Abstract
Although reductive metabolism of Cr(VI) always results in the production of Cr(III) and extensive Cr-DNA binding, cellular studies have indicated that different reduction processes are not equivalent in the induction of mutagenic events. Here we examined mutagenicity and formation of Cr-DNA damage by Cr(VI) activated in vitro by one of its important reducers, glutathione (GSH). Our main focus was on reactions containing 2 mM GSH, corresponding to its average concentration in CHO (1.8 mM) and V79 (2.6 mM) mutagenicity models. We found that Cr(VI) reduction by 2 mM GSH produced only weak mutagenic responses in pSP189 plasmids replicated in human fibroblasts. Reductive activation of Cr(VI) with 5 mM GSH resulted in approximately 4-times greater DNA adduct-normalized yield of mutations. Mutagenic DNA damage formed in GSH-chromate reactions was caused by nonoxidative mechanisms, as blocking of Cr-DNA adduction led to a complete loss of mutagenesis. All GSH-mediated reactions also lacked significant DNA single-strand breakage. We developed a sensitive HPLC procedure for the detection of GSH-Cr-DNA crosslinks based on dissociation of DNA-conjugated GSH by Cr(III) chelation and its derivatization with monobromobimane. Weak mutagenicity of 2 mM GSH reactions was associated with a low production of mutagenic GSH-Cr-DNA crosslinks (5.0% of total Cr-DNA adducts). In agreement with their greater mutation-inducing ability, 5 mM GSH reactions generated 4-5 times higher levels of GSH-DNA crosslinking. Overall, our results indicate that chromate reduction by physiological concentrations of GSH is a weakly mutagenic process, which is consistent with low mutagenicity of Cr(VI) in ascorbate-deficient cells.
Introduction
Hexavalent Cr compounds most commonly encountered in the forms of various chromate salts are firmly recognized as human carcinogens (1–3). Toxic and genotoxic effects of Cr(VI) result from its cellular activation via one- or two-electron reduction processes yielding thermodynamically stable Cr(III). Reductive metabolism of Cr(VI) in mammalian cells is largely nonenzymatic and principally involves ascorbate and nonprotein thiols cysteine and glutathione (GSH) (3). Cr(VI) reduction is associated with the formation of transient Cr(V) and Cr(IV) species and organic radicals (4–6). Intermediate Cr forms are largely responsible for oxidative DNA damage which, depending on the coordinated ligands and the presence of other catalytic metals or oxidants, can be caused by direct electron abstraction (7,8), Cr(V)-peroxo (9) or by reactive oxygen species produced in Fenton-like reactions with H2O2 (7,10). The final product of Cr(VI) reduction, Cr(III), forms stable coordinate complexes with DNA producing several types of Cr-DNA adducts including binary Cr-DNA adducts and various ligand-Cr-DNA crosslinks (ternary adducts) (11). The most common crosslinked ligands are usually unoxidized forms of Cr(VI) reducers, such as ascorbate, cysteine and glutathione (12–14). The relative importance of oxidative and adduct-mediated pathways in Cr(VI) genotoxicity and mutagenicity is expected to vary depending on the yield and stability of intermediate Cr forms and the concomitant formation of reactive oxygen species. Mutagenic responses in pSP189 plasmids induced by DNA damage during in vitro reduction of Cr(VI) with rigorously purified cysteine (15) and ascorbate (16,17) were completely dependent on Cr(III)-DNA binding, reflecting the formation of mutagenic cysteine-Cr-DNA and ascorbate-Cr-DNA crosslinks. The most abundant form of Cr-DNA damage, binary Cr-DNA adducts, were weak inducers of mutagenic events during replication of pSP189 plasmids in human cells (17,18).
Despite its complete conversion to Cr(III) (19,20), Cr(VI) was a weak mutagen at the Hprt locus in CHO and V79 cells grown under the standard tissue culture conditions (21,22). Significant increases in the mutation frequency were generally induced in the toxic dose ranges that corresponded to the appearance of oxidatively induced DNA single-strand breaks (23). As with all human and the majority of rodent cells, V79 and CHO cells contain very low ascorbate concentrations in normal tissue culture (22). In the absence of ascorbate, GSH is the most important cellular reducer due to its much higher concentrations than another Cr(VI)-reducing thiol cysteine (11). Restoration of physiological levels of ascorbate in V79 and CHO cells led to highly increased mutagenicity of Cr(VI) at doses that were completely nonmutagenic under ascorbate-deficient conditions (22). The reasons for weak mutagenicity of Cr(VI) reductively activated by cellular GSH is unclear since this reduction process contains both one- and two-electron electron transfer reactions (24) and consequently, generates both Cr(V) and Cr(IV) intermediates (25,26). As with other in vitro reduction systems, Cr(VI) metabolism by GSH also led to the formation of Cr-DNA adducts (23,27). Cr(VI) uptake-normalized Cr-DNA binding in ascorbate-deficient and supplemented cells was very similar (22,27), indicating that the switch from a slow reduction by GSH to a rapid reduction by ascorbate did not affect the overall yield of Cr-DNA adducts. Since reduction of Cr(VI) in cells even under the conditions of the predominant metabolism by GSH is likely to involve reactions with some other reducers and ligands, it is unclear whether GSH-driven metabolism is intrinsically poorly mutagenic or some cellular processes inhibit the production of mutagenic DNA damage.
In this work, we examined the mutagenicity and the spectrum of DNA damage arising during Cr(VI) reduction by GSH under defined in vitro conditions. Our results indicate that Cr(VI) metabolism by physiologically relevant GSH concentrations is a weakly mutagenic process due to inefficient formation of GSH-Cr-DNA crosslinks.
Experimental Section
Materials
Chelex-100 resin and Bio-Gel P-30 columns were purchased from Bio-Rad (Hercules, CA). Potassium chromate (A.C.S. reagent) was from Aldrich (Milwaukee, WI), 35S-labeled GSH was from Amersham (Arlington Heights, IL) and supercoiled preparations of ΦX174 DNA were obtained from New England Biolabs (Beverly, MA). All other reagents were from Sigma (St. Louis, MO). Stock solutions of 10 mM chromate and 0.5 M stocks of MOPS and phosphate buffers were purified from the trace amounts of iron using Chelex-100 columns as described previously (13). To remove redox-active metal contaminants from GSH, 10 mM stock solutions was mixed with the equal volume of the Chelex resin adjusted to pH 7.0 by 5 washes with 0.5 ml of 0.5 M MOPS or phosphate buffers (pH 7.0). After 10 min incubation at room temperature, GSH was recovered by centrifugation of Chelex columns at 2000g for 5 min. Purified GSH solutions were kept on ice and used within 30 min after preparation. The pSP189 vector was a generous gift from Dr. M. Seidman. A large-scale purification of pSP189 plasmids was carried out using a kit from Qiagen (Valencia, CA). Caution: Cr(VI) compounds are human carcinogens, and appropriate precautions should be taken in handling of these materials.
