Abstract
Understanding the regulation of fat synthesis and the consequences of its misregulation is of profound significance for managing the obesity epidemic and developing obesity therapeutics. Recent work in the roundworm Caenorhabditis elegans has revealed the importance of evolutionarily conserved pathways of fat synthesis and nutrient sensing in adiposity regulation. The powerful combination of mutational and reverse genetic analysis, genomics, lipid analysis, and cell-specific expression studies enables dissection of complicated pathways at the level of a whole organism. This review summarizes recent studies in C. elegans that offer insights into the regulation of adiposity by conserved transcription factors, insulin and growth factor signaling, and unsaturated fatty acid synthesis. Increased understanding of fat-storage pathways might lead to future obesity therapies.
Introduction
The number of people afflicted with obesity has reached epidemic proportions worldwide. Chronic weight gain is a cause for concern because excess body weight is associated with increased incidence of type II diabetes, cardiovascular disease, hypertension, stroke, sleep apnea and some types of cancer [1]. Unlike the conditions under which humans evolved, today most people have access to ample energy-rich food and partake in low levels of physical activity.
Obesity is essentially a disorder in energy homeostasis that develops when energy intake exceeds energy expenditure. Energy balance is highly regulated and involves complex interplay between food sensation, nutrient intake signals, transport and storage of nutrients, eating behavior, growth, reproduction, and energy expenditure via basal metabolic rate and physical activity [2]. Whereas modern living conditions have certainly exacerbated the obesity epidemic, it is notable that a portion of the population is resistant to excess weight gain [3]. It is likely that variants in genes encoding nutrient-sensing systems, metabolic enzymes and central nervous system regulators are responsible in part for the weight gain differences observed in humans. Indeed, human obesity is estimated to be 40–70% determined by genes [4].
The obesity epidemic has spurred exciting research that has resulted in important advances in the understanding of energy homeostasis, including the discovery of adipocyte hormones (such as leptin) that regulate energy homeostasis, and peptides synthesized and secreted from the gastrointestinal tract (such as cholecystokinin and ghrelin) [5]. However, human systems that control appetite, energy partitioning and integration of metabolic processes have proven to be highly complex and redundant. Therefore, the development of safe and effective drugs for long-term use is challenging, and several previously approved drugs have been withdrawn from the market because of unanticipated side-effects [6]. Therefore, there is a great need for further understanding the systems that regulate adiposity and for expanded research on how these molecules influence energy homeostasis, growth, reproduction and lifespan.
The nematode Caenorhabditis elegans has become a popular model for exploring the genetic basis of fatty acid synthesis and regulation of fat storage [7]. Many aspects of fatty acid synthesis and adiposity regulation by central nervous system regulators are conserved in this simple organism. Powerful genetic and behavioral tools are available to study energy homeostasis in the context of the whole organism, enabling examination of the relationship of metabolic processes with growth, reproduction and lifespan. This review highlights recent studies in C. elegans focusing on the synthesis of fatty acids and the regulation of adiposity.
Fatty acid synthesis in C. elegans
Fatty acids comprise the hydrophobic portion of all cellular membranes and have crucial roles in selective permeability, membrane fluidity and signaling. The other major role for fatty acids is as storage molecules; they are esterified to glycerol to form triacylglycerides (TAGs), which are stored in lipid droplets and yolk. TAGs are a vital energy source during embryogenesis, during periods of low food availability and for the specialized non-feeding dauer stage.
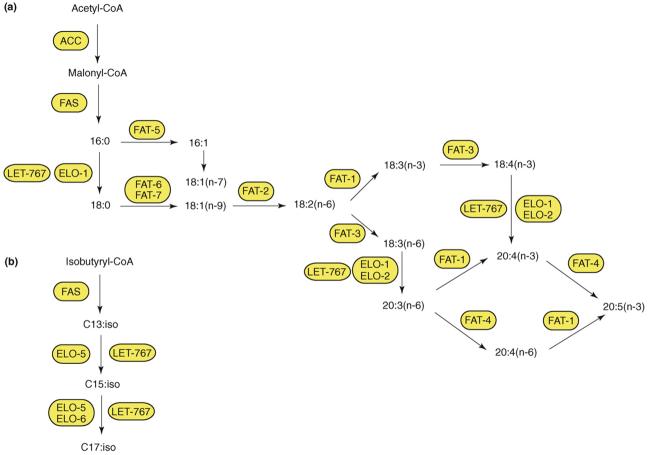
C. elegans obtains fatty acids from its bacterial diet and also synthesizes them de novo from acetyl CoA. For de novo synthesis, all of the enzyme activities necessary for the synthesis of palmitic acid (16:0) are encoded by two multi-functional enzymes: acetyl CoA carboxylase and fatty acid synthase [8]. Palmitic acid can be integrated into TAGs or phospholipids or can be modified by fatty acid elongases and desaturases to form a variety of long-chain polyunsaturated fatty acids (PUFAs) [9] (Figure 1a). In addition, monomethyl branched-chain fatty acids (mmBCFAs), which are essential for growth, are synthesized de novo starting from a branched-chain CoA primer [10] (Figure 1b). The biosynthesis and functions of PUFAs and mmBCFAs are described further in Box 1.
