.93 | 0.99898 | 1.04234 | NS | 0.09 |
| 12 | 2.98 | 3.76 | 4.63 | 5.68 | 0.98906 | 0.89979 | NS | 0.99 |
| 13 | 2.59 | 3.80 | 4.71 | 5.66 | 0.99493 | 1.01562 | NS | 0.73 |
| 14 | 4.08 | 5.20 | 6.10 | 7.25 | 0.99789 | 1.04226 | 1.4 | <0.0001 |
| 15 | 2.04 | 3.20 | 4.26 | 5.56 | 0.99848 | 1.17756 | −0.47 | 0.03 |
| 16 | 2.71 | 3.68 | 4.73 | 5.67 | 0.99835 | 1.02046 | NS | 0.08 |
| 17 | * | 3.45 | 4.14 | 4.75 | 0.99851 | 0.64993 | NS | 0.10 |
| 18 | 2.51 | 3.50 | 4.62 | 5.63 | 0.99952 | 1.04313 | −0.14 | 0.02 |
| 19 | * | 3.29 | 4.30 | 4.97 | 0.99929 | 0.83920 | −0.60 | 0.02 |
| 20 | 2.60 | 3.68 | 4.72 | 5.68 | 0.99907 | 1.03937 | NS | 0.25 |
| 21 | 2.53 | 3.54 | 4.46 | 5.47 | 0.99992 | 1.01326 | −0.26 | 0.0001 |
| 22 | 2.30 | 2.95 | 3.74 | 4.83 | 0.96067 | 0.86626 | −0.82 | 0.01 |
| 23 | 3.41 | 4.30 | 5.04 | 5.45 | 0.97437 | 0.69326 | NS | 0.20 |
| Mean | 2.96 | 3.81 | 4.77 | 5.66 | ||||
Samples with undetectable CMV were excluded from the analysis.
Figure 1.
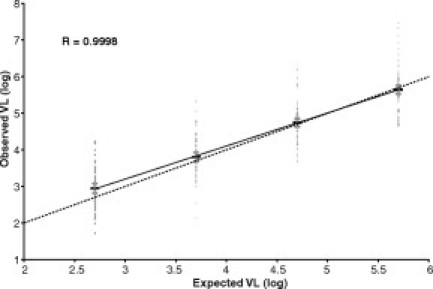
The mean viral load value for each panel member is shown (thick horizontal line) and is flanked by error bars (shorter horizontal lines) reflecting 95% confidence intervals. Raw viral load values (n = 343) are also shown. The best-fit linear regression line (r = 0.9998; slope = 0.90) is shown as a solid line connecting the four data groups. The dotted diagonal line reflects equivalence of X- and Y-values.
The relative quantification of CMV DNA, as judged by the regression line slope of each laboratory's dilution curve, showed an R-squared value of >0.99 in 14 laboratories (64%), and >0.97 in all but two laboratories (91%) (Table 2). Twelve laboratories (55%) had regression line slopes within 5% of the ideal 1.0 value (0.95 to 1.05), while 9 laboratories (41%) had slopes outside of the ± 10% range (0.9 to 1.1). Assay accuracy, as judged by bias (mean difference) relative to expected values, showed that, of the 23 participating laboratories, 13 (57%) reported CMV viral loads that were not significantly different from expected values. Of the 10 laboratories with a statistically significant assay bias, six laboratories consistently reported lower VLs (mean 0.46 logs) and four laboratories consistently reported higher VLs (mean 0.81 logs) than the expected values. The magnitude of this assay bias was variable, but was greater than 1-log in only one laboratory. The significant bias in these 10 laboratories suggests a possible assay accuracy issue, which would be correctable with a recalibration using “gold standard” viral load standards (if any were available). In transplant centers, however, the predominant clinical application of CMV viral load values is monitoring the efficacy of anti-viral therapy by confirming a relative (not absolute) post-therapy decline in viral loads—which requires only a linear assay, and not necessarily an accurate one. As assay linearity, as judged by the regression line slope, was not significantly different in the 10 laboratories with significant assay bias (mean slope = 0.982) as compared with the 13 laboratories without assay bias (mean slope = 0.899; P > 0.1), there was no evidence to suggest that this analytical assay bias would negatively impact clinical care. Conversely, if clinical decisions, such as when to initiate anti-viral therapy, were being made based on absolute CMV viral load thresholds, the analytical assay bias in these 10 laboratories could perhaps be negatively impacting clinical care. The laboratories with significant assay bias also could not be distinguished (P > 0.05) from those without assay bias with respect to any of the three “technical” variables listed in Table 1 (PCR platform, extraction method, and calibrator).
