Abstract

The α subunit of the replicative DNA polymerase III of Escherichia coli is the active polymerase of the 10-subunit bacterial replicase. The C-terminal region of the α subunit is predicted to contain an oligonucleotide binding (OB-fold) domain. In a series of optical tweezers experiments, the α subunit is shown to have an affinity for both double- and single-stranded DNA, in distinct subdomains of the protein. The portion of the protein that binds to double-stranded DNA stabilizes the DNA helix, because protein binding must be at least partially disrupted with increasing force to melt DNA. Upon relaxation, the DNA fails to fully reanneal, because bound protein interferes with the reformation of the double helix. In addition, the single-stranded DNA binding component appears to be passive, as the protein does not facilitate melting but instead binds to single-stranded regions already separated by force. From DNA stretching measurements we determine equilibrium association constants for the binding of α and several fragments to dsDNA and ssDNA. The results demonstrate that ssDNA binding is localized to the C-terminal region that contains the OB-fold domain, while a tandem helix-hairpin-helix (HhH)2 motif contributes significantly to dsDNA binding.
Cellular DNA replicases are asymmetric dimers (1,2) or trimers (3) that efficiently and accurately copy millions of base pairs of DNA in every cell cycle. The Escherichia coli replicase DNA polymerase III is a complex of 10 subunits that tightly coordinates leading and lagging strand synthesis (2). The polymerase subunit of this complex is the 1160-residue α protein, encoded by the dnaE gene (2). E. coli Pol III contains a separate 3′−5′ exonuclease proofreading protein, ε, that together with the θ subunit is tightly bound to the α subunit (2). The β processivity clamp encircles DNA, binds the Pol III α subunit, and facilitates highly processive DNA replication by Pol III (2). The clamp loader is a dynamic complex that includes combinations of the γ, δ, δ′, τ, χ, and ψ subunits and has roles in loading the β clamp onto DNA, dimerization of the polymerase core, and polymerase recycling on the lagging strand (2,4).
DNA polymerases are classified as members of the A, B, C, D, X, and Y families (5). E. coli pol III α protein is a member of the C family (5,6). In many Gram-negative bacteria, including E. coli, DNA pol III is responsible for genomic DNA replication (2). Protein sequence analysis indicates that the E. coli Pol III α subunit has little or no detectable sequence similarity to eukaryotic or archaeal polymerases of known structure (7). A recent crystal structure of residues 1−917 of E. coli α reveals that the protein adopts a right-hand shape typical of DNA polymerases (8). The Thermus aquaticus full-length Pol III crystal structure shows that this DNA polymerase is similar overall to that of E. coli(9). One of the major differences between the structures of Pol III and those of many other DNA polymerases is that the fingers domain, which binds the incoming nucleotide, is much larger in Pol III than in most polymerases (8). Three conserved acidic residues in the palm domain are responsible for polymerase activity (10). The palm domain of Pol III is similar to that of X family member Pol β (8–10). The thumb, fingers, and palm domains form a deep cleft that could bind DNA, although the only available structure of E. coli Pol III lacks DNA substrates (8). The extreme N-terminus of Pol III harbors a polymerase and histidinol phosphatase (PHP) domain, which may be involved in pyrophosphatase or exonuclease activity (8). The PHP domain of Thermus thermophilus has been shown to exhibit Zn2+-dependent 3′−5′ exonuclease activity (11); however, the metal-ion-binding residues are not conserved in the E. coli PHP domain, suggesting a different function for this domain in E. coli Pol III (9).
The 10-subunit DNA polymerase III holoenzyme has been shown to bind approximately 30 nucleotides of double-stranded DNA in a primer:template complex (12). Pol III α subunit has at least two distinct domains that may be important for binding DNA (Figure 1). A “helix-hairpin-helix” (HhH) motif, initially predicted in the α subunit by sequence analysis (13), is a widespread motif involved in non-sequence-specific binding of either ds- or ss-DNA. Crystal structures of the Pol III α subunits of E. coli(8) and T. aquaticus(9) confirm the presence of the HhH motif slightly N-terminal to the internal β-processivity clamp binding motif. The HhH motif in both structures may be considered as part of a distinct (HhH)2 domain formed by two consecutively duplicated HhH motifs (14). (HhH)2 domains are present in a majority of HhH-containing proteins and provide a symmetric way of binding to dsDNA, as in DNA polymerase β (14). HhH domains are also known to mediate protein−protein interactions (15). The second putative DNA binding domain in the α subunit is an oligonucleotide/oligosaccharide binding (OB-fold) domain (16,17). The OB-fold domain is located near the C-terminus (7–9,18), which is not present in the E. coli structure. OB-domains are functionally diverse, as they are involved in binding oligonucleotides, oligosaccharides, or metal ions and also in mediating protein−protein interactions (18,19). Thus, pol III possesses two domains outside of the polymerase core that are often associated with DNA binding. Yet the DNA binding activity of either domain has not been directly demonstrated.