Figure 1.
Fatty acid synthesis pathways in C. elegans. (a) De novo synthesis of polyunsaturated fatty acids (PUFAs) begins with acetyl-CoA and utilizes acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS). Dietary fatty acids such as palmitic acid (16:0) can also enter the pathway and be converted to PUFAs by various desaturase and elongase activities. (b) Monomethyl branched-chain fatty acid synthesis uses a branched-chain primer as a substrate, FAS and specialized elongase activities to synthesize C15:iso and C17:iso, which are essential for growth. Fatty acid nomenclature: X:Yn-Z, fatty acid chain of X carbon atoms and Y methylene-interrupted cis double bonds; Z indicates the position of the terminal double bond relative to the methyl end of the molecule. Gene activities: FAT-1, omega-3 desaturase; FAT-2, Δ12 desaturase; FAT-3, Δ6 desaturase; FAT-4, Δ5 desaturase; FAT-5, Δ9 desaturase; FAT-6, Δ9 desaturase; FAT-7, Δ9 desaturase; ELO, fatty acid elongase; LET-767, 3-ketoacyl-CoA reductase.
Box 1. Biosynthesis and functions of PUFAs and mmBCFAs.
Fatty acid desaturase enzymes introduce double bonds at specific carbons in a fatty acid chain to produce unsaturated fatty acids. Mammals cannot convert oleic acid (18:1n-9) to linoleic acid (18:2n-6), nor can they convert omega-6(n-6) fatty acids to omega-3(n-3) fatty acids, because they lack the Δ12 and omega-3 fatty acid desaturases. Therefore, mammals require linoleic acid (18:2) and linolenic acid (18:3) in their diet. These essential fatty acids are precursors for long-chain polyunsaturated fatty acids (PUFAs), such as arachidonic acid (20:4n-6) and docosahexaenoic acid (22:6n-3). By contrast, C. elegans is capable of synthesizing all of its PUFAs from dietary or de novo synthesized 16:0 (Figure 1a) [53]. Seven fatty acid desaturases (FAT-1 through FAT-7), one 3-ketoacyl-CoA reductase (LET-767) [54] and several fatty acid elongases (ELO-1 and ELO-2) have been shown to be necessary for PUFA biosynthesis [9,19,55].
C. elegans mutants defective in fatty acid desaturation and PUFA elongation have been isolated [9,19,55]. Use of these mutants together with dietary fatty acid supplementation has revealed roles of specific fatty acid species in determining growth rate and brood size [19,20,56], neurotransmission and synaptic vesicle recycling [57,58], sensory signaling [59], germ-cell maintenance [11], sperm attraction to oocytes [60], and innate immune response [61].
The monomethyl branched-chain fatty acids (mmBCFAs) C15iso and C17iso are saturated tetradecanoic and hexadecanoic fatty acids with a single methyl group attached to the penultimate carbon atom. In bacteria, these molecules are synthesized starting from a primer derived from precursors of branched-chain amino acids, such as leucine, to form mmBCFA acyl-CoA primers that can be elongated by fatty acid synthase [62] (Figure 1b). In C. elegans, the production of C15iso and C17iso from C13iso requires specialized elongase activity encoded by elo-5 and elo-6 [10], as well as 3-ketoacyl-CoA reductase encoded by let-767 [54]. The mmBCFAs are essential in C. elegans because mutants that cannot synthesize them arrest as L1 larvae, and this growth arrest can be bypassed by dietary supplementation of C17iso [10]. Indeed, C17iso has a unique regulatory role that acts in parallel with the insulin pathway to sense nutrients and regulate larval growth [63]. The vital function of mmBCFAs might be due to their role as structural components of sphingolipids. These membrane components have important roles in signal transduction and cell recognition. C. elegans sphingolipids contain a single branched-chain C17 sphingoid base, and let-767 and elo-5 are required for its synthesis [54,64].
Quantification of fat stores and fat synthesis
To study the mechanisms that regulate adiposity in C. elegans, it is essential to quantify the amount of TAG stores in mutants or in worms grown under specified conditions. Quantification of fat stores can be achieved by several methods after biochemical extraction of lipids from populations of nematodes [11-14]. In addition, researchers visualize fat stores in individual worms using Sudan Black staining [15], Nile Red staining [16] and a specialized microscopic technique called coherent anti-Stokes Raman scattering [17]. The advantages and limitations of the various methods are described in Box 2.
Box 2. Comparison of methods for quantification of fat stores in C. elegans.
Lipid extraction and quantification
C. elegans lipids are extracted using a chloroform:methanol mixture. Phospholipid and TAG fractions are then separated using thin-layer chromatography [11] or solid-phase-exchange chromatography [12]. The separated lipid fractions can be quantified by sulfuric acid charring and densitometric scanning of thin-layer chromatography plates [13] or by converting the lipid fractions to fatty acid methyl esters and analyzing them with gas chromatography [11]. Alternatively, TAGs can be analyzed from nematode extracts using an enzymatic triacylglyceride assay [14]. The advantage of these methods is that they are readily quantifiable, and the fatty acid composition of various lipid fractions can also be determined. The main limitation of these methods is that they are not suitable for high-throughput genetic screens because a large number of nematodes are required for lipid extraction.
