Abstract
The killer cell immunoglobulin-like receptor (KIR) repertoire of natural killer (NK) cells determines their ability to detect infected or transformed target cells. Although epigenetic mechanisms play a role in KIR gene expression, work in the mouse suggests that other regulatory elements may be involved at specific stages of NK-cell development. Here we report the effects of the transcription factor c-Myc on KIR expression. c-Myc directly binds to, and promotes transcription from, a distal element identified upstream of most KIR genes. Binding of endogenous c-Myc to the distal promoter element is significantly enhanced upon interleukin-15 (IL-15) stimulation in peripheral blood NK cells and correlates with an increase in KIR transcription. In addition, the overexpression of c-Myc during NK-cell development promotes transcription from the distal promoter element and contributes to the overall transcription of multiple KIR genes. Our data demonstrate the significance of the 5′ promoter element upstream of the conventional KIR promoter region and support a model whereby IL-15 stimulates c-Myc binding at the distal KIR promoter during NK-cell development to promote KIR transcription. This finding provides a direct link between NK-cell activation signals and KIR expression required for acquisition of effector function during NK-cell education.
Introduction
Killer immunoglobulin-like receptors (KIR) constitute a polymorphic gene family containing 15 genes and 2 pseudogenes located on chromosome 19q13.4. Although inhibitory KIR recognize human leukocyte antigen (HLA) class I molecules, the natural ligands for activating KIR are less clear, even though some activating KIR fusion proteins bind class I with low affinity.1 Despite their divergent function, both types of KIR are expressed in a variegated manner on the surface of natural killer (NK) cells and distinct subsets of T cells.2 Because NK cells can be triggered by either down-regulation of HLA molecules or the induction of stress-related molecules on the surface of tumor targets, NK cell–based strategies hold promise for the successful treatment of both hematopoietic and solid tumors.3 Genetic studies have also shown that particular combinations of KIR and their HLA ligands can impact the course of HIV-1 and hepatitis C virus (HCV) infections.4,5 Therefore, an elucidation of the factors that influence KIR gene transcription and a more thorough understanding of how KIR signaling affects NK-cell development are needed to understand how to manipulate the innate immune system for therapeutic purposes
Progress in the elucidation of how KIR genes are regulated has been limited because of the complexity of the KIR gene locus and the fact that KIR genes are not present in model rodent species, which are amenable to genomic manipulation. The conventional 250-bp core promoter located in the 5′ region just proximal to the translational start site has been characterized in detail for many KIR genes.6–8 However, an entire 2-kb intergenic region exists upstream of the translational start site for each KIR gene, with the exception of KIR2DL4, which has a 14-kb upstream intergenic region.9 A recent report has identified the presence of active distal KIR promoter elements and spliced transcripts originating from these elements.10
Because the distal promoter contains a Myc-binding site,10 we hypothesized that c-Myc can bind to the distal promoter element and directly affect KIR expression. c-Myc is a basic helix-loop-helix leucine zipper transcription factor that binds E-box DNA motifs as a heterodimer with Max, resulting in transcriptional activation or silencing of target genes.11–13 Many major cellular processes, including cell cycle entry,14 proliferation,15 cell size regulation,16 and apoptosis,17 are influenced by c-Myc.18,19
c-Myc is particularly interesting in the context of KIR transcriptional regulation because c-Myc functions as a downstream component of the interleukin-15 (IL-15) signaling pathway during CD8+ T-cell homeostasis,20 and the IL-15 pathway is critical for NK-cell maturation,21 activation on infection in the periphery,22 and homeostasis.23 In the present study, we demonstrate a direct, functional interaction between c-Myc induced by IL-15 and the distal KIR promoter element and show that full-length KIR transcripts are transcribed from the distal promoter element early during development of the NK-cell KIR repertoire.
Methods
Electric mobility shift assay of c-Myc binding to the distal KIR promoter element
Nuclear extracts were prepared from YT-Indy cells using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich, St Louis, MO). Protein concentration was measured with a Bio-Rad protein assay (Hercules, CA), and samples were stored at −70°C until use. Six double-stranded DNA oligonucleotide probes corresponding to the predicted c-Myc–binding sequence of the distal KIR promoter alleles were synthesized (Figure 1A, sense strand shown). Sense and antisense oligonucleotides were annealed to generate double-stranded oligonucleotides and labeled with [α-32P]deoxycytidine triphosphate (3000 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) by fill-in using the Klenow fragment of DNA polymerase I (Invitrogen, Carlsbad, CA). 32P-labeled double-stranded oligonucleotides were purified using mini Quick Spin Oligo Columns (Roche Diagnostics, Mannheim, Germany). DNA-protein binding reactions were performed in a 10-μL mixture containing 5 μg nuclear protein and 1 μg poly(dI-dC)poly(dI-dC) (Sigma-Aldrich) in 4% glycerol, 1 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5). After a 10-minute incubation on ice, samples were incubated with 1 μL 32P-labeled oligonucleotide probe (10 000 cpm) at room temperature for 20 minutes and then loaded on a 5% polyacrylamide gel (37:5:1). Electrophoresis was performed in 0.5× Tris borate ethylenediaminetetraacetic acid buffer for 2 hours at 130 V, and the gel was visualized by autoradiography after 2 days exposure at −70°C. For inhibition of complex formation by antibody, nuclear extracts were incubated with 1 μL antibody for 30 minutes on ice before the addition of 32P-labeled DNA probe. After addition of the labeled DNA probe, the binding reaction was incubated for an additional 20 minutes at room temperature. The antibodies used were Myc (9E11; Abcam, Cambridge, MA), Max (H-2; Santa Cruz Biotechnology, Santa Cruz, CA), CREB (24HB4; Santa Cruz Biotechnology), and YY1 (C-20; Santa Cruz Biotechnology).