Figure 1.
Catalytic α subunit of E. coli replicative DNA pol III. a) Domain architecture of α subunit and five additional constructs studied in this work. Polymerase core indicates palm, thumb, and fingers domains; HhH denotes helix-hairpin-helix DNA-binding motif; β-binding motif denotes internal canonical and C-terminal β-binding motif; and OB-domain denotes predicted OB-fold domain. Residue numbers at the boundaries of major domains and at the ends of the fragments are noted according to ref (8). b) Homology model of the OB-fold domain, encompassing residues 978−1078 and derived as described in the text. ssDNA is predicted to interact with the highly conserved residue F1031 shown in the figure. The coordinates for the model are available at http://www.ibt.lt/bioinformatics/models/polIII_OB. (c) dsDNA binds nonspecifically to the tandem HhH motif in this model of residues 833−889.
To investigate DNA-binding properties of the E. coli Pol III α subunit and its individual regions, we first modeled the 3D structure of the OB-domain and found that it is consistent with ssDNA binding. Next, we used single molecule DNA stretching experiments to show that the full-length E. coli Pol III α subunit binds both double-stranded DNA and single-stranded DNA. Further, we show that the dsDNA binding domain maps to the N-terminal 917 residues that harbor the (HhH)2 domain, whereas the C-terminal 182 residues including the OB-domain bind preformed ssDNA without actively melting the DNA. Therefore, this domain may interact with ssDNA created by another process during DNA replication, such as with the template strand or ssDNA created during proofreading.
Results and Discussion
Homology Modeling of the OB-Fold Domain
Homology modeling is a powerful molecular modeling technique that allows complex structures to be deduced by comparison to known structures with similar sequences (20–23). In this process, sequences of multiple known structures, “templates”, are compared to the OB-fold domain sequence of the α subunit (residues 978−1078). Sections of the sequence that show strong similarity may be used to construct a 3D model based upon the structures of the templates. The aligned sequences for the templates and α are shown in Supplementary Figure 1, and the final model from this iterative process is shown in Figure 1, panel b. Additional modeling details are included in Supporting Information.
DNA−Protein Binding Experiments
Force spectroscopy experiments (Figure 2) probe the elasticity of dsDNA and convert it into ssDNA at a critical melting force (Figure 3). These experiments may be repeated in the presence of DNA binding ligands. Whereas binding to dsDNA is observed to stabilize the helical form, binding to ssDNA destabilizes the double helix and is manifested in the data as a decrease in either the melting force or the length of the overstretching plateau (24–29). Thus monitoring the force versus length of a cycle of DNA extension and relaxation serves to assay double- and single-stranded DNA binding. Additional experimental details are included in Supporting Information.
Figure 2.
Dual beam optical tweezers experiment schematic with accompanying microscope images. a) Streptavidin-coated beads (5.5 μm diameter) are fixed upon a micropipette tip and held within a dual beam confocal trap (∼1 μm diameter, which appears as the bright spot in the accompanying photograph). Biotinylated phage λ-DNA (16.5 μm contour length) is extended between these beads, and the subsequent increase in tension is determined by the deflection of the trapping lasers upon a pair of photodiode detectors. b) Solutions of varying concentrations of buffer, salt, and DNA binding protein (shown in green and yellow) are introduced. c) Measuring the force required for DNA extension in the presence of protein reveals information about binding to the double strand. d) Force-induced DNA melting (and subsequent relaxation) allows protein binding to ssDNA, which is observed as a change in the measured length.
Figure 3.
Force extension and relaxation data for phage λ-DNA are shown as solid and open circles, respectively. As the DNA is extended and unwound, the tension increases. In the overstretching transition, base stacking is disrupted and dsDNA is melted, becoming ssDNA tethered by a few remaining G−C-rich regions. Force-induced melting is reversible, though there is some hysteresis due to the relatively slow observed time scale of reannealing (blue circles). Complete strand separation is observed at much higher forces (red and green symbols). The solid black line is a fitted polymer model of dsDNA, known as the worm-like chain model, while ssDNA elasticity is described by the freely jointed chain model (green line). Models of composite DNA (1/3 ssDNA−2/3 dsDNA and 2/3 ssDNA−1/3 dsDNA) are shown as intermediates (blue lines) and are described in the text. Thus force extension measurements reveal the fractional DNA melting. The experimental buffer contains 10 mM HEPES (pH 7.5) and 100 mM Na+.