Cells
SV40-immortalized human fibroblasts (HF/SV) were grown in 90% DMEM-10% fetal bovine serum at 37°C in the humidified atmosphere containing 95% air and 5% CO2. Transfections with pSP189 DNA were performed at 60–70% confluency.
Cr(VI) reduction
Reduction of chromate was monitored by recording its characteristic absorbance at 372 nm. Reaction mixtures contained 25 mM MOPS or phosphate buffer (pH 7.0), and indicated concentrations of GSH and chromate. The reductions were initiated by the addition of the equal volume of pre-warmed Cr(VI) solutions to buffer-GSH mixtures, which ensured a rapid mixing of the reagents. All reduction reactions were performed at 37°C using electronically controlled temperature unit. The presence or absence of DNA in the reaction mixtures did not change reduction kinetics. Absorbance measurements were recorded with Schimadzu UV1601 spectrophotometer.
Detection of DNA single-strand breaks (SSB)
Formation of DNA single-strand breaks was measured by the conversion of supercoiled ΦX174 DNA into a nicked, open circular from. A standard reaction contained 0.3 µg supercoiled plasmid DNA, 25 mM MOPS or phosphate buffer (pH 7.0) and indicated concentrations of GSH and chromate in the total volume of 25 µl. After incubation at 37°C for 60 min, the samples were placed on ice, mixed with 6× Ficoll buffer (15% Ficoll 400, 0.25% bromphenol blue, 0.25% xylene cyanol) and loaded onto 1% agarose-TAE gels. DNA bands were stained with ethidium bromide and analyzed using Bio-Rad GelDoc 2000 gel documentation system. The amounts of DNA in individual bands were determined by the Quantity One computer program.
Measurements of chromium-DNA adducts
Reaction mixtures (final volume=50 µl) contained 25 mM MOPS or phosphate (pH 7.0), 2 µg pSP189 DNA, 2 or 5 mM GSH and 0–200 µM chromate. Samples were incubated for 60 min at 37°C and DNA was purified from the unbound components by the size-exclusion chromatography using Bio-Gel P-30 spin columns. Recovered DNA was incubated for 20 min at room temperature in the presence of 200 mM NaCl to dissociate ionically bound Cr(III) (13) and passed through P-30 columns again. The amount of DNA-bound Cr was determined by graphite furnace atomic absorption spectroscopy (GF-AAS) using Zeeman background correction (Perkin-Elmer GF-AAS, model 41002L). Specific absorbance was measured at 357.5 nm wavelength with a bandwidth of 0.7 nm. The following 5-step graphite furnace program was used: 1)110°C: ramp 1s, hold 20 s 2) 130°C: ramp 5 s, hold 30 s 3) 1500°C: ramp 10 s, hold 20 s 4) 2300°C: hold 5 s 5) 2500°C: ramp 1 s, hold 2 s.
Determination of DNA-crosslinked GSH
Experimental conditions for Cr(VI) reductions and purification of Cr-modified DNA were as described for analysis of total Cr-DNA adducts. In the initial experiments, DNA-crosslinked GSH was quantified by the addition of trace quantities of radioactive S35-GSH (~3×106 cpm/reaction). The amounts of DNA-bound 35S-GSH were measured using a scintillation counter. For the determination of DNA-conjugated GSH by a nonradioacive method, GSH was first released from crosslinks by chelation of Cr(III) and then converted into a highly fluorescent conjugate with monobromobimane (mBBr). To dissociate crosslinks, DNA samples were incubated for 24 hr at 37°C (4°C in a set of the initial studies) in the solution containing 50 mM phosphate (pH 7.0), 5 mM diethylenetriaminepentaacetic acid (DTPA) and 0.1 mM dithiotreitol (DTT). The derivatization reactions contained 50 mM HEPES, pH 8.0, 5 mM DTPA, 15 mM NaOH, and 0.1 mM mBBr. Samples were incubated for 10 min at room temperature and the reactions were terminated by the addition of 25 mM methanesulfonic acid. HPLC detection of the fluorescent GSH-mBBr conjugate was performed using Shimadzu LC-10ADvp equipped with Ultrasphere ODS column (5 µm, 250 × 4.6 mm) and RF-10AxL fluorescence detector. The chromatographic conditions were as described previously (29). The final values for GSH-Cr-DNA crosslinks included corrections for the incomplete dissociation of DNA-bound GSH (79.8 % recovery).
Measurements of cellular GSH
Cellular concentrations of GSH were determined by HPLC as described previously (29). In brief, cells were suspended in a 40 mM methanesulfonic acid-1 mM DTPA solution, lysed by two cycles of freezing/thawing and GSH-containing supernatants were recovered by centrifugation at 12,000g for 10 min. The cell extracts were reacted with 2 mM mBBr and the fluorescent GSH-mBBr conjugates were measured by HPLC.
Shuttle-vector mutagenesis
The pSP189 vector containing the supF gene as a mutagenic target (30) was used to examine the production of mutagenic DNA lesions during chromate reduction by GSH. This vector contains the SV40 origin of replication and encodes large T-antigen, which permits replication of this plasmid in a broad variety of human cell lines. Untreated and Cr-modified plasmids were replicated in SV40-immortalized human HF/SV fibroblasts. The frequency of supF mutants was scored using E.coli MBL50 strain (a gift from Dr. C. Pueyo). This indicator strain contains dual araC and lacZ amber mutations suppressed by the supF gene (31). Inactivating mutations in the araC gene allow selection of the colonies with a mutated supF gene on plates containing L-arabinose. The pSP189 DNA was transfected into the cells using TransFast reagent (Promega) according to manufacturer's recommendations. Plasmids were propagated for 48 hr and their replicated progeny was isolated by a plasmid isolation kit from Qiagen. DNA was precipitated with ethanol and dissolved in deionized H2O. Recovered plasmids were transfected into the E.coli MBL50 by electroporation. The total number of bacterial transformants was determined on the minimal agar plates containing 30 µg/ml ampicillin and 0.5 µg/ml chloramphenicol. Mutants were selected on plates additionally containing 2 mg/ml L-arabinose and mutation frequency was calculated as the ratio of ampicillin/arabinose dual resistant colonies to ampicillin only resistant colonies.
