Abstract
Along with unequivocal hits produced by matching multiple MS/MS spectra to database sequences, LC-MS/MS analysis often yields a large number of hits of borderline statistical confidence. To simplify their validation, we propose to use rapid de novo interpretation of all acquired MS/MS spectra and, with the help of a simple software tool, display the candidate sequences together with each database search hit. We demonstrate that comparing hit database sequences and independent de novo interpretations of the same MS/MS spectra assists in rapid examination of ambiguous matches.
Keywords: De novo sequencing, Hit validation, MASCOT, MS BLAST, MS, MS
LC-MS/MS experiments typically produce a large number of protein hits with varying statistical confidence. While hits with several matched peptides are usually undisputed within any statistical framework, database searches also yield many hits with borderline confidence. Statistical estimates, if applied to individual MS/MS spectrum, might only indicate the probability that its match to a database sequence is a random event (algorithms and their software implementation in database searches are reviewed in [1]; for recent advances in statistical confidence assessments see [2–5]). However, relatively poor scoring does not necessarily mean a false-positive hit. It is often caused by a relatively small number of detected fragments and high chemical noise that are both typical for MS/MS spectra acquired from low abundant peptides or reflect their specific structural features. Therefore, if exhaustive interpretation of the dataset is desired, further consideration of borderline hits typically proceeds via a case-by-case inspection of corresponding MS/MS spectra by an MS expert or by applying additional empirical confidence criteria.
It was also suggested that complementary de novo interpretation of MS/MS spectra could validate borderline database searching hits [6–8]. Indeed, de novo interpretation deduces a stretch of the peptide sequence directly from the interpreted MS/MS spectrum and is fully independent of a database sequence resource. However, it seldom yields a single unequivocal peptide sequence. Instead, it is far more common that a series of redundant, degenerate, incomplete and partially accurate sequence candidates is produced. Arbitrary sequence quality scores only provide a rough idea of which might be the most trusted candidate, or if none of them is sufficiently accurate for validating the search hit. Therefore, it was further suggested to combine de novo interpretation of doubtful spectra with sequence-similarity database searches [9]. A combination of de novo sequencing and MS-driven BLAST (MS BLAST) [10, 11] was applied to validate borderline hits in organisms with both known and unknown genomes [12–16]. Linear ion trap MS/MS spectra that produced matches with borderline significance were interpreted de novo by PepNovo software [17] (version Pep- Novo2MSB is available at http://proteomics.bioprojects.org/Software/PepNovo.html). Several candidate sequences per each spectrum were assembled into a single search string, irrespective of their expected quality, and submitted to MS BLAST server at http://genetics.bwh.harvard.edu/msblast/index.html [9, 12]. De novo interpretation of a single IT spectrum took less than 0.5 s on a desktop PC and the output format conformed to MS BLAST conventions [11]. Considering several partially redundant sequence candidates in parallel and tolerating multiple mismatches within produced sequence alignments increased the chances of identifying source peptide sequences and compensated for several minor de novo-sequencing inaccuracies [12]. Statistical confidence of MS BLAST hits was determined using an alternative scoring scheme that did not rely upon E-values or p-values of the high scoring segment pairs (HSPs) [11]. Being complementary to MASCOT scoring, it is fully independent of matching m/z of individual fragment ions to the ones pre-computed from the presumed peptide sequence. Because of their high speed, MS BLAST searches were always performed against a comprehensive (all species) database, which readily identified unanticipated protein contaminants originating from cell media, protein expression host organisms, etc. In a series of computational experiments, MS BLAST identified more than 50% of bona fide source peptide sequences from MS/MS spectra, in which PepNovo was expected to determine at least six amino acid residues. While the statistical significance thresholds for single-peptide hits depends on the database size the proposed method was able to validate >70% of borderline hits in searches against a comprehensive protein database [9, 12].
Although the validation procedure described in detail in [9] is simple, the examination of many borderline MASCOT hits remained tedious. Each time the operator needed to select the questioned hits, identify relevant spectra in the MS/MS query, submit them to de novo sequencing, collect sequence candidates from the output file and then compare them to sequences of examined MASCOT hits.