Pol III α Subunit Binds to ssDNA Created by Force-Induced Melting
A sequence of extension and relaxation curves for DNA in the presence of 100 nM full-length Pol III α subunit is shown in Figure 4, panel a. Upon the addition of 100 nM α protein, the extension curve is largely unaffected. The slight deviations at low stretching forces are due to protein−protein and protein−DNA aggregation (24). After force-induced melting, the melted DNA does not appear to reanneal on the ∼5 min time scale of relaxation. The melted regions of DNA do not reanneal because ssDNA has been stabilized by the binding of full-length α protein. Subsequent extension data shows that some reannealing occurs while the DNA is relaxed, though a significant fraction remains single-stranded. After a few cycles, the extension and relaxation data nearly overlap, suggesting that protein binding is in equilibrium.
Figure 4.
Force extension (solid) and relaxation (dotted) data for DNA in the presence of 100 nM full-length α protein in a 100 mM Na+ solution, compared to a reference of DNA in the absence of protein (black). a) In this series, the initial cycle (blue) shows a slight increase in the melting plateau and reveals substantial hysteresis. Only a fraction of the melted DNA returns to the double helical form (the solid and dotted blue lines do not overlap), as protein bound to ssDNA inhibits annealing over the time scales of these experiments. Subsequent cycles (in the sequence order green, yellow, red) show that additional regions become stabilized by protein binding, until an apparent equilibrium length of ∼1/2 of the double strand is melted. Complete strand separation occurs in the last cycle at ∼140 pN. b) A sequence of constant extension experiments in the presence of 100 nM full-length α protein. DNA is stretched to the extension marked by each arrow, held for 30 min, and then relaxed. Relaxation data is shown as open circles, and the extension data has been removed for clarity. As DNA is held to greater extensions, correspondingly larger fractions of the force-melted DNA are stabilized in the single-stranded form through protein binding to the single-stranded regions, though some unbinding and DNA reannealing is evident upon relaxation. Solid lines represent the best fits to this data, according to eq 1, and yield stabilized ssDNA fractions of 0.00 ± 0.01 (violet), 0.14 ± 0.02 (blue), 0.24 ± 0.02 (green), 0.40 ± 0.02 (yellow), and 0.42 ± 0.03 (red). Though the fixed extensions are progressively increased, the fraction of stabilized ssDNA saturates at ∼1/2, as noted in the main text.
To consistently melt a defined fraction of DNA, a series of constant extension experiments was performed. Protein was introduced into solution, and the translation stage was held at a fixed position for 30 min and then relaxed. For relatively low fixed extensions, (<1/2 of the length of the overstretching plateau), a fraction of the double helix remains melted upon relaxation, as shown in Figure 4, panel b. In these regions, ssDNA has been stabilized by the binding of full-length α protein. The value of the fraction of stabilized ssDNA (γss) may be found by fitting the measured force-dependent contour length (b) to a linear combination of the contour length of double-stranded (bds) and single-stranded (bss) DNA:
| 1 |
Results for these fits are shown as solid lines in Figure 4, panel b, where the fits are confined to data below 30 pN to minimize effects due to protein unbinding described below. The fitting results are then extrapolated to higher forces for clarity. As the fixed extension is increased, the fraction of ssDNA bound increases, as evidenced by a decrease in the length of the melting plateau. However, full-length α binds only up to a maximum of ∼1/2 of the full DNA length. Unbinding is also noted but is only complete upon full DNA relaxation (F ≤ 1 pN) for at least 20 min (see Supporting Information).
Proteins that actively bind to ssDNA generally lower the observed melting force as the protein helps to stabilize ssDNA relative to dsDNA (27–29). Full-length α protein does not lower the melting force but inhibits reannealing of DNA that has been melted by force (Figure 4, panel b). Therefore this protein passively binds to regions that have been melted by force and have long-lived single-stranded regions and is unable to stabilize small regions of ssDNA created during short-lived thermal fluctuations. As the melted regions that are subsequently bound by protein appear relatively stable even as DNA is relaxed to forces below the melting transition, dissociation from ssDNA must be relatively slow. Thus the inhibition of reannealing is likely due to long regions of stabilized ssDNA. The fact that only one-half of the melting transition is affected indicates that the protein cannot saturate the ssDNA lattice. This may be due to variations in the stability of regions with different base composition (30,31) or other effects related to the ability of the protein to access ssDNA created by force-induced melting.