Staining of fixed worms
Fat can be visualized in individuals by staining fixed worms with Sudan Black, which stains lipid stores a blue–black color [15] (Figure Ia). The lipid droplets are visible in intestinal and hypodermal tissues. The limitation of this method is that fixation can be variable and difficult to quantify. However, variable fixation can be overcome by fixing and staining both mutant and wild-type strains in the same tube, distinguishing the strains by labeling one of them with fluorescent dye [42].
Staining of live worms
The lipophilic dye Nile Red fluoresces in hydrophobic environments and is used to stain intracellular lipid droplets. It can be fed to C. elegans without affecting their growth rate, brood size or lifespan. The advantages of Nile Red are that it can be used with live animals and is convenient for high-throughput screens designed to identify gene inactivations that cause fat reduction or accumulation [16]. The limitation is that, in live worms, it only fluoresces in gut granules, which are lysosome-related organelles with birefringent and autofluorescent content [65] (Figure Ib). Mutants deficient in formation of gut granules, such as pgp-2, apt-7 and glo-1, show no detectable Nile Red staining, yet quantification of TAGs from lipid extracts reveals normal fat stores in these strains. This indicates that fat stored outside of gut granules cannot be readily visualized in live worms with Nile Red. It is, therefore, necessary to confirm actual fat stores in mutants showing altered Nile Red staining by Sudan Black staining of fixed animals or by quantification of extracted lipids.
Coherent anti-Stokes Raman scattering microscopy
Coherent anti-Stokes Raman scattering (CARS) microscopy takes advantage of the characteristic vibrational properties of lipid molecules to visualize fat stores in live animals without the need for labeling [17]. Both hypodermal and intestinal fat stores are visualized using this technique. The disadvantage of CARS microscopy is that specialized equipment is required.
Figure I.
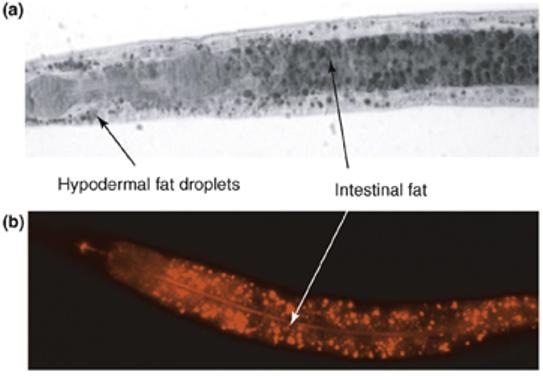
Fat staining of wild-type C. elegans. (a) Sudan Black staining of a fixed L4-stage larva. Black-stained fat droplets are visible in the intestinal and hypodermal cells. (b) Nile Red staining of a live L4-stage larva. Intestinal gut granules fluoresce red. In both pictures, anterior is to the left.
Fat synthesis can be quantified using a technique described in a recent study by Perez and Van Gilst [12]. They developed a 13C isotope assay to determine the contribution of dietary fat absorption and de novo synthesis to fat stores and membrane lipids in C. elegans. The assay entails feeding a mixture of 13C-enriched E. coli and unlabeled E. coli to worms and analyzing worm triglycerides and phospholipids with gas chromatography–mass spectrometry. Using these methods, the investigators determined that the relative proportion of absorbed versus synthesized molecules varies depending on the fatty acid. For example, in wild-type worms, only 7% of palmitic acid (16:0) is derived from de novo synthesis, whereas the remaining 16:0 is directly absorbed from the bacteria. However, de novo synthesis plays a larger part in the accumulation of C18 monounsaturated fatty acids (MUFAs) and PUFAs, the levels of which approach 20%. The mmBCFAs C15iso and C17iso are >99% derived from de novo synthesis because these fatty acids are not present in the E. coli diet and cannot be produced from modification of bacterial fatty acids. Other dietary fatty acids, including 16:0, 16:1 and 18:1n-7, are found in both phospholipid and TAG fractions. 13C isotope assays indicate similar amounts of synthesis and absorption of these fatty acids in phospholipids and TAG fractions [12].
A role for Δ9 desaturase in adiposity regulation
A crucial control point regulating lipid synthesis and breakdown is the production of MUFAs. The Δ9 desaturases catalyze the insertion of the first double bond into a saturated fatty acid at the C9 position. The MUFAs produced by Δ9 desaturases are abundant components of TAGs and phospholipids. C. elegans encodes three Δ9 desaturases: FAT-5, FAT-6 and FAT-7. The FAT-5 desaturase is specific for palmitic acid (16:0), whereas the FAT-6 and FAT-7 desaturases mainly desaturate stearic acid (18:0) [18,19] (Figure 1). The fatty acid composition changes and physiological consequences of the single desaturase mutants are subtle because increased expression of the remaining desaturases in the single mutants compensates for the lost activity [19]. By contrast, the fat-5;fat-6;fat-7 triple-mutant strain is not viable, revealing that endogenous production of MUFAs is essential for survival [19]. Examination of double-mutant strains reveals additional roles for MUFA synthesis in lipid partitioning and the regulation of fat storage. The fat-6;fat-7 double mutant is unable to carry out normal PUFA synthesis because it cannot synthesize the oleic acid precursor that is the substrate for the remaining PUFAs. However, unusual PUFAs are formed from the FAT-5 product palmitoleic acid (16:1) and can apparently partially substitute for vital functions performed by normal PUFAs [20].