Results and Discussion
CHO and V79 are two commonly used cell lines for testing of mutagenicity at the endogenous chromosomally located Hprt gene. Studies in these cells under the normal tissue culture conditions found only a weak mutagenicity of Cr(VI) that was detectable in the toxic dose range (21,22). To select appropriate GSH concentrations for our experiments, we determined cellular levels of this thiol in CHO and V79 cells. A specific HPLC assay quantifying the amounts of the fluorescent mBBr-GSH conjugate showed that the average GSH concentration in V79 cells was 2.6±0.3 mM and in CHO cells −1.8±0.1 mM (n=3). Thus, the use of 2 mM GSH in chromate reactions in vitro would closely approximate cellular levels of this reducer in the two cellular test systems. To assess the impact of elevated rates of reduction, we also examined the formation of DNA damage in reactions containing 5 mM GSH concentration.
Reduction of Cr(VI) by GSH and formation of DNA damage
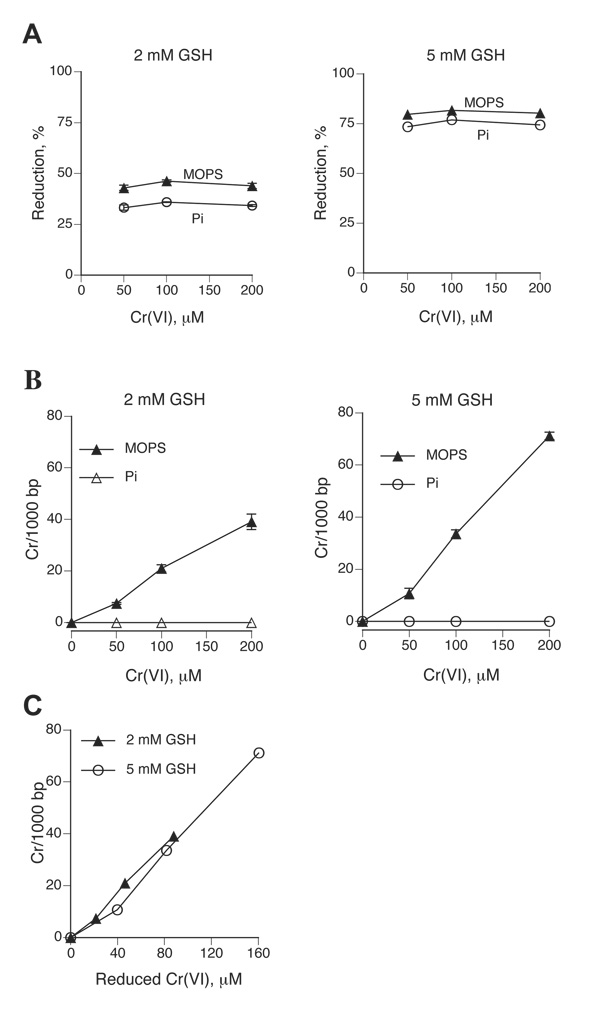
Cr(VI) reduction by physiological concentrations of GSH has two components: a fast reaction during the first 10–15 min followed by a slow reduction phase (32). To minimize the induction of nonspecific oxidative damage, Cr(VI) and GSH were reacted for 1 hr as this incubation time was sufficiently long to include both reduction phases. Under the selected reaction conditions, 2 mM GSH reduced on average 44.1% Cr(VI) in MOPS buffer and slightly less in phosphate buffer (Fig. 1A). The amount of reduced Cr(VI) in MOPS-5 mM GSH reactions was about 2-times greater −80.5%. The extent of reduction did not vary in the range of the employed Cr(VI) concentrations (Fig. 1A), indicating that reactions proceeded under the pseudo first-order conditions and that the expected modest depletion of GSH due to the formation of Cr(III)-GSH complexes and consumption of GSH in reduction reactions had no significant impact on the overall reduction process. MOPS-based reduction reactions containing 2 or 5 mM GSH produced Cr-DNA adducts as a linear function of Cr(VI) concentration (Fig. 1B). Higher levels of Cr-DNA binding in the presence of 5 mM relative to 2 mM GSH appears to simply reflect greater amounts of reduced Cr(VI), as plots of Cr-DNA binding vs. reduced Cr(VI) concentrations showed similar abilities of both reactions to generate Cr-DNA adducts (Fig. 1C). In agreement with results from other reactions (13,16,33), reduction of Cr(VI) by 2 or 5 mM GSH failed to produce any detectable amounts of Cr-DNA adducts in samples containing Cr(III) chelating phosphate ions (Fig. 1B).
Figure 1. Reduction of Cr(VI) and formation of Cr-DNA adducts in reactions containing 2 and 5 mM GSH.
Reaction mixtures were incubated for 1 hr at 37°C followed by immediate absorbance measurements or purification of DNA by Bio-Gel P-30 chromatography. Cr-DNA adducts were measured by GF-AAS. Where not seen, error bars were smaller than symbols. (A) Reduced amounts of Cr(VI) after incubation with 2 or 5 mM GSH in 25 mM MOPS or phosphate (Pi) buffer. Data are means from 3 independent determinations. (B) Formation of Cr-DNA adducts in reactions containing 2 or 5 mM GSH, 0–200 µM Cr(VI), 2 µg pSP189 DNA and 25 mM MOPS or phosphate (Pi) buffer. Means±SD from four independent determinations. (C) Yield of Cr-DNA adducts as a function of reduced Cr(VI) in reactions containing 2 or 5 mM GSH.