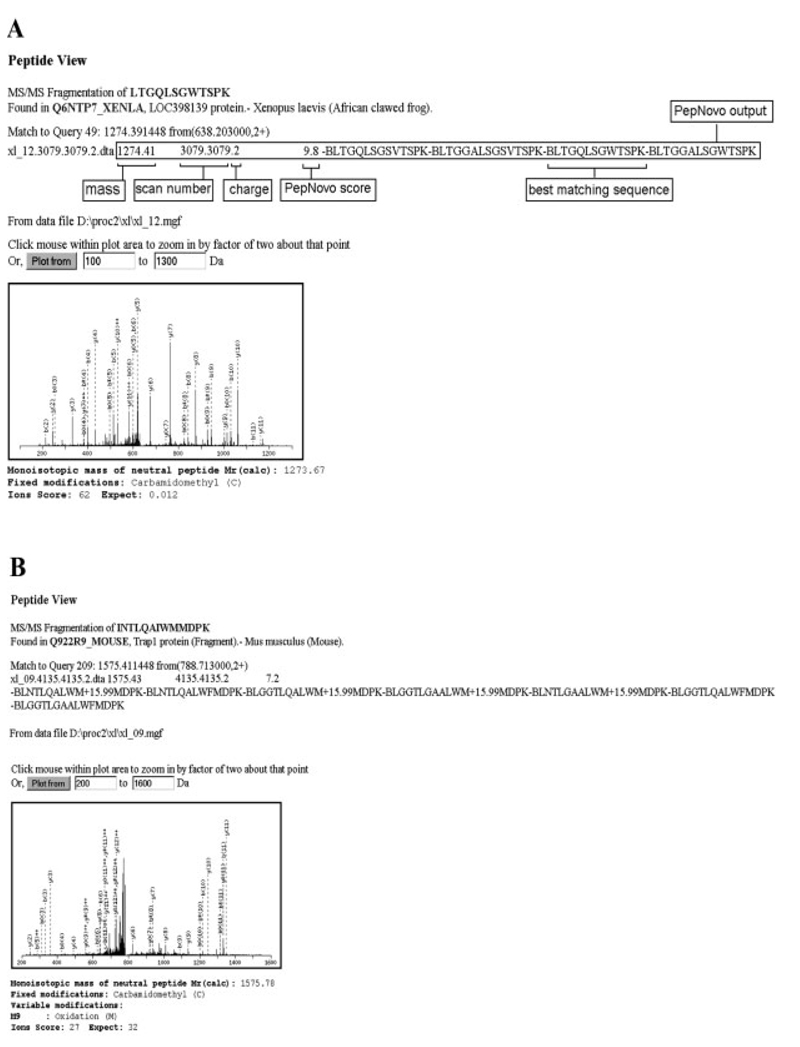
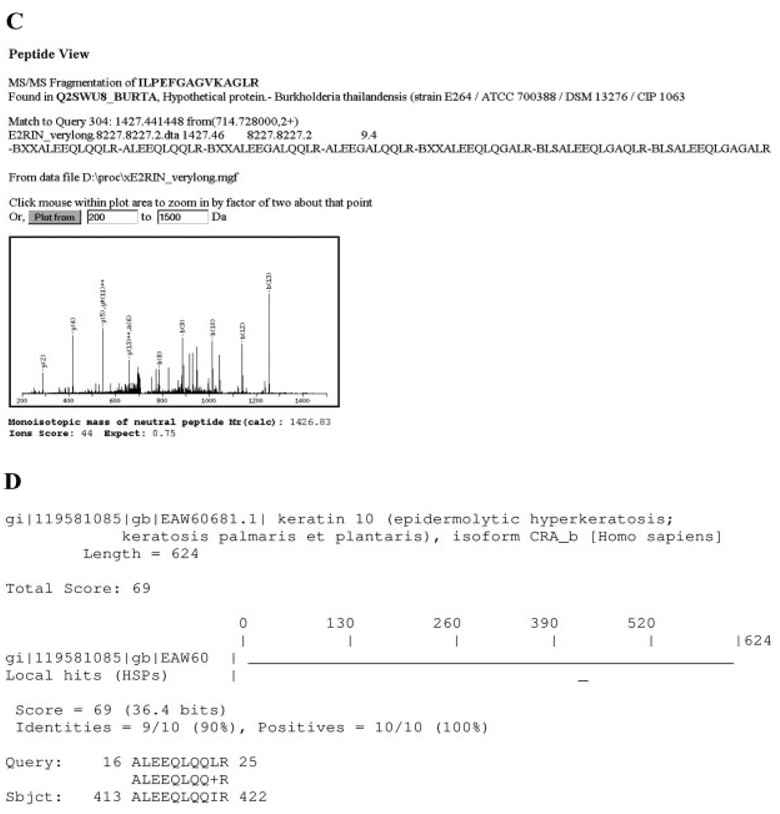
Because of the speed of PepNovo, we found it more practical to attempt with de novo sequencing all acquired MS/MS spectra prior to MASCOT searches. To simplify further analysis, we used a software tool developed inhouse that wrote de novo sequence candidates directly into the title line of each MS/MS spectrum within MASCOT (.mgf) query. Upon completing the search, candidate sequences were displayed, together with the MASCOT results, on the same Peptide View page. They could be also viewed in a pop-up window containing the ranked list of peptide hits and it was easy to find out if other lower ranked MASCOT hits agreed better with the de novo predictions. If no meaningful similarity to any MASCOT assignments was apparent, all de novo candidates were selected for the MS BLAST search against a comprehensive nr database (regularly updated, currently comprises 3 295 137 protein-sequence entries). We found that this simple approach was useful in sorting out false positives, which were rather common when MASCOT searches were performed against species-restricted sequence databases (Fig. 1). The presented MS/MS spectra were acquired on a LTQ IT mass spectrometer (Thermo Fisher Scientific, Waltham, MA) coupled with Ultimate Plus nanoLC system (Dionex, Amsterdam, The Netherlands) as described in [12]. Raw files were converted to .mgf files using extra-ct_ msn.exe and merge.pl scripts from Xcalibur software, (Thermo Fisher Scientific), which were then searched against an MSDB database (updated in September 2006 and comprising 3 239 079 sequence entries) using MAS-COT v.2.1 software installed on a local 2CPU server.
Figure 1.
Reporting de novo sequence candidates directly in Peptide View panels facilitates the validation of borderline hits. Searches were performed against a full MSDB database and the threshold score for statistically confident matching of a single peptide was equal to 55. Note that according to MS BLAST conventions, “-” spaces candidate sequences within the search string; B stands for a generic trypsin cleavage site (R or K amino acid residues); X stands for unidentified amino acid residue; M + 15.99 stands for the residue of oxidized methionine [11, 