Pol III α Subunit Stabilizes dsDNA
In addition to ssDNA binding, a very slight increase in the melting force is observed in Figure 4, panel a. This increase is attributed to protein binding to dsDNA, in which case the protein must be removed for melting to occur. This effect is most clear in low salt conditions, which destabilize dsDNA by reducing the screening between the charged backbones. Figure 5, panels a and b, shows dsDNA melted in 100 mM Na+ and the same strands melted in 10 mM Na+. In the absence of protein, the expected destabilization of the dsDNA with decreasing salt concentration is observed (32). To quantify this effect, we measured the melting force at the approximate midpoint of the transition (averaging the observed force over the extension range of 0.44−0.46 nm per base pair). Then, 100 nM full-length α protein (Figure 5, panel a) or 500 nM α1−917 (Figure 5, panel b) was added, resulting in a strong increase in the melting force, indicating dsDNA binding, whereas α1−917 shows little binding to ssDNA (see below). Figure 5, panel c summarizes this data over a range of salt concentrations and shows dsDNA stabilization that is independent of salt concentration within uncertainty. Moreover, α protein is active in DNA polymerization under this range of salt concentrations (Supplementary Figure 2). Different protein concentrations were used as different constructs displayed different equilibrium association constants (see below).
Figure 5.
Stabilization of the double helix is shown in a series of extension (solid lines) and relaxation (dotted lines) cycles at varying salt concentrations. a, b) dsDNA is extended (black), and the helical form is disrupted at 62.6 ± 1.0 pN (32). As Na+ concentration is decreased, this transition occurs at lower forces and greater hysteresis is observed as DNA is relaxed (purple). As full-length protein is added (to the low-salt solution), the force required to disrupt the double helix increases again (blue). Furthermore, after a constant extension experiment in which the extension is fixed for 30 min, significant binding to the single strand appears (green), but only for the full-length protein (panel a) and not for the N-terminal α1−917 (panel b). c) The DNA melting force varies with Na+ concentration (purple), while melting force in the presence of 100 nM α and 500 nM α1−917 was observed to be independent of salt concentration. Uncertainties are determined from averages over three experiments. The average melting force for DNA, over the full range of Na+ concentrations studied, is 65.2 ± 0.4 pN in the presence of α protein and 65.1 ± 0.6 pN for α1−917. Higher concentrations of the α1−917 construct were necessary because of weaker binding (see Figure 5, panel b and Figure 8, panel a).
C-Terminal Constructs Bind ssDNA Created by Force-Induced Melting
To further isolate the dsDNA and ssDNA binding sites, we constructed fragments of the α subunit and performed stretching experiments in the presence of these protein fragments. C-terminal protein fragments consisted of the residues 917−1160, 978−1160, and 1076−1160. The first two include the putative OB-fold domain region (residues 978−1078). Force-extension experiments with all three proteins indicate that there is little stabilization of dsDNA, even in low salt (10 mM Na+, data not shown). However, binding to ssDNA is evident for α917−1160 and α978−1160 (in 100 mM Na+), as shown in Figure 6, panels a and b, respectively. Strong aggregation effects also appear in the extension curves for α978−1160, indicated by jagged extension curves (as opposed to jagged relaxation curves, which occur normally) at low to moderate extension, as seen with other small proteins (24,33). The final construct, α1076−1160, shows no binding to ssDNA, as the DNA completely reanneals at high forces (∼40 pN) upon relaxation (Figure 6, panel c). Our results suggest that an ssDNA binding site is contained within the OB-fold (978−1078) domain (we were unable to purify the isolated OB-domain). Our modeling, in agreement with other observations (7), suggests that the OB-domain of α protein is most similar to a subclass of OB-fold domains that includes archaeal/eukaryotic ssDNA binding proteins and anticodon binding domains rather than to eubacterial SSBs. Notably, the OB-fold domain of the α protein has fewer aromatic residues lining the putative ssDNA binding path than either the tRNA anticodon-binding domain of tRNA synthetases or ssDNA-binding OB-fold domains. Instead, the OB-domain features a highly conserved phenylalanine (F1031), structurally equivalent to an aromatic residue that makes a base stacking interaction in the human RPA/ssDNA complex (34). A corresponding aromatic residue also makes a similar base-stacking contact in the DNA-bound RecG structure (35), while in bacteriophage gene 32 protein, which also possesses an OB-fold domain, the residue F183 also stacks onto ssDNA (36). These results suggest that the OB-domain of the α subunit may similarly bind ssDNA. However, unlike in the case of the RPA/DNA structure, there are no other aromatic residues lining the putative ssDNA binding path (Figure 1), suggesting weaker interaction.
Figure 6.
DNA binding of C-terminal α constructs isolates the binding site of ssDNA in this series of extension/relaxation cycles (solid and dotted lines, respectively). Both a longer construct (α917−1160, panel a) and a shorter construct (α978−1160, panel b) show binding to ssDNA. A series of extension and relaxation cycles (in the sequence order blue, green, red) show significant binding to the single strand, similar to that of full-length protein (Figure 4). Upon initial relaxation in the presence of protein (blue), a significant fraction remains single-stranded as bound protein inhibits formation of the double helix. The rough appearance of the extension data for α978−1160 is evidence of aggregation and of protein unbinding from the DNA as it is stretched. c) The smallest construct of α1076−1160 shows no binding to ssDNA, effectively isolating the observed ssDNA binding region to the OB-domain.