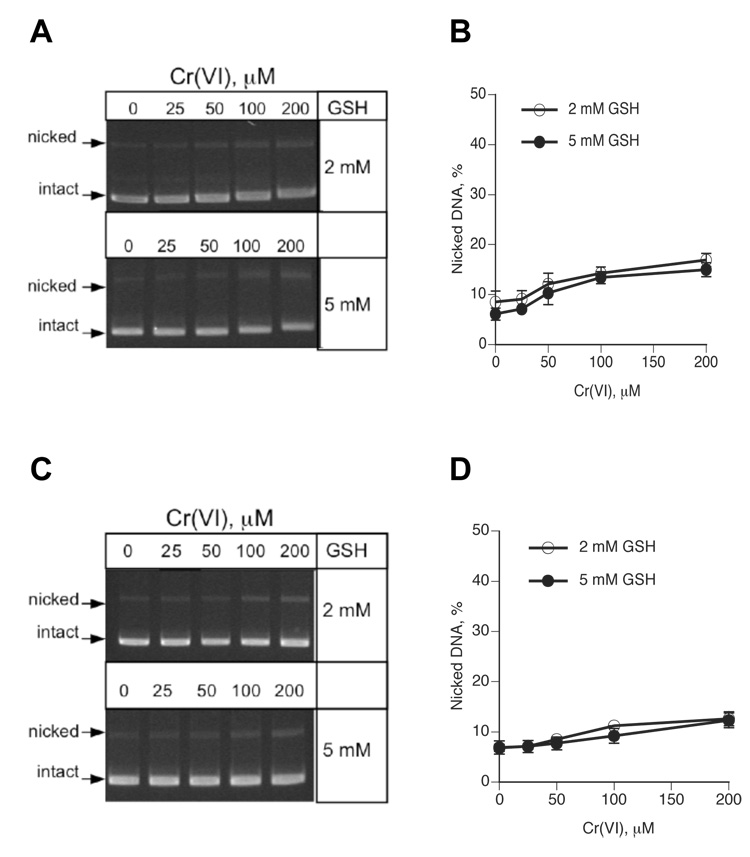
To examine a potential induction of oxidative DNA damage in chromate-GSH reactions, we monitored SSB induction by the plasmid relaxation procedure. The sensitivity of SSB detection was increased by lowering the amount of DNA/sample to 0.3 µg from the standard 2 µg in other experiments. Incubation of ΦX174 DNA with Cr(VI) in the presence of 2 or 5 mM GSH in MOPS buffer led a modest unwinding of supercoiled (intact) plasmid molecules, as evidenced by their slower mobility in agarose gels (Fig. 2A). Plasmid DNA incubated in the phosphate-based reactions showed no changes in the electrophoretic behavior (Fig. 2C), consistent with the absence of Cr-DNA binding in these samples (Fig. 1B). Quantitative analysis of DNA bands from three independent experiments found only a marginal increase in the percentage of nicked plasmids in the reactions containing high 100 and 200 µM Cr(VI) concentrations (Fig. 2B,D). The observed changes in the amount of nicked molecules correspond to approximately 1–2 SSB per 100,000 bp DNA, which is close to the detection sensitivity of the plasmid relaxation assay (32). For comparison, reactions containing 200 µM Cr(VI) generated 3910 and 7120 Cr adducts/100,000 bp DNA in the presence of 2 and 5 mM GSH, respectively. Thus, the formation of Cr-DNA adducts is quantitatively a far more predominant form of DNA damage, exceeding the production of SSB by about 2000:1 for 2 mM GSH and 3500:1 for 5 mM GSH reactions. While SSB is one of many forms of oxidative DNA damage, a minimal production of breaks is indicative of the overall weak oxidative DNA damage as chromate-GSH reactions have previously been shown to lack oxidized base products and form abasic sites in approximately 1:1 ratio to SSB (9,34). The induction of oxidative damage to the sugar-phosphate backbone in GSH-chromate samples required H2O2 that originated in the reactions of adventitious Fe with O2 (9,23). Therefore, low levels of SSB in Cr-treated plasmids indicate that secondary oxidative processes related to impurities with other metals were limited in our reduction reactions.
Figure 2. Detection of DNA single-strand breaks by a plasmid relaxation assay.
Supercoiled ΦX174 DNA was incubated with GSH-chromate for 1 hr at 37°C followed by immediate separation of samples on agarose gels. (A) Representative agarose gels of plasmids reacted with Cr(VI)-GSH in MOPS buffer. (B) Quantitation of DNA nicks generated in Cr(VI)-GSH reactions performed in MOPS buffer. Means±SD from three independent reactions (gels). (C) Representative agarose gels of plasmids treated with Cr(VI)-GSH in phosphate buffer. (D) Amounts of nicked DNA generated in Cr(VI)-GSH reactions performed in MOPS buffer. Means±SD from three independent reactions (gels).
Mutagenic responses in the pSP189 vector
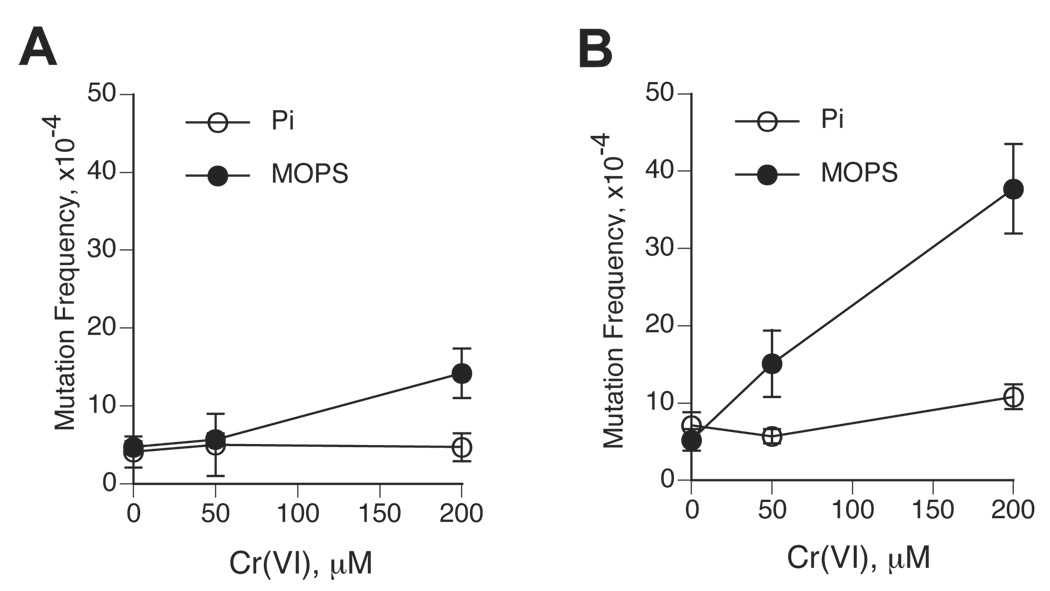
The ability of GSH-chromate reactions to generate mutations-inducing DNA damage was assessed using the pSP189 shuttle vector containing the supF gene as the mutagenic target. To allow easier comparisons with other Cr(VI) reduction reactions, pSP189 plasmids were replicated in the same cell line as in our previous studies – HF/SV fibroblasts. HS/SV cells are proficient in removal of Cr-DNA adducts with the repair rate similar to that of other SV40-immortalized and primary human cells (35). Thus, mutagenic responses induced at the supF gene during replication in HF/SV cells integrate both pre-mutagenic potential and repair susceptibility of specific forms of DNA damage. The pSP189 system is not biased toward any particular type of mutational events, as it is capable of detecting both point mutations and frameshifts (16,30). We found that replication of pSP189 DNA modified in MOPS-based chromate-GSH reactions led to only a modest increase in the supF mutations at the highest dose of Cr(VI) – 200 µM (Fig. 3A). Significantly higher although still relatively modest increases in mutation frequencies were observed in pSP189 plasmids damaged in the reaction of Cr(VI) with 5 mM GSH (Fig. 3B). In agreement with previous studies (17), our control experiments detected very strong mutagenic responses in pSP189 vectors modified in the presence of chromate and 0.2 mM ascorbate (210.8±30.7×10−4 mutation frequency for 200 µM Cr-treated vectors, for example). When induced mutation frequencies were analyzed as a function of Cr-DNA binding, we found that vectors incubated in the presence of 2 mM GSH required 4.1 times higher number of Cr-DNA adducts relative to 5 mM GSH-treated plasmids to generate a 3-fold increase in the number of mutants above background. This result indicates that reduction of Cr(VI) by higher GSH concentrations leads to the shift towards the formation of more mutagenic Cr-DNA damage.