Figure 1.
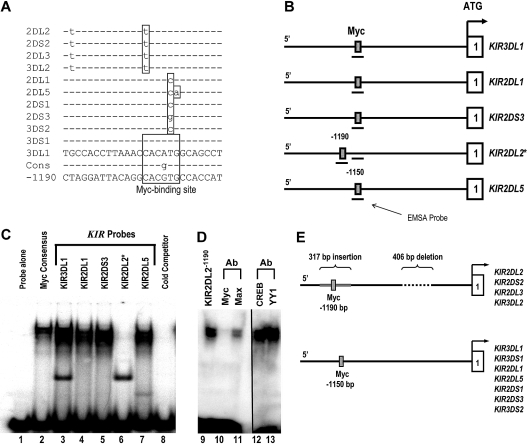
c-Myc binds to the distal KIR promoter element in NK cells. (A) The intergenic region preceding each of the KIR genes on chromosome 19 was analyzed for transcription factor binding sites. The sequence of the distal promoter element, −1150 bp upstream of the translational start site, was aligned for each KIR gene, and a predicted Myc-binding site was identified. The sequence of the putative Myc-binding region of the KIR3DL1 gene is shown, and polymorphisms found in the other genes are highlighted, as well as the consensus Myc probe used for EMSA analysis (Cons). The sequence of the distal promoter element −1190 bp upstream of the translational start site for the KIR2DL2/2DS2/2DL3/3DL2 is also shown. (B) The location of the EMSA probes spanning the Myc-binding sequence 1150 bp upstream of selected KIR translational start sites. The location of the EMSA probe spanning the consensus Myc site 1190 bp upstream of the KIR2DL2 translational start site is also shown. (C) EMSA analysis of the predicted Myc-binding sites. Probes corresponding to the nucleotide sequences shown in panel A were used for an EMSA with YT cell extracts. A Myc consensus probe alone control is shown in lane 1. Complexes formed by individual 32P-labeled KIR probes are shown in lanes 2 to 7. A cold competitor control for probe specificity is shown in lane 8. (D) Inhibition of the complex formed by the KIR2DL2−1190 probe with anti-c-Myc and anti-Max antibodies is shown in lanes 10 and 11. The vertical line has been inserted between lanes 11 and 12 to indicate a repositioned gel lane. Inhibition of the complex formed by the KIR2DL2−1190 probe with anti-CREB and anti-YY1 control antibodies is shown in lanes 12 and 13. (E) Because of a 406-bp deletion and a 317-bp Alu insertion in the intergenic region of KIR2DL2/2DS2/2DL3/3DL2, a Myc-binding consensus is located at position −1190 relative to the transcriptional start site instead of −1150, where the site is located for the rest of the KIR genes.
Cell lines
NK92 cells were cultured at 37°C with 5% CO2 in alpha medium containing 12.5% fetal calf serum, 12.5% horse serum (HyClone Laboratories, Logan, UT), 0.2 mM inositol, 0.1 mM β-mercaptoethanol, 0.02 mM folic acid (Sigma-Aldrich), 100 U/mL penicillin, 100 U/mL streptomycin (Invitrogen), and 500 U/mL recombinant human IL-2 (Chiron, Emeryville, CA). NKL cells were cultured 37°C with 5% CO2 in RPMI media supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin (Invitrogen), and 200 U/mL recombinant human IL-2 (Chiron).