12]. (A) Confident MASCOT hit (peptide ions score of 62, E-value of 0.012) that was unequivocally confirmed by de novo sequencing. (B) False negative match (peptide ions score of 27, E-value of 32) that was, however, confirmed by de novo sequencing, which produced a candidates sequence with one mismatched amino acid residue. (C) Borderline hit (peptide ions score of 44; E-value of 0.75) that looked ambiguous since the matched peptide sequence poorly corroborated de novo predictions. In this case, the search string was directly submitted to MS BLAST search, which revealed its significant similarity to the sequence of human keratin peptide (D). Confidence of MS BLAST hits was determined according to its scoring scheme [11].
The software tool combining MASCOT and PepNovo outputs is a system of Windows shell scripts and a program written in FreeBasic and is available at http://www.mpicbg.de/~thomas/pepnovo-mascot-tool/. It is only required that .mgf files are located within the specified input folder. The program folder can be copied/renamed/moved and no program installation is required. Several .mgf files can be processed in batch mode and complete interpretation of a full LC-MS/MS run (comprising ca. 7000 IT MS/MS spectra) took less than 60 min on a desktop PC with a 2 GHz Intel Pentium 4 processor. In our experience, the time required for complete de novo interpretation could be further reduced by pre-processing .mgf queries using the filtering software, which removes non-annotated spectra originating from typical protein (trypsin autolysis, keratins, etc.) and chemical background [18]. We note, however, that a prudent user should always consider that de novo sequencing accuracy strongly depends on the charge, mass and amino-acid composition of fragmented peptide precursors. It is therefore possible that even highly informative spectra (as judged by their apparently rich fragment patterns and good S/N) acquired from triply or higher charged ions might not be amenable for de novo sequencing. Although sequence similarity searches relax the accuracy requirements [12], we anticipate that complementary peptide fragmentation methods [19] together with high-mass resolution of instruments [8, 20] might improve the overall quality of de novo interpretations.