The fat-6;fat-7 double mutants display slow growth, reduced fertility and reduced fat stores compared to wild type. The TAG stores in fat-6;fat-7 mutants are reduced by 20% compared to wild type. That MUFAs are preferred substrates for TAG synthesis suggests that this low-fat phenotype might be due to reduced synthesis. However, real-time quantitative RT-PCR analysis of fatty acid oxidation genes reveals that a considerable proportion of genes encoding components of mitochondrial fatty acid oxidation are expressed at higher levels in the fat-6;fat-7 double mutant compared to wild type [20]. The fat-storage phenotypes observed in C. elegans parallel those in Δ9-desaturase-deficient mice – which have low fat stores, increased metabolic rate and increased expression of fatty acid oxidation genes [21] – indicating the suitability of C. elegans as a model for mammalian lipid metabolism. Pharmacological manipulation of Δ9 desaturase activity might, therefore, benefit treatments of obesity, diabetes and other diseases of metabolic syndrome [22].
Regulation of Δ9 desaturation and fat synthesis
Regulation of Δ9 desaturation in C. elegans has been most extensively studied at the transcription level, although regulation at the translation level and of protein stability is likely to occur, based on studies in other organisms [21]. For example, the fat-7 gene seems to be influenced by various environmental conditions, such as temperature and food availability, as well as by at least five transcriptional regulators (Figure 2). The expression of fat-7 is upregulated ∼20-fold upon shifting to cold temperature [23]. When nematodes are separated from food, fat-7 is repressed tenfold in adults and to a greater extent in some larval stages [24]. The observation that fat-7 expression is altered to such a large extent in response to various environmental conditions, together with the requirement for multiple transcription factors, indicates that precise regulation of Δ9 desaturation is crucial for proper membrane fluidity and nutrient utilization.
Figure 2.
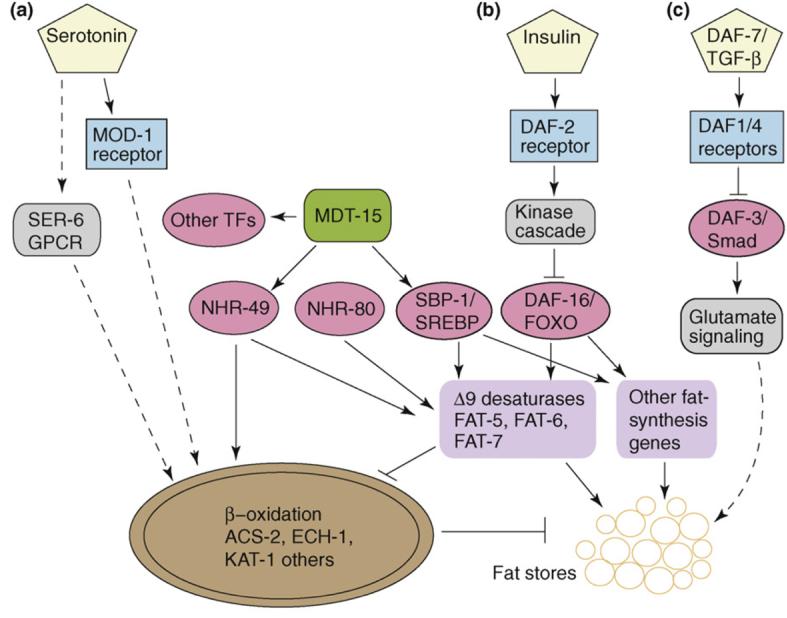
Proposed pathways influencing adiposity in C. elegans. Sensory neurons secrete (a) serotonin, (b) insulin and (c) TGF-β in response to nutritional cues. These signals are perceived by receptors (blue boxes), and signals are transduced by kinases and other signaling pathways (grey boxes). At least four transcription factors (pink ovals), in addition to the transcriptional mediator MDT-15 (green), are required for efficient transcription of the Δ9 desaturase genes fat-5, fat-6 and fat-7. These and other fat-synthesis genes (purple boxes) promote fat storage, whereas expression of genes encoding β-oxidation enzymes in mitochondria (brown oval) promote increased fat oxidation, culminating in decreased fat stores. Experiments performed to determine the downstream pathway from serotonin used exogenous serotonin. Abbreviations: TGF-β, transforming growth factor-β; SREBP, sterol regulatory element-βinding protein; GPCR, G-protein-coupled receptor; TF, transcription factor; NHR, nuclear hormone receptor.