Figure 3. Mutagenic responses generated in Cr(VI)-treated pSP189 vectors following their replication in human cells.
The pSP189 DNA was incubated in Cr(VI)-GSH mixtures, purified by P-30 chromatography and propagated in HF/SV cells for 48 hr. Replicated progeny was electroporated into E.coli MBL50 cells and the frequency of supF mutants was scored on arabinose-containing plates. Data are means±SD from four (MOPS) or two (phosphate) independent transfections. (A) Frequency of supF mutants in pSP189 samples treated in Cr(VI)-2 mM GSH reactions containing MOPS or phosphate (Pi) buffer. (B) As panel A with the exception that reduction reactions contained 5 mM GSH.
Blocking of Cr-DNA binding due to the presence of Cr(III)-sequestering phosphate ions completely abolished mutagenic responses in chromate reactions containing either 2 or 5 mM GSH (Fig. 3A,B). Thus, consistent with our results on the absence of significant DNA nicking, Cr(VI) reductions by either 2 or 5 mM GSH generated mutation-inducing DNA damage via a nonoxidative process involving the formation of Cr-DNA adducts. Cr-DNA adducts were also responsible for all mutagenic responses detected in pSP189 vector damaged in the reactions of Cr(VI) with two other biological reducers, cysteine and ascorbate (15,16). Despite this similarity, the number of supF mutants induced by DNA damage generated during Cr(VI) reduction by ascorbate or cysteine was dramatically higher. For example, reduction of 100 µM Cr(VI) by 1 mM ascorbate produced a net increase of 193.8×10−4 mutants (16) vs. 9.5×10−4 mutants for reduction of 88 µM Cr(VI) by 2 mM GSH in this work, a 18.0-fold difference after correction for the amounts of reduced Cr(VI). Yield of mutations divided by the number of Cr-DNA adducts showed a 15.2-times higher mutagenicity of ascorbate reactions relative to 2 mM GSH. Reduction of 100 µM Cr(VI) with 1 mM cysteine (15) generated 7.2-times higher number of the supF mutations in comparison to the equivalent reactions with 2 mM GSH. Because of its abundancy, GSH is the most important Cr(VI)-reducing thiol and its reactions dominate reductive metaboslim of Cr(VI) in cultured cells due to the absence or severely limited amounts of ascorbate in the standard tissue cultures (11). Overall, our shuttle-vectors studies found a very weak mutagenicity of DNA damage arising during chromate reduction by the physiological 2 mM GSH concentration, which is consistent with poor mutagenesis of Cr(VI) at the Hprt locus in V79 and CHO cells grown without ascorbate supplementation (21,22).
Formation of GSH-Cr-DNA crosslinks during chromate reduction by GSH
Differences in the mutagenicity of DNA damage generated in ascorbate- and GSH-based reactions can be at least in part attributed to the different spectra of Cr-DNA adducts produced in these reactions. About 25% of Cr-DNA complexes generated in Cr(VI)-1 mM ascorbate reactions are ascorbate-Cr-DNA crosslinks (14) which are the most potent mutagenic form of Cr-DNA damage with 31-times higher mutagenicity than binary Cr-DNA adducts (16). Poor mutagenicity of DNA damage produced in Cr(VI)-2 mM GSH reactions could also be caused by a low yield of GSH-Cr-DNA crosslinks that exhibit several times stronger pre-mutagenic potential than binary Cr-DNA adducts (18).
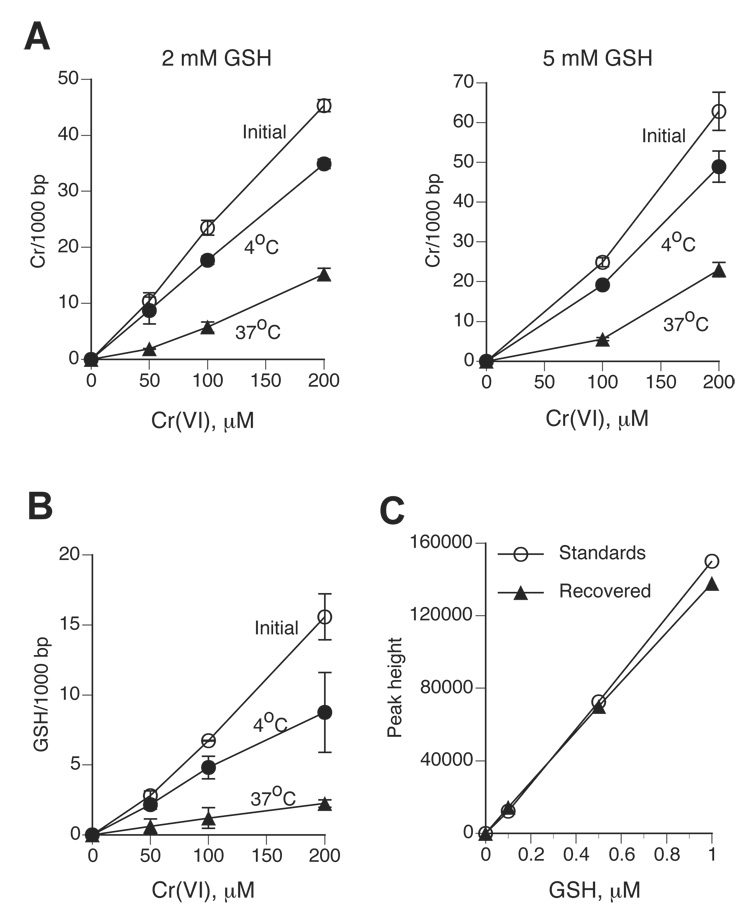
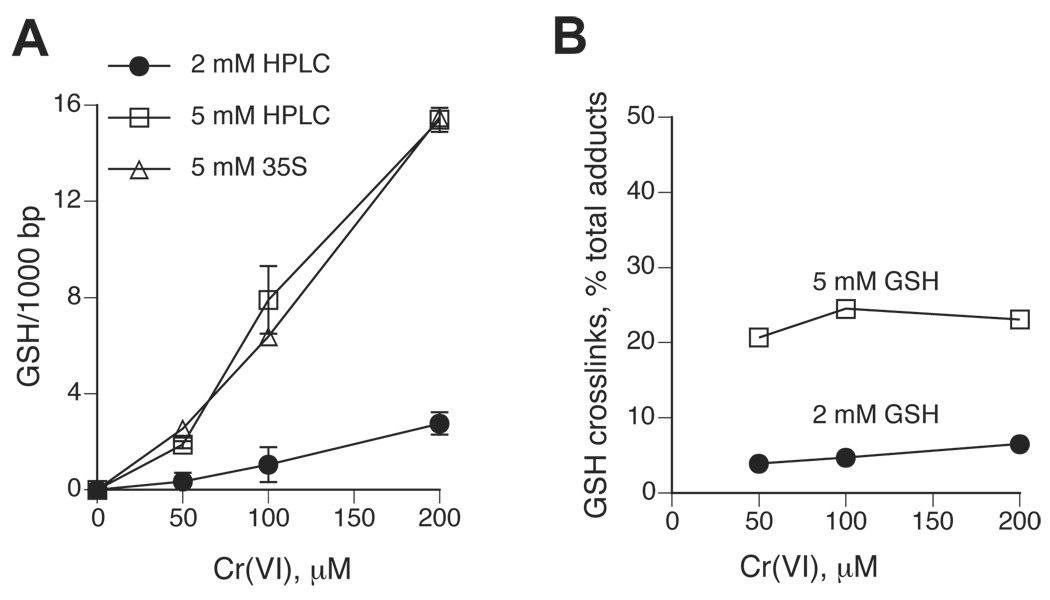