N-Terminal Constructs Do Not Bind to ssDNA
N-terminal constructs consisting of either residues 1−917 (including the polymerase core and the HhH motifs) or 1−835 (lacking the tandem HhH motifs) showed no evidence of binding to ssDNA (Figure 7), as complete DNA reannealing occurs at high forces (∼40 pN) upon relaxation. Thus our observations allow us to rule out ssDNA binding by the (HhH)2 motifs or other regions of the N-terminus of α. Furthermore, the α1−917 construct shows substantial binding to dsDNA. However, the α1−835 construct does not show strong dsDNA binding, and therefore the tandem HhH motifs contribute significantly to dsDNA binding.
Figure 7.
Binding of the N-terminal constructs α1−917 (panel a) and α1−835 (panel b) shows no stabilization of ssDNA through the sequence of extension/relaxation cycles (in the sequence order blue, green, red), as indicated by the complete reannealing of DNA at high forces upon relaxation in both cases. A slight change in the melting force, indicating some dsDNA binding, is evident for α1−917 but not for α1−835. The strong overlap of the extension curves (solid lines) emphasizes the reproducibility of the data, whereas some variability is evident upon relaxation (dotted lines), though this is typical of these experiments with DNA even in the absence of protein (black).
Equilibrium Binding
To quantify the binding of α protein to double- and single-stranded DNA, a series of experiments were carried out using varying concentrations of the full-length α subunit and the fragments α1−835, α1−917, and α917−1160. Binding to dsDNA was characterized by observing the change in the melting force (37):
| 2 |
The fractional binding to dsDNA (γds) is determined by measuring the melting force at a given protein concentration (Fm) relative to the melting force in the absence of protein 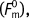 and the melting force when the DNA molecule appears saturated
and the melting force when the DNA molecule appears saturated 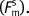 This method assumes that the protein is in equilibrium only with dsDNA, since it does not actively denature DNA (as discussed above). These forces are determined by finding the force at the midpoint of the melting transition, averaging over the extensions of 0.44−0.46 nm per base pair to minimize any variations due to subtle changes in the slope of the transition. These data are then fit to a site exclusion binding isotherm that describes the fractional binding as a function of free protein in solution (c) (38):
This method assumes that the protein is in equilibrium only with dsDNA, since it does not actively denature DNA (as discussed above). These forces are determined by finding the force at the midpoint of the melting transition, averaging over the extensions of 0.44−0.46 nm per base pair to minimize any variations due to subtle changes in the slope of the transition. These data are then fit to a site exclusion binding isotherm that describes the fractional binding as a function of free protein in solution (c) (38):
| 3 |
For simplicity, the binding site size (n) was fixed, and the equilibrium association binding constant (K) was the only free parameter used to obtain the best fit. While the structure of Figure 1 suggests a large occluded site size, the fits shown in Figure 8 are minimized for n = 1, while fits with n = 2 are also consistent with the data. This may simply result from the relative weakness and nonspecificity of protein binding, which could allow additional binding as dsDNA is stretched. Fits of γ and K have the appropriate subscript added, depending on the type of binding measured. The observed data and best fits are shown in Figure 8, panel a, and the equilibrium association constants are in Table 1.
Figure 8.
Binding isotherms for full-length protein and selected α constructs to DNA. a) Binding to dsDNA is determined by measuring the change in the melting force for varying protein concentrations for full-length α (blue), the N-terminal constructs α1−917 (green) and α1−835 (yellow), and the C-terminal construct α917−1160 (red). Error bars represent the standard error from four experiments. Fits are to binding isotherms described within the text (eq 2 and eq 3), for n = 1 (solid lines) and n = 2 (dotted lines). For n = 1, Kds = 28 ± 7 × 106 M−1 for full-length α, 3.5 ± 0.9 × 106 M−1 for α1−917. and 0.4 ± 0.1 × 106 M−1 for α1−835. b) Binding to ssDNA is determined by measuring the fraction of ssDNA stabilized by protein binding, as found from eq 1, for varying protein concentrations for full-length α (blue), the N-terminal constructs α1−917 (green) and α1−835 (yellow), and the C-terminal construct α917−1160 (red). Error bars are determined from errors in the fits. Fitting this data to the binding isotherms of the text (eq 3 and eq 4) determines Kss = 22 ± 6 × 106 M−1 for full-length α and 10 ± 3 × 106 M−1 for α917−1160. These results are summarized in Table 1.