As each laboratory tested four independent replicate samples at each of the four dilutions, intra-assay precision, as determined by the CV, could be assessed. Precision data (Table 3) showed that the CV values for each laboratory, at each dilution level, were heterogeneous, but averaged 7.47% at 500 copies/ml, 4.66% at 5000 copies/ml, 2.83% at 50,000 copies/ml, and 2.35% at 500,000 copies/ml. Predictably, as with other quantitative assays, precision progressively worsened at analyte concentrations approaching the assay's lower limit of detection. These intralaboratory CV values of ∼2% to 5% (at VLs above the lower-level detection limit) were used to calculate a 95% confidence interval for any measured viral load value that encompassed a 0.52 to 0.70 log (3.3 to fivefold) range (Table 3). From a practical perspective, this means that within any one laboratory, any two independently-determined VL values must differ by more than 3 to fivefold (non-log scale) to be considered statistically “different” (with 95% confidence). Conversely, VL changes of less than 3 to fivefold must be interpreted cautiously and may represent intrinsic analytical assay imprecision rather than a true biological difference.
Table 3.
Intra-laboratory Variability of the CMV Viral Load Assay
| CMV viral load intra-lab coefficient of variation (CV) (%) |
||||
|---|---|---|---|---|
| Member 1 | Member 2 | Member 3 | Member 4 | |
| Laboratory | 2.70 log copies/ml | 3.70 log copies/ml | 4.70 log copies/ml | 5.70 log copies/ml |
| 1 | 3.85 | 3.60 | 2.60 | 0.96 |
| 2 | * | * | 2.74 | 1.51 |
| 3 | 6.14 | 5.58 | 2.37 | 4.07 |
| 4 | 9.62 | 2.51 | 0.30 | 6.94 |
| 5 | 7.35 | 4.31 | 5.07 | 5.94 |
| 6 | 7.20 | 6.67 | 2.81 | 1.15 |
| 7 | 6.39 | 1.01 | 0.85 | 2.20 |
| 8 | 5.19 | 2.29 | 1.70 | 2.38 |
| 9 | 3.45 | 3.40 | 0.64 | 0.30 |
| 10 | 4.28 | 2.22 | 0.95 | 1.83 |
| 11 | 14.05 | 0.56 | 0.95 | 1.07 |
| 12 | 5.30 | 12.33 | 6.67 | 3.46 |
| 13 | 5.97 | 1.94 | 1.89 | 1.41 |
| 14 | 6.11 | 3.14 | 2.11 | 3.30 |
| 15 | 16.16 | 1.76 | 4.02 | 3.53 |
| 16 | 14.87 | 8.81 | 3.52 | 4.68 |
| 17 | * | 3.27 | 1.85 | 2.04 |
| 18 | 2.54 | 0.53 | 2.29 | 2.02 |
| 19 | * | 3.78 | 11.56 | 2.71 |
| 20 | 9.16 | 1.28 | 3.07 | 1.07 |
| 21 | 14.35 | 9.42 | 4.25 | 0.25 |
| 22 | 2.88 | 22.84 | 2.21 | 0.72 |
| 23 | 4.50 | 1.30 | 0.72 | 0.62 |
| Average CV (%) | 7.47 | 4.66 | 2.83 | 2.35 |
| Mean VL (log) | 2.96 | 3.81 | 4.77 | 5.66 |
| 95% CI of VL (log) | 2.53–3.39 | 3.46–4.16 | 4.51–5.03 | 5.40–5.92 |
| Log Change: 95% CI | 0.87 | 0.70 | 0.53 | 0.52 |
| Fold Change: 95% CI | 7.4 | 5.0 | 3.4 | 3.3 |
Samples with undetectable CMV were excluded from the analysis.
Each laboratory-developed test varied with respect to the PCR platform, extraction method, and calibrators that were used to measure the CMV viral load. In addition, laboratories may have optimized the testing systems for different sample types, such as whole blood or plasma. The resulting small number of laboratories within each technical subgroup yielded insufficient statistical power for definitive conclusions regarding the effects of these technical variables on assay performance characteristics. Expectedly, with one borderline exception, none of the three technical variables listed in Table 1 (PCR platform, extraction method, and calibrator) had a detectably significant effect (P > 0.05) on assay linearity, as measured by either the slope or the R2 value of the laboratory-specific regression line. The one possible exception was a trend toward significantly improved slope (P = 0.08) and R2 (P = 0.05) parameters for the five laboratories using calibrators made in-house (mean slope = 1.03; mean R2 = 0.998) compared with the six laboratories using the commercially-available plasmid calibrator from TIB (mean slope = 0.885; mean R2 = 0.980).