The amount of DNA-crosslinked GSH can be measured by the inclusion of 35S-GSH in the reduction reactions, however, large quantities of radiaoactivity/samples need to be used and the radiochemical purity of 35S-GSH preparations quickly deteriorates during the storage. Poor quality of commercial 35S-GSH and/or 51Cr(VI) preparations was probably responsible for the overestimated ability of Cr(III) atom to cause DNA crosslinking of numerous GSH molecules (27). Therefore, we decided to develop a sensitive nonradioactive procedure for the detection of GSH-DNA crosslinks by HPLC. Our strategy was to dissociate GSH-Cr-DNA crosslinks by incubation with Cr(III)-chelating agents and then quantitatively conjugate released GSH with low mBBr concentrations, with the latter being important for maintaining low background. We tested several different conditions for dissociation of Cr-DNA adducts while conducting parallel analyses of the recovery of spiked GSH. Although incubations in 50 mM phosphate buffer at 4°C resulted in a very good recovery of added GSH and were previously found to release DNA-crosslinked ascorbate (14), this approach was ineffective in dissociation of Cr-DNA adducts or GSH-DNA crosslinks formed in reactions of Cr(VI) with 2 or 5 mM GSH (Fig. 4A,B). However, incubation of Cr-adducted DNA with 50 mM phosphate at 37°C did lead to the release of the majority of DNA-bound Cr: 74.4±6.3% and 70.6±7.2% for 2 and 5 mM GSH-based samples, respectively (Fig. 4A). The dissociation of DNA-crosslinked 35S-GSH at this temperature was also very efficient and averaged 79.8±5.0% (Fig. 4B). The inclusion of the iron-chelator DTPA and the thiol group reducer DTT to the phosphate buffer led to essentially 100% recovery of the spiked GSH standards after 24 hr-long incubations at 37°C (Fig. 4C). Determination of GSH-Cr-DNA crosslinks formed in chromate-5 mM GSH reactions using HPLC and 35S-GSH methods gave very similar results (Fig. 5A), further validating the accuracy of our nonradiaoctive assay. The detection sensitivity for DNA-crosslinked GSH by the current version of our HPLC procedure was about 0.4 pmol, which is similar to the detection sensitivity for DNA-bound Cr by GF-AAS (14).
Figure 4. Dissociation of Cr-DNA adducts by phosphate chelation of Cr(III).
Cr(VI) reductions were performed in MOPS buffer and pSP189 DNA was purified by two rounds of P-30 chromatography. Cr-modified plasmids were then incubated for 24 hr in 50 mM phosphate (pH 7.0) and passed through P-30 columns again. (A) Stability of total Cr-DNA adducts formed in 2 or 5 mM GSH-containing reaction to dissociation by phosphate ions at different temperature. Data are means±SD from four independent measurements. (B) Dissociation of DNA-crosslinked GSH by phosphate ions. Reduction reactions contained 5 mM GSH and trace amounts of 35S-GSH. Data are means±SD from four independent analyses. (C) Efficient recovery of GSH standards incubated for 24 hr in phosphate buffer. Standards – freshly prepared GSH solutions, recovered – GSH amounts measured after 24 hr incubations in phosphate buffer at 37°C. Data are means from duplicate measurements.
Figure 5. Formation of GSH-Cr-DNA crosslinks during Cr(VI) reduction by GSH.
Cr(VI) reduction was performed in 25 mM MOPS buffer, pH 7.0. Total Cr-DNA adducts were measured by GF-AAS and the amount of DNA-crosslinked GSH was determined either by the inclusion of 35S-GSH or by a phosphate dissociation-HPLC procedure. (A) Levels of GSH-DNA crosslinks generated in Cr(VI) reactions containing 2 or 5 mM GSH. HPLC – GSH crosslinks were quantified by HPLC, 35S – GSH crosslinks were measured using radioactive GSH. Data are means±SD from 4–8 independent determinations. (B) Relative yield of GSH-Cr-DNA crosslinks in reactions containing 2 and 5 mM GSH. Data are GSH-DNA crosslinks as the percentage of total Cr-DNA adducts.