Nuclear hormone receptors (NHRs) are transcription factors that reside outside of the nucleus. Upon binding their specific ligands, which are often small hydrophobic molecules, NHRs dimerize and enter the nucleus to bind specific sequences of promoters containing hormone-response elements. Two NHRs, NHR-49 and NHR-80, are important for expression of fat-5 and fat-7. In an nhr-49-deletion mutant, the expression levels of fat-5 and fat-7 are greatly decreased and the ratio of 18:0 to 18:1(n-9) increases more than twofold [25]. Similar changes in fatty acid composition, as well as in fat-5 and fat-7 expression levels, are observed in nhr-80 mutants [19]. In fact, NHR-80 is required for the fat-7 upregulation needed to compensate for fat-6-deletion mutants [19]. Despite the similarities in their effects on Δ9 desaturation, nhr-49-deletion mutants display more severe growth and reproductive phenotypes than nhr-80 mutants, probably because NHR-49 also regulates β-oxidation and expression of other genes that are necessary to respond to dietary input [24]. Even though the mammalian nuclear receptor family with the highest similarity to NHR-49 is the hepatocyte nuclear factor 4 family, its function might be closer to mammalian peroxisome proliferator-activated receptors (PPARs). Indeed, a comparative metabolomic study of NHR-49 and mouse liver PPAR-α demonstrated that the role of NHR-49 in regulating lipid synthesis, β-oxidation of fatty acids, glycolysis and gluconeogenesis is similar to that of PPAR-α [26].
The sterol-regulatory-element-binding protein (SREBP) family of basic helix–loop–helix zipper transcription factors comprises crucial regulators of cholesterol and fatty acid homeostasis in mammals [27,28]. Newly synthesized SREBPs reside in the endoplasmic reticulum membrane and are inactive. When specific cellular lipid levels are low, the proteins are transported to the Golgi, where they are acted on by site 1 and site 2 proteases released from the N-terminal fragment of SREBP, which then enters the nucleus and activates transcription of lipogenic genes. C. elegans possesses one SREBP homolog, sbp-1, and RNAi or mutants corresponding to this gene have been shown to contain low fat stores, high saturated fatty acid content, impaired growth, and reduced expression of lipogenic genes including fat-5 and fat-7 [10,16,29,30]. These studies indicate that SREBP is a crucial regulator of fatty acid synthesis and lipid homeostasis in C. elegans.
After binding to target sequences in specific promoters, SREBP and NHRs recruit multi-protein co-regulator complexes containing transcriptional cofactors that link the transcriptional activator to the transcription initiation machinery [31]. One component, the Mediator subunit MDT-15, interacts directly and specifically with the activation domain of SREBP [30], and the C. elegans MDT-15 also binds specifically to NHR-49 [32]. RNAi reduction of mdt-15 expression results in slow growth, low fat stores and reduced expression of fat-7. Supplementation with oleic acid, a common dietary MUFA, rescues many of the defects of sbp-1 and mdt-15 RNAi animals, indicating that the Δ9 desaturases are key targets of these regulators [30]. Recently, MDT-15 has been shown to be important for broader functions, including induction of detoxification genes in response to certain ingested xenobiotics and heavy metals [33]. Thus, a key role of MDT-15 is to integrate multiple transcriptional responses to ingested materials, including nutrients and environmental toxins.
Increased fat synthesis in dauer pathway mutants
In response to harsh environmental conditions, C. elegans larvae often enter an alternative larval stage called dauer. The decision to enter this stage depends on temperature, food availability and presence of an excreted pheromone that cues population density. Dauer larvae do not feed, yet are remarkably long-lived and stress resistant [34]. Because they do not feed, however, dauers must utilize lipid stores that are synthesized during the larval stage leading to dauer formation. Oxidation of fatty acids produces acetyl-CoA, which C. elegans, in contrast to mammals, can use as a substrate for gluconeogenesis to produce carbohydrates and amino acids. Because C. elegans encodes enzymes for the glyoxylate pathway, acetyl-CoA can be converted into oxaloacetate, which can enter the gluconeogenesis pathway [35] (Figure 3).
Figure 3.