Table 1. Association binding constants for full-length α subunit and fragments α1−917, α1−835, and α917−1160 to double- and single-stranded DNA.
| Protein | Kds (× 106 M−1) | Kss (× 106 M−1) |
|---|---|---|
| α | 28 ± 7 | 22 ± 6 |
| α1−917 | 3.5 ± 0.9 | <0.1 |
| α1−835 | 0.4 ± 0.1 | <0.01 |
| α917−1160 | <0.03 | 10 ± 3 |
The binding to ssDNA was determined by finding the relative fraction of ssDNA stabilized by protein (γss) from the fits to eq 1 for varying protein concentrations. This fraction may also be expressed in terms of the observed contour length (b) at a given protein concentration and the contour lengths of double- and single-stranded DNA in the absence of protein. Thus a fractional occupancy may be constructed:
| 4 |
The fractional occupancy saturates at a value of approximately 1/2. The fits to the data are shown in Figure 8, panel b, and the equilibrium association constants are in Table 1. Equilibrium association constants for binding to dsDNA are found to be Kds = 28 ± 7 × 106 M−1 for full-length α, 0.4 ± 0.1 × 106 M−1 for α1−835, and 3.5 ± 0.9 × 106 M−1 for α1−917. Binding to ssDNA revealed Kss = 22 ± 6 × 106 M−1 for full-length α and 10 ± 3 × 106 M−1 for α917−1160. Upper limits for the dsDNA binding of α917−1160 and ssDNA binding of α1−917 show very weak binding. Thus, the contribution to dsDNA binding by other polymerase domains outside the region containing the (HhH)2 motif is weak. Clearly, the full-length protein shows strong binding to both dsDNA and ssDNA, whereas the fragments α1−917 and α917−1160 show strong binding only to dsDNA and ssDNA, respectively. However, binding for both fragments is somewhat weaker than for the full-length protein, suggesting that the fragments may be less stable or that other domains may make modest contributions to binding affinity.
Conclusions
Using single molecule force spectroscopy, we have demonstrated that the full-length Pol III α subunit binds to both dsDNA and ssDNA and have quantified the affinity for both types of binding. On the basis of modeling and structural (9) studies that located an OB-domain within the C-terminus, we isolated the putative dsDNA and ssDNA binding sites by constructing fragments that contain only the dsDNA binding site (α1−917) or only the ssDNA binding site (α917−1160 and α978−1160). Further experiments with α1−835 and α1076−1160 fragments show substantially weaker binding, limiting strong DNA binding sites to the (HhH)2 domain and the OB-domain for double- and single-stranded DNA binding, respectively. Although the existence of a dsDNA binding site on the polymerase is not unexpected, the role of the ssDNA binding site is unknown but may be required for processes involving ssDNA during replication, such as binding template DNA or ssDNA during proofreading. In previous studies, a chimeric RB69 DNA polymerase fused to SSB exhibited higher processivity and a better ability to polymerize through pause sites in the template that are attributed to secondary structure compared to the wild-type DNA polymerase, without altering the fidelity of the polymerase (39). The location and orientation of the OB-fold domain in the T. aquaticus Pol III structure suggest that the OB-domain may be able to undergo a domain rearrangement in order to bind downstream template DNA (9). Moreover, it has been shown that in the case of the E. coli Klenow fragment, four base pairs are unpaired to allow the terminal nucleotide of the primer to enter the exonuclease proofreading site (40). The results from previous studies leave open the possibility that the OB-fold domain of α may be involved in binding ssDNA generated during proofreading.
The characteristics of ssDNA binding by both the full-length protein and C-terminal binding fragment differ substantially from that of classical SSB proteins such as E. coli SSB (41), T4 gene 32 protein (gp32) (42), and T7 gene 2.5 protein (gp2.5) (29). In particular, all three of these SSB proteins are capable of actively destabilizing dsDNA. For example, both T4 gp32 and T7 gp2.5 lower the DNA melting force in real time as the DNA is being stretched, such that the observed melting transition force is lowered in the presence of the protein (28,29). This indicates that these SSB proteins are capable of binding to and stabilizing ssDNA that is created by transient thermal fluctuations that induce short-lived bubbles of melted DNA. In contrast, ssDNA binding by the α subunit occurs only after ssDNA has been fully melted by force. This unusual behavior may be functionally important, as ssDNA binding will likely only occur after other replication processes create ssDNA.