Reduction of Cr(VI) by either 2 or 5 mM GSH resulted in a dose-dependent GSH-DNA crosslinking, however, the formation of these crosslinks in 2 mM GSH-based reactions was dramatically lower (Fig. 5A). On average, GSH-Cr-DNA crosslinks made up only 5±1.1% of all Cr-DNA adducts in 2 mM GSH reactions, while the yield of crosslinks with 5 mM GSH was substantially higher and reached 22.8±1.6% (Fig. 5B). The relative yield of GSH-Cr-DNA crosslinks did not vary significantly as a function of Cr(VI) concentration for reactions with 5 mM GSH while 2 mM GSH reactions showed a modest trend for increased GSH-DNA crossliking at higher Cr(VI) doses: 3.9, 4.7 and 6.5% for 50, 100 and 200 µM Cr(VI), respectively. A low GSH-DNA crosslinking is likely to be responsible for poor mutagenicity of Cr-DNA damage generated in chromate-2 mM GSH reactions because GSH-Cr-DNA crosslinks were several times more mutagenic at the supF gene than weakly mutagenic binary Cr-DNA adducts (18). Consistent with this conclusion, the comparison of 2 mM and 5 mM GSH reactions showed that Cr(VI) doses (50 µM Cr for 5 mM GSH and 200 µM Cr for 2 mM GSH) that generated similar levels of GSH-Cr-DNA crosslinks also produced similar mutagenic responses (Fig. 3A,B and Fig. 5A). Importantly, the total number of Cr-DNA adducts in these reactions was about 4-times different (Fig. 1B).
Conclusions
Reductive metabolism of Cr(VI) by the physiological 2 mM GSH concentration generates weakly mutagenic Cr-DNA damage, which is consistent with low mutagenicity of chromate in CHO and V79 cells under the conditions of its predominant reductive activation by GSH (21,22). Poor mutagenicity of this reduction process is associated with low yields of mutagenic GSH-Cr-DNA crosslinks (5.0% of total Cr-DNA adducts). Reduction of Cr(VI) with the higher 5 mM GSH concentration increased both mutagenic responses and GSH-Cr-DNA crosslinking. Although GSH-Cr-DNA crosslinks were found to be about 2.5–3-times more mutagenic than cysteine-Cr-DNA crosslinks (18), a significantly lower mutagenicity of 2 mM GSH-Cr(VI) reactions relative to those with 1 mM cysteine (15) probably reflects about 10-times higher yield of cysteine-Cr-DNA crosslinks (36). Mutagenic responses in plasmids damaged during Cr(VI) reduction by physiological concentrations of ascorbate (16) were 15–20-times greater than in GSH-based reactions, which was caused by a higher formation of ascorbate-Cr-DNA crosslinks and their several times stronger pre-mutagenic potential relative to GSH-containing crosslinks. Since the supF gene is a more sensitive target for the detection of single base substitutions than chromosomal Hprt gene (30), the predominance of point mutations in the mutational spectrum of GSH-Cr-DNA crosslinks (18) should make these lesions less efficient inducers of phenotypically detectable mutagenic events in cellular genes relative to ascorbate-Cr-DNA crosslinks that generate approximately equal frequency of point and frameshift mutations (16).
Acknowledgements
This work was supported by grants R01 ES012915 and P42 ES013660 from the National Institute of Environmental Health Sciences. We thank Jana Jarolimova for the initial work on the optimization of GSH measurements by HPLC.
Abbreviations
- DTPA
diethylenetriaminepentaacetic acid
- DTT
dithiotreitol (DTT)
- GSH
glutathione
- mBBr
monobromobimane
References
- 1.Park RM, Bena JF, Stayner LT, Smith RJ, Gibb HJ, Lees PS. Hexavalent chromium and lung cancer in the chromate industry: a quantitative risk assessment. Risk Anal. 2004;24:1099–1108. doi: 10.1111/j.0272-4332.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 2.Costa M, Klein CB. Toxicity and carcinogenicity of chromium compounds in humans. Crit. Rev. Toxicol. 2006;36:155–163. doi: 10.1080/10408440500534032. [DOI] [PubMed] [Google Scholar]
- 3.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic and chromium. Chem. Res. Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose RN, Moghaddas S, Gelerinter E. Long-lived chromium(IV), chromium(V) metabolites in the chromium(VI)- glutathione reaction: NMR, ESR, HPLC, and kinetic characterization. Inorg. Chem. 1992;31:1987–1994. [Google Scholar]
- 5.Stearns DM, Wetterhahn KE. Reaction of Cr(VI) with ascorbate produces chromium(V), chromium(IV), and carbon based radicals. Chem. Res. Toxicol. 1994;7:219–230. doi: 10.1021/tx00038a016. [DOI] [PubMed] [Google Scholar]
- 6.Lay PA, Levina A. Activation of molecular oxygen during the reactions of chromium(VI/V/IV) with biological reductants: implications for chromium-induced genotoxicities. J. Am. Chem. Soc. 1998;120:6704–6714. [Google Scholar]
- 7.Sugden KD, Wetterhahn KE. Direct and hydrogen peroxide-induced chromium(V) oxidation of deoxyribose in single-stranded and double-stranded calf thymus DNA. Chem. Res. Toxicol. 1997;10:1397–1406. doi: 10.1021/tx970135r. [DOI] [PubMed] [Google Scholar]
- 8.Slade PG, Hailer MK, Martin BD, Sugden KD. Guanine-specific oxidation of double-stranded DNA by Cr(VI) and ascorbic acid forms spiroiminodihydantoin and 8-oxo-2'-deoxyguanosine. Chem. Res. Toxicol. 2005;18:1140–1149. doi: 10.1021/tx050033y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall M, da Cruz Fresco P, Kortenkamp A. Chromium(VI)-mediated DNA damage: oxidative pathways resulting in the formation of DNA breaks and abasic sites. Chem. Biol. Interact. 1999;123:117–132. doi: 10.1016/s0009-2797(99)00128-3. [DOI] [PubMed] [Google Scholar]
- 10.Shi X, Mao Y, Knapton AD, Ding M, Rojanasakul Y, Gannett PM, Dalal N, Liu K. Reaction of Cr(VI) with ascorbate and hydrogen peroxide generates hydroxyl radicals and causes DNA damage: role of a Cr(IV)-mediated Fenton-like reaction. Carcinogenesis. 1994;15:2475–2478. doi: 10.1093/carcin/15.11.2475. [DOI] [PubMed] [Google Scholar]
- 11.Zhitkovich A. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI) Chem. Res. Toxicol. 2005;18:3–11. doi: 10.1021/tx049774+. [DOI] [PubMed] [Google Scholar]