Results from this study demonstrated that a heterogeneous spectrum of different laboratory-developed CMV viral load assays maintained linearity with a serial dilution panel of samples—implying accurate relative quantification within each laboratory. However, due to each laboratory developing its own assay parameters and calibration systems, a substantial minority of laboratories (43%) generated numeric VLs significantly different from expected values. The specific technical variables associated with diagnostic accuracy could not be confidently ascertained by this small study. The resulting interlaboratory variability of numeric VL values is problematic for clinical laboratories, as well as for physicians and patients, particularly when serial CMV monitoring is performed at multiple sites. To better standardize numeric CMV VL quantification and assist laboratories in calibrating their diverse laboratory-developed assays, an accurately quantitated, large-volume, stable, widely available analytic standard material is greatly needed. Plans for producing such a CMV reference standard have recently been undertaken by the National Institute of Standards and Technology.
Acknowledgements
We acknowledge the participation of Advocate Lutheran General Hospital (Jan Nowak); Children's Mercy Hospital (Rangaraj Selvarangan); Baylor University Medical Center (Rana Saad); Carolinas HealthCare System (John Longshore); Children's Hospital (Doug Salamon); Loma Linda Mercantile (Sonia Neidigh); Mayo Clinic (Thomas F. Smith); Medical College of Virginia (Michael R. Langley); Medical University of South Carolina (Daynna Wolff); University of Rochester Medical Center (Paul G. Rothberg); Molecular Pathology Laboratory Network, Inc. (Teresa Webb-Martin); Oregon Health & Sciences University (Mary Hawley); Sharp Memorial Hospital (Cathy Woerle); Spectrum Health (Thomas J. Monroe); St. Barnabas Medical Center (Marlene Sabbath-Solitare); The Methodist Hospital (Winnie Wu); The Nebraska Medical Center (Shirley Shepherd); UMDNJ/NJ Medical School (Helen Fernandes); University of Florida (Herbert J. Houck); University of North Carolina (Margaret Gulley); University of Pittsburgh/Magee-Women's Hospital (Jeanne Jordan); University of Texas SW Medical Clinic (Shamira Nilasena); and University of Washington Medical Center (Ka Wing Ng); Washington Hospital Center (Raghunath P. Agarwal).
Footnotes
The CMV Working Group is a subcommittee of the AMP Clinical Practice Committee.
The 2003–2008 Association for Molecular Pathology (AMP) Clinical Practice Committee consisted of Jan Nowak (Chair 2003 to 2004), Elaine Lyon (Chair 2005–2006), Vicky Pratt (Chair 2007–2008), Jean Amos Wilson, Aaron Bossler, Michele Caggana, Domnita Crisan, Deborah Dillon, William Funkhouser, Julie Gastier-Foster, Meera Hameed, Dan Jones, Daniel Sabath, Antonia Sepulveda, Kathleen Stellrecht, Gregory Tsongalis, and Daynna Wolff.
Standard of practice is not being defined by this article, and there may be alternatives.
Financial disclosures: AcroMetrix Corp. provided reagents for these studies. P.N. is an employee of AcroMetrix Corp.
Current address for D.L.H. is Roche Diagnostics Corporation, 10480 Cranchester Way, Alpharetta, GA.