Methods
Optical tweezer experiments immobilize oppositely labeled phage λ-DNA between a pair of spheres; one is fixed upon a micropipette tip and another held in an optical trap, as shown in Figure 2(25,43–45). As the fixed end is moved, the increase in tension is measured by the deflection of the trapping lasers. Figure 3 shows this increase, due first to entropic and then enthalpic elasticity of dsDNA (46). The increase in force versus extension ends abruptly as DNA is observed to lengthen at nearly constant force, as dsDNA is converted to ssDNA in a force-induced melting transition (25,47). If the DNA molecules are allowed to relax, the bases are observed to reanneal and restack, though some hysteresis is observed. Both single-stranded and double-stranded forms of DNA have been characterized by polymeric models that describe the flexibility of the backbone for various solution conditions (see Supporting Information) (32,48). Standard experimental solutions incorporate 10 mM HEPES buffer (pH 7.5) and 100 mM Na+, unless noted. Additional experimental details may be found in Supporting Information.
Acknowledgments
This work was funded by NIH (GM75965) and NSF (MCB-0744456) to M.C.W. and a Camille and Henry Dreyfus Foundation New Faculty Award to P.J.B. Č.V. was supported in part by a Howard Hughes Medical Institute International Research Scholar grant. We thank M. Lamers and J. Kuriyan (U. C. Berkeley) for the plasmids expressing full-length α and α1−917. We gratefully acknowledge D. Barsky (Lawrence Livermore National Laboratory) for helpful discussions. We thank N. West for assistance.
Supporting Information Available
This material is available free of charge via the Internet.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Glover B. P.; McHenry C. S. (2001) The DNA polymerase III holoenzyme: an asymmetric dimeric replicative complex with leading and lagging strand polymerases. Cell 105, 925–934. [DOI] [PubMed] [Google Scholar]
- Kornberg A., and Baker T. A. (1992) DNA Replication, 2nd ed.,W. H. Freeman & Company, New York. [Google Scholar]
- McInerney P.; Johnson A.; Katz F.; O’Donnell M. (2007) Characterization of a triple DNA polymerase replisome. Mol. Cell 27, 527–538. [DOI] [PubMed] [Google Scholar]
- Johnson A.; O’Donnell M. (2005) Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315. [DOI] [PubMed] [Google Scholar]
- Rothwell P. J.; Waksman G. (2005) Structure and mechanism of DNA polymerases. Adv. Protein Chem. 71, 401–440. [DOI] [PubMed] [Google Scholar]
- Kelman Z.; O’Donnell M. (1995) DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64, 171–200. [DOI] [PubMed] [Google Scholar]
- Leipe D. D.; Aravind L.; Koonin E. V. (1999) Did DNA replication evolve twice independently?. Nucleic Acids Res. 27, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M. H.; Georgescu R. E.; Lee S. G.; O’Donnell M.; Kuriyan J. (2006) Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell 126, 881–892. [DOI] [PubMed] [Google Scholar]
- Bailey S.; Wing R. A.; Steitz T. A. (2006) The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126, 893–904. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E.; McHenry C. S. (1999) Identification of the acidic residues in the active site of DNA polymerase III. J. Mol. Biol. 285, 1067–1080. [DOI] [PubMed] [Google Scholar]
- Stano N. M.; Chen J.; McHenry C. S. (2006) A coproofreading Zn2+-dependent exonuclease within a bacterial replicase. Nat. Struct. Mol. Biol. 13, 458–459. [DOI] [PubMed] [Google Scholar]
- Reems J. A.; McHenry C. S. (1994) Escherichia coli DNA polymerase III holoenzyme footprints three helical turns of its primer. J. Biol. Chem. 269, 33091–33096. [PubMed] [Google Scholar]
- Doherty A. J.; Serpell L. C.; Ponting C. P. (1996) The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X.; Grishin N. V. (2000) Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 28, 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T.; Komori K.; Ishino Y.; Morikawa K. (2005) Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure 13, 1183–1192. [DOI] [PubMed] [Google Scholar]
- Zhao X.-Q.; Hu J.-F.; Yu J. (2006) Comparative analysis of eubacterial DNA polymerase III alpha subunits. Genomics Proteomics Bioinf. 4, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A. G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald D. L.; Mitton-Fry R. M.; Wuttke D. S. (2003) Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus V. (2002) OB-fold domains: a snapshot of the evolution of sequence, structure, and function. Curr. Opin. Struct. Biol. 12, 794–801. [DOI] [PubMed] [Google Scholar]
- Venclovas Č.; Zemla A.; Fidelis K.; Moult J. (2001) Comparison of performance in successive CASP experiments. Proteins: Struct., Funct., Genet. 45, 163–170. [DOI] [PubMed] [Google Scholar]