- 12.Zhitkovich A, Voitkun V, Costa M. Glutathione and free amino acids form stable adducts with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis. 1995;16:907–913. doi: 10.1093/carcin/16.4.907. [DOI] [PubMed] [Google Scholar]
- 13.Zhitkovich A, Messer J, Shrager S. Reductive metabolism of Cr(VI) by cysteine leads to the formation of binary and ternary Cr-DNA adducts in the absence of oxidative DNA damage. Chem. Res. Toxicol. 2000;13:1114–1124. doi: 10.1021/tx0001169. [DOI] [PubMed] [Google Scholar]
- 14.Quievryn G, Messer J, Zhitkovich A. Carcinogenic chromium(VI) induces cross-linking of vitamin C to DNA in vitro and in human lung A549 cells. Biochemistry. 2002;41:3156–3167. doi: 10.1021/bi011942z. [DOI] [PubMed] [Google Scholar]
- 15.Zhitkovich A, Song Y, Quievryn G, Voitkun V. Nonoxidative mechanisms are responsible for the induction of mutagenesis by reduction of Cr(VI) with cysteine: role of ternary DNA adducts in Cr(III)-dependent mutagenesis. Biochemistry. 2001;40:549–560. doi: 10.1021/bi0015459. [DOI] [PubMed] [Google Scholar]
- 16.Quievryn G, Peterson E, Messer J, Zhitkovich A. Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry. 2003;42:1062–1070. doi: 10.1021/bi0271547. [DOI] [PubMed] [Google Scholar]
- 17.Quievryn G, Messer J, Zhitkovich A. Lower mutagenicity but higher stability of Cr-DNA adducts formed during gradual chromate activation with ascorbate. Carcinogenesis. 2006;27:2316–2321. doi: 10.1093/carcin/bgl076. [DOI] [PubMed] [Google Scholar]
- 18.Voitkun V, Zhitkovich A, Costa M. Cr(III)-mediated crosslinks of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic Acids Res. 1998;26:2024–2030. doi: 10.1093/nar/26.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillon CT, Lay PA, Cholewa M, Legge GJ, Bonin AM, Collins TJ, Kostka KL, Shea-McCarthy G. Microprobe X-ray absorption spectroscopic determination of the oxidation state of intracellular chromium following exposure of V79 Chinese hamster lung cells to genotoxic chromium complexes. Chem. Res. Toxicol. 1997;10:533–535. doi: 10.1021/tx970010m. [DOI] [PubMed] [Google Scholar]
- 20.Ortega R, Fayard B, Salome M, Deves G, Susini J. Chromium oxidation state imaging in mammalian cells exposed in vitro to soluble or particulate chromate compounds. Chem. Res. Toxicol. 2005;18:1512–1519. doi: 10.1021/tx049735y. [DOI] [PubMed] [Google Scholar]
- 21.Cohen MD, Kargacin B, Klein CB, Costa M. Mechanisms of chromium carcinogenicity and toxicity. Crit. Rev. Toxicol. 1993;23:255–281. doi: 10.3109/10408449309105012. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds M, Stoddard L, Bespalov I, Zhitkovich A. Ascorbate acts as a highly potent inducer of chromate mutagenesis and clastogenesis: linkage to DNA breaks in G2 phase by mismatch repair. Nucleic Acids Res. 2007;35:465–476. doi: 10.1093/nar/gkl1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messer J, Reynolds M, Stoddard L, Zhitkovich A. Causes of DNA single-strand breaks during reduction of chromate by glutathione in vitro and in cells. Free Radical Biol. Med. 2006;40:1981–1992. doi: 10.1016/j.freeradbiomed.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 24.O’Brien P, Wang G, Wyatt PB. Studies on the kinetics of the reduction of chromate by glutathione and related thiols. Polyhedron. 1992;11:3211–3216. [Google Scholar]
- 25.Borges KM, Boswell JS, Liebross RH, Wetterhahn KE. Activation of chromium(VI) by thiols results in chromium(V) formation, chromium binding to DNA and altered DNA conformation. Carcinogenesis. 1991;12:551–561. doi: 10.1093/carcin/12.4.551. [DOI] [PubMed] [Google Scholar]
- 26.Levina A, Lay PA. Mechanistic studies of relevance to the biological activities of chromium. Coord. Chem. Rev. 2005;42:281–298. [Google Scholar]
- 27.O’Brien T, Xu J, Patierno SR. Effect of glutathione on chromium-induced DNA crosslinking and DNA polymerase arrest. Mol. Cell. Biochem. 2001;222:173–182. [PubMed] [Google Scholar]
- 28.Reynolds M, Zhitkovich A. Cellular vitamin C increases chromate toxicity via a death program requiring mismatch repair but not p53. Carcinogenesis. 2007;28:1613–1620. doi: 10.1093/carcin/bgm031. [DOI] [PubMed] [Google Scholar]
- 29.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteasome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 30.Parris CN, Seidman MM. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992;117:1–5. doi: 10.1016/0378-1119(92)90482-5. [DOI] [PubMed] [Google Scholar]
- 31.Ariza RR, Roldan-Arjona T, Hera C, Pueyo C. A method for selection of forward mutations in supF gene carried by shuttle-vector plasmids. Carcinogenesis. 1993;14:303–305. doi: 10.1093/carcin/14.2.303. [DOI] [PubMed] [Google Scholar]
- 32.Quievryn G, Goulart M, Messer J, Zhitkovich A. Reduction of Cr(VI) by cysteine: significance in human lymphocytes and formation of DNA damage in reactions with variable reduction rates. Mol. Cell. Biochem. 2001;222:107–118. [PubMed] [Google Scholar]
- 33.Levina A, Lay PA, Dixon NE. Disproportionation of a model chromium(V) complex causes extensive chromium(III)-DNA binding in vitro. Chem. Res. Toxicol. 2001;14:946–950. doi: 10.1021/tx010077g. [DOI] [PubMed] [Google Scholar]
- 34.Kortenkamp A, Casadevall M, Faux SP, Jenner A, Shayer RO, Woodbridge N, O'Brien P. A role for molecular oxygen in the formation of DNA damage during the reduction of the carcinogen chromium (VI) by glutathione. Arch. Biochem. Biophys. 1996;329:199–207. doi: 10.1006/abbi.1996.0209. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 36.Zhitkovich A, Quievryn G, Messer J, Motylevich Z. Reductive activation with cysteine represents a chromium(III)-dependent pathway in the induction of genotoxicity by carcinogenic chromium(VI) Environ. Health Persp. 2002;110:729–731. doi: 10.1289/ehp.02110s5729. [DOI] [PMC free article] [PubMed] [Google Scholar]