Generation of luciferase reporter constructs
The full-length KIR3DL1 and KIR2DL3 promoter and the distal KIR3DL1 promoter element were amplified from NK92 genomic DNA using the following primers: KIR3DL1 full promoter sense: 5′-AGTCGAGCTCTAGTGTGAGAATACGTTTAGATATAT, KIR3DL1 full promoter antisense: 5′-TCAGCTCGAGGGTGCTGCCGGTGCAGACAG, KIR3DL1 distal promoter sense: 5′-CATTGAGCTCACGAATAGTGAGGGATGACTGTA, KIR3DL1distal promoter antisense: 5′-GGTTCCTCGAGATACAAAAATTAGCCATGCCTG, KIR2DL3 full promoter sense: 5′-CACCAGGAGGATGTGCATGGGTTCTA, and KIR2DL3 full promoter antisense: 5′-CTGACGACCATGAGCGACAT. Polymerase chain reaction (PCR) fragments with the distal Myc site deletion were created using a PCR-bridging strategy. PCR products for the KIR3DL1 promoter were digested with XhoI (New England Biolabs, Ipswich, MA) and SstI (Invitrogen) and cloned into the pGL3-basic firefly luciferase reporter vector (Promega, Madison, WI). PCR products for the KIR2DL3 promoter were cloned into the pGL3 vector using the Invitrogen Gateway Cloning System (Invitrogen).
Cell transfection and luciferase assays
The NKL cell line was used for all transfection experiments. Cells were electroporated with 10 μg pGL3 constructs plus 100 ng Renilla luciferase pRL-SV40 vector using the Amaxa Nucleofector Kit V (Amaxa Biosystems, Gaithersburg, MD) according to a published method for the NKL line.24 Luciferase activity was assayed at 6 hours using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized relative to the Renilla luciferase activity for each transfection.
RT-PCR for full-length KIR2DL1, -2DL2, and -2DL3 transcripts
Umbilical cord blood mononuclear cells were depleted of CD3- and CD14-positive cells by microbead labeling and magnetic column separation (Miltenyi Biotec, Auburn, CA) and stained with allophycocyanin (APC)–conjugated NCAM16.2 (CD56), phycoerythrin (PE)–conjugated CD7, and fluorescein isothiocyanate (FITC)–conjugated CD34 (BD Biosciences, San Jose, CA). Cells were sorted into NK-cell precursor populations based on their CD34, CD7, and CD56 expression on a FACS DiVa. cDNA was synthesized and used for reverse-transcribed PCR (RT-PCR) to amplify full-length transcripts using Advantage II DNA Polymerase (Clontech, Mountain View, CA). The following cycling conditions were used: 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 60 seconds, and 60°C for 60 seconds for 40 cycles. Primers used were: distal sense: 5′-TGATGTGGTCAACATGTAAACTG, 2DL1/2/3 antisense: 5′-CATGGGCAGGAGACAACTT, GAPDH sense: 5′-GAGTCAACGGATTTGGTCGT, and GAPDH antisense: 5′TTGATTTTGGAGGGACTCCG.
Isolation of adult peripheral blood NK cells
Adult peripheral blood was collected from consenting adults at the Memorial Blood Center (Minneapolis, MN), and mononuclear cells were isolated by centrifugation using a Histopaque gradient (Sigma-Aldrich). NK cells were negatively selected using the magnetic-activated cell sorting (MACS) NK Cell Isolation Kit as per the manufacturer's protocol (Miltenyi Biotec). The purified population of NK cells was then stained with a cocktail of PE-conjugated DX9, EB6, GL183, and FES172 monoclonal antibodies and subsequently stained with anti-PE Microbeads (Miltenyi Biotec). KIR− NK cells were then isolated by negative selection by magnetic MACS separation. Cells were incubated with 10 ng/mL IL-15 for 4 to 48 hours before analysis.
Chromatin immunoprecipitation assay
For fresh or IL-15–stimulated adult peripheral blood KIR-negative cells, chromatin immunoprecipitation (ChIP) was performed with the EZ-ChIP Kit (Millipore, Billerica, MA). Formaldehyde cross-linked chromatin was immunoprecipitated with 2 μL rabbit antisera against c-Myc, USF-1 (Santa Cruz Biotechnology), histone H3, or purified rabbit Ig (Millipore). PCR (30-35 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 73°C for 1 second) was performed with primers specific for the KIR distal promoter region (5′-sense: GAGAAGACATTCTATGCCACCTTAAAC and 3′-antisense: AATACATCCGTGTACACACAGTC) resulting in an amplified fragment of 79 bp.
Isolation of progenitor cells from umbilical cord blood
The use of all human tissue was approved by the Committee on the Use of Human Subjects in Research at the University of Minnesota, and informed consent was obtained in accordance with the Declaration of Helsinki. Umbilical cord blood was obtained from full-term consenting mothers from the Memorial Blood Bank (Minneapolis, MN), Placental Blood Program of the New York Blood Center (New York, NY), St Louis Cord Blood Bank (St Louis, MO), or local obstetric units. Mononuclear cells were isolated using Histopaque (Sigma-Aldrich) density gradient centrifugation. CD34+ cells were then obtained by staining the mononuclear fraction with APC-conjugated anti-CD34 (BD Biosciences). The stained fraction was purified using the MACS magnetic bead selection system (Miltenyi Biotec).