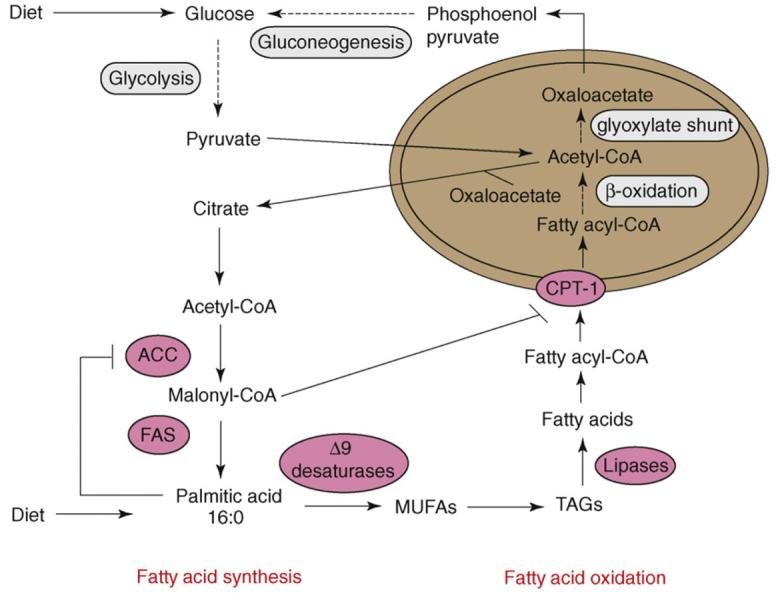
Schematic representation highlighting the relationship between fatty acid synthesis and fatty acid oxidation in C. elegans. Dietary carbohydrates that are not immediately used are converted to acetyl-CoA, which can be used to synthesize fatty acids. Along with dietary fatty acids, these molecules can be stored as triacylglycerides (TAGs) for future use. Fatty acids negatively regulate acetyl-CoA carboxylase (ACC) by feedback inhibition, which results in reduced levels of malonyl-CoA. This reduction increases the activity of carnitine palmitoyl transferase (CPT), which is sensitive to levels of malonyl-CoA, enabling increased uptake of fatty acids into mitochondria (brown oval), where they are oxidized. In C. elegans, dauer-stage larvae do not feed and rely on fat stores for energy. Unlike most other animals, they possess enzymes of the glyoxylate shunt, which enables the fat breakdown product acetyl-CoA to be converted into carbohydrates via gluconeogenesis. Key lipid synthesis enzymes are depicted as pink ovals, and metabolic pathways of multiple enzymatic steps are depicted as gray ovals. Abbreviations: FAS, fatty acid synthase; MUFAs, monounsaturated fatty acids.
Mutant strains with reduced insulin signaling, such as the daf-2 insulin-receptor mutant, often display temperature-sensitive constitutive dauer formation, even under non-dauer-inducing conditions in which food is plentiful. When daf-2 nematodes are grown at permissive temperature, they bypass the dauer stage and adults display increased lifespan and increased fat stores [15,36]. The lifespan extension, constitutive dauer formation and increased fat stores in daf-2 mutants depend on the downstream FOXO transcription factor, DAF-16 [16,37]. Transcript profile data from wild-type dauers and daf-2 adults indicate that the increased lifespan and fat stores in daf-2 mutants might be a result of the misexpression of genes that ensure dauer larval longevity [35]. These misexpressed genes encode for proteins engaged in fat synthesis (fat-6 and fat-7), fat oxidation, storage carbohydrate metabolism, glycolysis and gluconeogenesis [35,38-40]. 13C isotope assays confirm that daf-2 mutant adults with higher fat stores contain higher levels of de novo synthesized fats in TAGs and phospholipids [12].
Dauer-constitutive mutants belonging to the transforming growth factor-β (TGF-β) family also show increased fat accumulation in adults compared to wild type. The TGF-β ligand is encoded by daf-7, whereas daf-1 and daf-4 encode TGF-β receptors and daf-3 encodes the Smad transcription factor regulating entry into dauer [41]. Greer and colleagues [42,43] recently published an extensive and elegant study examining the neuronal network responsible for TGF-β regulation of fat storage. The TGF-β ligand is expressed in a pair of sensory neurons, and even though daf-1 is expressed in various neurons, promoter-specific reconstitution showed that the DAF-1 receptor is necessary and sufficient for fat regulation by expression in just two interneurons. A suppressor screen identified G proteins, vesicular glutamate transporters and three glutamate receptors as downstream components of TGF-β control of fat storage (Figure 2). Again, 13C isotope assays confirm that daf-7 and daf-1 mutants have nearly threefold more de novo fatty acid synthesis than wild type [42]. The ability to dissect the neural circuits regulating feeding and fat accumulation to this level of resolution demonstrates the power of the C. elegans system to tease out subtleties and complexities of metabolic regulation. The TGF-β study offers a sophisticated example of an unbiased genetic screen successfully identifying previously unappreciated downstream effectors of adiposity regulation.
Regulation of fatty acid oxidation
The extent of adiposity in animals depends on rates of fat oxidation, as well as on rates of fat synthesis. As mentioned previously, the low fat stores in fat-6;fat-7 double mutants might be a consequence of increased expression of mitochondrial β-oxidation genes [20], whereas the high fat stores in nhr-49 mutants correlate with decreased expression of β-oxidation genes [25].
The neurotransmitter serotonin regulates fat stores in mammals; loss-of-function mutations in certain serotonin receptors cause obesity, and drugs that increase central nervous system serotonin levels cause weight loss [44]. In C. elegans, exogenous addition of serotonin reduces fat stores [13]. Whereas the effects of serotonin in mammals are thought to be primarily mediated by feeding behavior [44], the C. elegans study revealed that serotonergic fat regulation depends on a neuronal serotonin channel (MOD-1) and a G-protein-coupled receptor (SER-6) that ultimately signal increased fat oxidation in peripheral tissues [13] (Figure 2). These studies demonstrate that feeding behavior and fat metabolism are independent responses of the central nervous system to nutrient availability.