We found that, beyond validating borderline hits, complementary interpretation of all acquired MS/MS spectra provided a wealth of useful analytical information, especially important for proteomics in organisms with unsequenced genomes [12]. While interpreting MS/MS spectra, PepNovo computes a sequence quality score, which stands for the expected number of accurately determined aminoacid residues [17, 20]. Optionally, by setting the PepNovo score as a quality filter [9, 12], only spectra that yield quality de novo sequences could be further submitted to MAS-COT searches, which then produce hits with a close to zero false- positive rate [8, 20]. Alternatively, all spectra (along with their de novo interpretations) confidently matched by MASCOT could be removed from the query and only candidate sequences from unmatched spectra further submitted to MS BLAST searches, hence facilitating the identification of missed unknown or polymorphic peptide sequences or even entire proteins in crude mixtures with known proteins.
Acknowledgments
We are grateful for members of Shevchenko laboratory for their input and expert support and for Drs. Shamil Sunyaev and Ivan Adzhubey for their work on MS BLASTserver.We thank Ms Judith Nicholls for critical reading the manuscript. The work in the Shevchenko lab was in part supported by 1R01GM070986-01A1 grant from NIH NIGMS.
Footnotes
The authors have declared no conflict of interest.
References
- 1.Forner F, Foster LJ, Toppo S. Mass spectrometry data analysis in the proteomics era. Curr. Bioinformatics. 2007;2:63–93. [Google Scholar]
- 2.Choi H, Ghosh D, Nesvizhskii AI. Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. J. Proteome Res. 2008;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 3.Sadygov RG, Liu H, Yates JR. Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal. Chem. 2004;76:1664–1671. doi: 10.1021/ac035112y. [DOI] [PubMed] [Google Scholar]
- 4.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 5.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JA, Johnson RS. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 2001;73:2594–2604. doi: 10.1021/ac001196o. [DOI] [PubMed] [Google Scholar]
- 7.An Thieu V, Kirsch D, Flad T, Muller C, et al. Direct protein identification from nonspecific peptide pools by high-accuracy MS data filtering. Angew. Chem. Int. Ed. Engl. 2006;45:3317–3319. doi: 10.1002/anie.200503787. [DOI] [PubMed] [Google Scholar]
- 8.Savitski MM, Nielsen ML, Kjeldsen F, Zubarev RA. Proteomics-grade de novo sequencing approach. J. Proteome Res. 2005;4:2348–2354. doi: 10.1021/pr050288x. [DOI] [PubMed] [Google Scholar]
- 9.Wielsch N, Thomas H, Surendranath V, Waridel P, et al. Rapid validation of protein identifications with the borderline statistical confidence via de novo sequencing and MS BLAST searches. J. Proteome Res. 2006;5:2448–2456. doi: 10.1021/pr060200v. [DOI] [PubMed] [Google Scholar]
- 10.Shevchenko A, Sunyaev S, Loboda A, Shevchenko A, et al. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 11.Habermann B, Oegema J, Sunyaev S, Shevchenko A. The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol. Cell. Proteomics. 2004;3:238–249. doi: 10.1074/mcp.M300073-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Waridel P, Frank A, Thomas H, Surendranath V, et al. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics. 2007;7:2318–2329. doi: 10.1002/pmic.200700003. [DOI] [PubMed] [Google Scholar]
- 13.Katz A, Waridel P, Shevchenko A, Pick U. Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by Blue-Native gel electrophoresis and nanoLC-MS/MS analysis. Mol. Cell. Proteomics. 2007;6:1459–1472. doi: 10.1074/mcp.M700002-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Gache V, Waridel P, Luche S, Shevchenko A, et al. Purification and mass spectrometry identification of microtubule- binding proteins from Xenopus egg extracts. Methods Mol. Med. 2007;137:29–43. doi: 10.1007/978-1-59745-442-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J. Biol. Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- 16.Charneau S, Junqueira M, Costa CM, Pires DL, et al. The saliva proteome of the blood-feeding insect Triatoma infestans is rich in platelet-aggregation inhibitors. Intl. J. Mass Spectrom. 2007;268:265–276. [Google Scholar]
- 17.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 18.Junqueira M, Spirin V, Balbuena TS, Waridel P, et al. Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 2008;7:3382–3395. doi: 10.1021/pr800140v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn DM, Zubarev RA, McLafferty FW. Automated de novo sequencing of proteins by tandem high-resolution mass spectrometry. Proc. Natl. Acad. Sci. USA. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, et al. De novo peptide sequencing and identification with precision mass spectrometry. J. Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]