Address reprint requests to the Association for Molecular Pathology, 9650 Rockville Pike, Bethesda, MD 20814. E-mail: amp@amp.org
References
- 1.Aitken C, Barrett-Muir W, Millar CT, Thomas KJ, Sheridan F, Jeffries D, Yaqoob M, Breuer J. Use of molecular tests in diagnosis and monitoring of cytomegalovirus disease following renal transplantation. J Clin Microbiol. 1999;37:2804–2807. doi: 10.1128/jcm.37.9.2804-2807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen EF, Sabin CA, Wilson P, Griffiths PD, Davey CC, Johnson MA, Emery VC. Cytomegalovirus (CMV) viraemia detected by polymerase chain reaction identifies a group of HIV-positive patients at high risk of CMV disease. AIDS. 1997;11:889–893. doi: 10.1097/00002030-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Caliendo AM, St. George K, Allega J, Bullotta AC, Gilbane L, Rinaldo CR. Distinguishing cytomegalovirus (CMV) infection and disease with CMV nucleic acid tests. J Clin Microbiol. 2002;40:1581–1586. doi: 10.1128/JCM.40.5.1581-1586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassan-Walker AF, Kidd IM, Sabin C, Sweny P, Griffiths PD, Emery VC. Quantity of human cytomegalovirus (CMV) DNAemia as a risk factor for CMV disease in renal allograft recipients: relationship with donor/recipient CMV serostatus, receipt of augmented methylprednisolone and antithymocyte globulin (ATG) J Med Virol. 1999;58:182–187. [PubMed] [Google Scholar]
- 5.Humar A, Gregson D, Caliendo AM, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation. 1999;68:1305–1311. doi: 10.1097/00007890-199911150-00015. [DOI] [PubMed] [Google Scholar]
- 6.Paya CV, Sia IG, Wilson JA, Groettum CM, Espy MJ, Smith TF. Cytomegalovirus (CMV) DNA load predicts relapsing of CMV infection after solid organ transplantation. J Infect Dis. 2000;181:717–720. doi: 10.1086/315242. [DOI] [PubMed] [Google Scholar]
- 7.Roberts TC, Brennan DC, Buller RS, Gaudreault-Keener M, Schnitzler MA, Sternhell KE, Garlock KA, Singer GG, Storch GA. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis. 1998;178:626–635. doi: 10.1086/515383. [DOI] [PubMed] [Google Scholar]
- 8.Rollag H, Sagedal S, Kristiansen KI, Kvale D, Holter E, Degre M, Nordal KP. Cytomegalovirus DNA concentration in plasma predicts development of cytomegalovirus disease in kidney transplant recipients. Clin Microbiol Infect. 2002;8:431–434. doi: 10.1046/j.1469-0691.2002.00449.x. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez JL, Kruger RM, Paranjothi S, Trulock EP, Lynch JP, Hicks C, Shannon WD, Storch GA. Relationship of cytomegalovirus viral load in blood to pneumonitis in lung transplant recipients. Transplantation. 2001;72:733–735. doi: 10.1097/00007890-200108270-00030. [DOI] [PubMed] [Google Scholar]
- 10.Sia IG, Wilson JA, Groettum CM, Espy MJ, Smith TF, Paya CV. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J Infect Dis. 2000;181:717–720. doi: 10.1086/315242. [DOI] [PubMed] [Google Scholar]
- 11.Spector SA, Hsia K, Crager M, Pilcher M, Cabral S, Stempien MJ. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J Virol. 1999;73:7027–7030. doi: 10.1128/jvi.73.8.7027-7030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schvoerer E, Henriot S, Zachary P, Freitag R, Fuchs A, Fritsch S, Risch S, Meyer N, Caillard S, Lioure B, Stoll-Keller F. Monitoring low cytomegalovirus viremia in transplanted patients by a PCR on plasma. J Med Virol. 2005;76:76–81. doi: 10.1002/jmv.20326. [DOI] [PubMed] [Google Scholar]
- 13.Pancholi P, Wu F, Della-Latta P. Rapid detection of cytomegalovirus infection in transplant patients. Expert Rev Mol Diagn. 2004;4:231–242. doi: 10.1586/14737159.4.2.231. [DOI] [PubMed] [Google Scholar]
- 14.Cortez KJ, Fischer SH, Fahle GA, Calhoun LB, Childs RW, Barrett AJ, Bennett JE. Clinical trial of quantitative polymerase chain reaction for detection of cytomegalovirus in peripheral blood of allogeneic hematopoietic stem-cell transplant recipients. J Infect Dis. 2003;188:967–972. doi: 10.1086/378413. [DOI] [PubMed] [Google Scholar]
- 15.Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. Optimization of quantitative detection of cytomegalovirus DNA in plasma by PCR. J Clin Microbiol. 2004;42:1142–1148. doi: 10.1128/JCM.42.3.1142-1148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caliendo AM, Ingersoll J, Fox-Canale M, Pargman S, Bythwood T, Hayden MK, Bremer JW, Lurain NS. Evaluation of PCR laboratory-developed tests using analyte-specific reagents for cytomegalovirus quantification. J Clin Microbiol. 2007;45:1723–1727. doi: 10.1128/JCM.02558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Uncited references
- 17.Hong KM, Najjar H, Hawley M, Press RD. Quantitative PCR with automated sample preparation for diagnosis and monitoring of cytomegalovirus infection in bone marrow transplant patients. Clin Chem. 2004;50:846–856. doi: 10.1373/clinchem.2003.026484. [DOI] [PubMed] [Google Scholar]
- 18.Caliendo AM, Schuurman R, Yen-Lieberman B, Spector SA, Andersen J, Manjiry R, Crumpacker C, Lurain NS, Erice A. CMV Working Group of the Complications of HIV Disease RAC, AIDS Clinical Trials Group: Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J Clin Microbiol. 2001;39:1334–1338. doi: 10.1128/JCM.39.4.1334-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