One of the few single-gene mutations in mammals that causes obesity is tubby [45]. This gene is highly expressed in the hypothalamus and other tissues of the central nervous system, indicating that it might be involved in controlling satiety or regulating feeding behavior. C. elegans tub-1 mutants show increased lifespan and increased Nile Red staining compared to wild type. The lifespan extension depends on the FOXO transcription factor DAF-16; however, unlike in upstream insulin-signaling mutants, the increased Nile Red staining does not [46]. Instead, the increased Nile Red phenotype of tub-1 depends on the Rab GTPase-activating protein RGB-3, its substrate GTPase RAB-7, and genes that regulate Rab membrane localization and nucleotide recycling, indicating a role for endocytic recycling in the process [46,47]. An enhancer of tub-1 was identified in a genetic screen in which multiple alleles of the 3-ketoacyl-CoA thiolase gene encoded by kat-1 were isolated [48]. The KAT-1 protein localizes to intestinal mitochondria and is likely to be a key player in fatty acid oxidation.
Regulation of feeding behavior
C. elegans feeds by pumping a bacterial slurry into its pharynx, where cells are ruptured and pushed into the lumen of the intestine. Pumping rates are modulated by food availability (i.e. they are high in the presence of food and low in food absence) [49]. Mutants that affect pharyngeal pumping rates have been isolated [50]. One of these mutants, eat-2, is defective in pharyngeal pumping because of a mutation in a pharyngeal nicotinic acetylcholine receptor subunit. This mutant pumps at a greatly reduced rate compared to wild type and has been used as a genetic model of caloric restriction, which results in increased lifespan owing to lower ingestion of nutrients [51]. Consistent with reduced food uptake, eat-2 mutants produce fewer progeny and display a pale, thin morphology, as well as reduced Nile Red staining [52,13].
However, fat accumulation is not simply a passive outcome of feeding rate. In fact, exogenous serotonin increases feeding rates even though fat stores decrease [13], whereas daf-7 TGF-β mutants show decreased pharyngeal pumping but increased fat stores [42]. Distinct neurons and proteins responding to TGF-β are responsible for the independent regulation of feeding and fat storage. Therefore, unfavorable nutrient availability perceived by the nervous system results in activation of fat-storage pathways without an increase in feeding rate [42]. A recent study reported a non-feeding behavior called quiescence, which is defined as a complete cessation of feeding and moving [52]. When fed certain bacterial strains, up to 90% of worms on a plate were found to be quiescent, perhaps representing satiety. This behavior is regulated by insulin and TGF-β signaling; mutants of these pathways show reduced quiescence when feeding on some bacterial strains [52]. It is also possible that the time spent in quiescent versus non-quiescent states will have a considerable effect on fat stores, but this has not yet been investigated.
Concluding remarks and future challenges
Metabolic demands on living organisms are dynamic, and energy levels in cells must be constantly monitored so that fat-storage and fat-oxidation pathways, as well as eating behavior, are increased or decreased based on the present need. Recent work in C. elegans has identified many regulatory proteins and downstream effector genes responsible for lipid homeostasis. These studies verify that C. elegans is a powerful model for dissecting the molecular mechanisms of gene function at a level of resolution not possible in humans and other mammals.
Some of the challenging questions awaiting future studies include the following: What are the nutrient signals that instruct sensory neurons to secrete TGF-β, insulin and other hormones? What are the ligands of the NHRs that regulate fatty acid composition? Which specific lipids affect SREBP activation? How are newly synthesized lipids transported to peripheral tissues? To build on our knowledge of adiposity regulation in C. elegans and other animals, it will be important to develop new tools, such as reliable techniques to study oxygen consumption and metabolic rate, as well as antibodies to monitor regulation of synthesis and stability of key metabolic enzymes and regulators. In addition, biochemical studies of lipid synthesis enzymes and carbon flux will shed light on the pathways of lipid remodeling and TAG synthesis. Considering the paucity of studies pertaining to fat synthesis and regulation in C. elegans that were published at the turn of the millennium, rapid progress has been made toward establishing C. elegans as a powerful model to unravel the complicated pathways of adiposity regulation.
Acknowledgements
The author thanks National Institutes of Health grant DK074114 for research support and members of her laboratory and John Browse for comments on the manuscript.