- Venclovas Č.; Margelevičius M. (2005) Comparative modeling in CASP6 using consensus approach to template selection, sequence-structure alignment, and structure assessment. Proteins: Struct., Funct., Genet. 61, 99–105. [DOI] [PubMed] [Google Scholar]
- Venclovas Č.; Ginalski K.; Kang C. (2004) Sequence-structure mapping errors in the PDB: OB-fold domains. Protein Sci. 13, 1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schaffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley M.; Hardwidge P. R.; Maher L. J. 3rd; Williams M. C. (2005) Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys. J. 89, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley M. J.; Williams M. C. (2007) Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers 85, 154–168. [DOI] [PubMed] [Google Scholar]
- Vladescu I.; McCauley M.; Nunez M. E.; Rouzina I.; Williams M. C. (2007) Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods 4, 517–522. [DOI] [PubMed] [Google Scholar]
- Pant K.; Karpel R. L.; Rouzina I.; Williams M. C. (2004) Mechanical measurement of single molecule binding rates: kinetics of DNA helix-destabilization by T4 gene 32 protein. J. Mol. Biol. 336, 851–870. [DOI] [PubMed] [Google Scholar]
- Pant K.; Karpel R. L.; Rouzina I.; Williams M. C. (2005) Salt dependent binding of T4 gene 32 protein to single- and double-stranded DNA: single molecule force spectroscopy measurements. J. Mol. Biol. 349, 317–330. [DOI] [PubMed] [Google Scholar]
- Shokri L.; Marintcheva B.; Richardson C. C.; Rouzina I.; Williams M. C. (2006) Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J. Biol. Chem. 281, 38689–38696. [DOI] [PubMed] [Google Scholar]
- Rouzina I.; Bloomfield V. A. (2001) Force-induced melting of the DNA double helix. 1. Thermodynamic analysis. Biophys. J. 80, 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri L., McCauley M. J., Rouzina I., and Williams M. C. (2008) DNA overstretching in the presence of glyoxal: structural evidence of force-induced melting, Biophys. J. Epub ahead of print. DOI:10.1529 /biophysj.1108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenner J. R.; Williams M. C.; Rouzina I.; Bloomfield V. A. (2002) Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 82, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu M.; Urbaneja M. A.; Hixson C. V.; Johnson D. G.; Datta S. A.; Fivash M. J.; Stephen A. G.; Fisher R. J.; Gorelick R. J.; Casas-Finet J. R.; Rein A.; Rouzina I.; Williams M. C. (2006) Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E.; Belegu V.; Korolev S.; Bochkarev A. (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M. R.; Scaife S.; Wigley D. B. (2001) Structural analysis of DNA replication fork reversal by RecG. Cell 107, 79–89. [DOI] [PubMed] [Google Scholar]
- Shamoo Y.; Friedman A. M.; Parsons M. R.; Konigsberg W. H.; Steitz T. A. (1995) Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature 376, 362–366. [DOI] [PubMed] [Google Scholar]
- McCauley M. J.; Zimmerman J.; Maher L. J. 3rd; Williams M. C. (2007) HMGB binding to DNA: single and double box motifs. J. Mol. Biol. 374, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D.; von Hippel P. H. (1974) Theoretical aspects of DNA-protein interactions: cooperative and non-cooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86, 469–489. [DOI] [PubMed] [Google Scholar]
- Sun S.; Geng L.; Shamoo Y. (2006) Structure and enzymatic properties of a chimeric bacteriophage RB69 DNA polymerase and single-stranded DNA binding protein with increased processivity. Proteins: Struct., Funct., Bioinf. 65, 231–238. [DOI] [PubMed] [Google Scholar]
- Freemont P. S.; Friedman J. M.; Beese L. S.; Sanderson M. R.; Steitz T. A. (1988) Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc. Natl. Acad. Sci. U.S.A. 85, 8924–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T. M.; Ferrari M. E. (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570. [DOI] [PubMed] [Google Scholar]
- Karpel R. L. (1990) T4 Bacteriophage Gene 32 Protein, CRC Press, Boca Raton. [Google Scholar]
- Smith S. B.; Cui Y.; Bustamante C. (2003) Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 361, 134–162. [DOI] [PubMed] [Google Scholar]
- Williams M. C.; Rouzina I. (2002) Force spectroscopy of single DNA and RNA molecules. Curr. Opin. Struct. Biol. 12, 330–336. [DOI] [PubMed] [Google Scholar]
- Wang M. D.; Yin H.; Landick R.; Gelles J.; Block S. M. (1997) Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F.; Siggia E. D. (1995) Stretching DNA. Macromolecules 28, 8759–8770. [Google Scholar]
- Williams M. C.; Rouzina I.; Bloomfield V. A. (2002) Thermodynamics of DNA interactions from single molecule stretching experiments. Acc. Chem. Res. 35, 159–166. [DOI] [PubMed] [Google Scholar]
- Smith S. B.; Cui Y. J.; Bustamante C. (1996) Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.