References
- 1.Lawrence VJ, Kopelman PG. Medical consequences of obesity. Clin. Dermatol. 2004;22:296–302. doi: 10.1016/j.clindermatol.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Badman MK, Flier JS. The adipocyte as an active participant in energy balance and metabolism. Gastroenterology. 2007;132:2103–2115. doi: 10.1053/j.gastro.2007.03.058. [DOI] [PubMed] [Google Scholar]
- 3.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 4.Herbert A, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 5.Stanley S, et al. Hormonal regulation of food intake. Physiol. Rev. 2005;85:1131–1158. doi: 10.1152/physrev.00015.2004. [DOI] [PubMed] [Google Scholar]
- 6.Cooke D, Bloom S. The obesity pipeline: current strategies in the development of anti-obesity drugs. Nat. Rev. Drug Discov. 2006;5:919–931. doi: 10.1038/nrd2136. [DOI] [PubMed] [Google Scholar]
- 7.Ashrafi K. Obesity and the regulation of fat metabolism. WormBook. 2007;9:1–20. doi: 10.1895/wormbook.1.130.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappleye CA, et al. Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev. Biol. 2003;3:8. doi: 10.1186/1471-213X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kniazeva M, et al. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004;2:e257. doi: 10.1371/journal.pbio.0020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts JL, Browse J. Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans. Dev. Biol. 2006;292:381–392. doi: 10.1016/j.ydbio.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan S, et al. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008;7:533–544. doi: 10.1016/j.cmet.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans lifespan by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Kimura KD, et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 16.Ashrafi K, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 17.Hellerer T, et al. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts JL, Browse J. A palmitoyl-CoA-specific Δ9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 19.Brock TJ, et al. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brock TJ, et al. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr. Opin. Lipidol. 2008;19:248–256. doi: 10.1097/MOL.0b013e3282f9b54d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes. Rev. 2005;6:169–174. doi: 10.1111/j.1467-789X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 23.Murray P, et al. An explicit test of the phospholipid saturation hypothesis of acquired cold tolerance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5489–5494. doi: 10.1073/pnas.0609590104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Gilst MR, et al. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Gilst MR, et al. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atherton HJ, et al. A comparative metabolomic study of NHR-49 in Caenorhabditis elegans and PPAR-α in the mouse. FEBS Lett. 2008;582:1661–1666. doi: 10.1016/j.febslet.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr. Opin. Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Raghow R, et al. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 29.McKay RM, et al. C. elegans: a model for exploring the genetics of fat storage. Dev. Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 31.Blazek E, et al. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 32.Taubert S, et al. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taubert S, et al. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu PJ. Dauer. WormBook. 2007;8:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McElwee JJ, et al. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 2006;127:458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 37.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 38.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 40.McElwee JJ, et al. Shared transcriptional signature in Caenorhabditis Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 41.Savage-Dunn C. TGF-β signaling. WormBook. 2005;9:1–12. doi: 10.1895/wormbook.1.22.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greer ER, et al. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watts JL. Fattening up without overeating. Cell Metab. 2008;8:95–96. doi: 10.1016/j.cmet.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tecott LH. Serotonin and the orchestration of energy balance. Cell Metab. 2007;6:352–361. doi: 10.1016/j.cmet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Carroll K, et al. Tubby proteins: the plot thickens. Nat. Rev. Mol. Cell Biol. 2004;5:55–63. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- 46.Mukhopadhyay A, et al. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2005;2:35–42. doi: 10.1016/j.cmet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Mukhopadhyay A, et al. An endocytic pathway as a target of tubby for regulation of fat storage. EMBO Rep. 2007;8:931–938. doi: 10.1038/sj.embor.7401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mak HY, et al. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 49.You YJ, et al. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3:237–245. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You YJ, et al. Insulin, cGMP, and TGF-β signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallis JG, et al. Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem. Sci. 2002;27:467. doi: 10.1016/s0968-0004(02)02168-0. [DOI] [PubMed] [Google Scholar]
- 54.Entchev EV, et al. LET-767 is required for the production of branched chain and long chain fatty acids in Caenorhabditis elegans. J. Biol. Chem. 2008;283:17550–17560. doi: 10.1074/jbc.M800965200. [DOI] [PubMed] [Google Scholar]
- 55.Kniazeva M, et al. Suppression of the ELO-2 FA elongation activity results in alterations of the fatty acid composition and multiple physiological defects, including abnormal ultradian rhythms, in Caenorhabditis elegans. Genetics. 2003;163:159–169. doi: 10.1093/genetics/163.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watts JL, et al. Deficiencies in C20 polyunsaturated fatty acids cause behavioral and developmental defects in Caenorhabditis elegans fat-3 mutants. Genetics. 2003;163:581–589. doi: 10.1093/genetics/163.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesa GM, et al. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. J. Cell Sci. 2003;116:4965–4975. doi: 10.1242/jcs.00918. [DOI] [PubMed] [Google Scholar]
- 58.Marza E, et al. Polyunsaturated fatty acids influence synaptojanin localization to regulate synaptic vesicle recycling. Mol. Biol. Cell. 2008;19:833–842. doi: 10.1091/mbc.E07-07-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kahn-Kirby AH, et al. Specific polyunsaturated fatty acids drive TRPV-dependent sensory signaling in vivo. Cell. 2004;119:889–900. doi: 10.1016/j.cell.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 60.Kubagawa HM, et al. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat. Cell Biol. 2006;8:1143–1148. doi: 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- 61.Nandakumar M, Tan M-W. Gamma-linolenic and stearidonic acids are required for basal immunity in Caenorhabditis elegans through their effects on p38 MAP kinase activity. PLoS Genet. 2008;4:e1000273. doi: 10.1371/journal.pgen.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oku H, Kaneda T. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J. Biol. Chem. 1988;263:18386–18396. [PubMed] [Google Scholar]
- 63.Kniazeva M, et al. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 2008;22:2102–2110. doi: 10.1101/gad.1692008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chitwood DJ, et al. The glycosylceramides of the nematode Caenorhabditis elegans contain an unusual, branched-chain sphingoid base. Lipids. 1995;30:567–573. doi: 10.1007/BF02537032. [DOI] [PubMed] [Google Scholar]
- 65.Schroeder LK, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol. Biol. Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]