Retroviral vectors and transduction
The full-length human c-Myc cDNA (provided by Robert Eisenman, Fred Hutchinson Cancer Research Center, Seattle, WA) was cloned into the murine stem cell virus (MSCV) enhanced green fluorescent protein (eGFP) vector upstream of the internal ribosomal entry sequence using EcoRI sites.
Isolated CD34+ cells were preactivated for 72 hours with Iscove medium supplemented with 20% fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin, and 20 ng/mL each of IL-7, c-kit ligand, Flt3 ligand, and thrombopoietin. After stimulation, 2 × 105 cells were placed in 6-well tissue-culture–treated transwells (0.4 μm pore size) coated with 100 μg of the recombinant CH-296 fibronectin fragment (Takara, Kyoto, Japan). c-Myc- or eGFP-containing viral supernatant was passed through the transwells twice in 48 hours. Two days after the last viral exposure, cells were harvested and stained with APC-conjugated CD34. CD34+ eGFP+ cells were selected using the fluorescence-activated cell sorter (FACS) Aria (BD Biosciences). eGFP+ cells were then cultured on the murine embryonic liver cell line EL08-1D2.25 Culture media consisted of a 2:1 (vol:vol) mix of Dulbecco modified Eagle medium (high glucose with sodium pyruvate)/Ham F12–based medium and supplemented with 24 μM 2-mercaptoethanol, 50 μM ethanolamine, 20 mg/L ascorbic acid, 50 μg/L sodium selenite, 100 U/mL penicillin, 100 U/mL streptomycin, and 20% heat-inactivated human AB serum in the presence of 10 ng/mL IL-15, 5 ng/mL IL-3, 20 ng/mL IL-7, 20 ng/mL c-kit ligand, and 10 ng/mL Flt3 ligand. Cultures were initiated with either 10 or 50 cells per well of a 96-well plate or 50 cells per well of a 24-well plate.
Flow cytometry and analysis
Phenotypic acquisition of cells was performed on the FACSCalibur (BD Biosciences) using CellQuest Pro Software (BD Biosciences). Cells were stained with the following monoclonal antibodies: APC-conjugated NCAM16.2 (CD56), PE-conjugated DX9 (anti-CD158e), EB6 (anti-CD158a/h), GL183 (CD158b/j), and FES172 (anti-CD158i) (BD Biosciences). Analysis was performed using FlowJo software (TreeStar, Ashland, OR).
Isolation of RNA and real-time quantitative PCR
Total RNA was extracted from cells using the RNeasy Mini Kit (QIAGEN, Valencia, CA), and genomic DNA was isolated using RNase-free DNase (Invitrogen). Quantitative RT-PCR was performed as previously described26 to quantify KIR gene expression. To detect transcripts originating from the distal promoter, primers were modified to include a sense primer approximately 100 bp upstream of the proximal promoter and an antisense primer located in the third exon of the indicated KIR gene. Additional primers used were: MYC-Applied Biosystems Hs99999003_m1, distal2D sense: 5′-TGATGTGGTCAACATGTAAACTG, antisense: 5′-AGGAGGGAAGGTTTTCTGTGGA, probe: 5′-ACTCCCTCATGTGGCCAG, distal2DL3 sense: 5′-TGATGTGGTCAACATGTAAACTG, antisense: 5′-AGGAGGGAAGGTTTTCTGTGGA, probe: 5′-CCAACACACACCATGCTGACGACCA, distal3DL1 sense: 5′-TGATGTGGTCAACATGTAAACTG, antisense: 5′-AGTGACACCGAAGAGTCACGTGTC, probe: 5′-TCCCTGTCTGCCTGC.
Results
The transcription factor c-Myc binds to the distal promoter element of multiple KIR genes
To investigate the functional significance of the distal promoter element, the 5′ intergenic region preceding each KIR gene was scanned for potential transcription factor–binding sites using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html). A putative Myc-binding site was identified within a L1 repeat region approximately 1.1 kb upstream of the classic transcriptional start site for 11 separate KIR genes (Figure 1A). Whereas the KIR3DL1 and KIR3DS1 promoters contain Myc sites that match the consensus sequence completely, the other 9 KIR genes in the alignment have polymorphisms inside of the Myc-binding region.
To test whether each of these polymorphic sites is still capable of binding Myc, we designed electric mobility shift assay (EMSA) probes for the Myc consensus sequence, KIR3DL1 (A-to-G change), KIR2DL1 (T-to-C change), KIR2DS3 (T-to-G change), KIR2DL2 (C-to-T change), and KIR2DL5 (TG-to-CA change). The position of each probe relative to the proximal transcriptional start site of each KIR gene is shown in Figure 1B. With the exception of KIR2DL2, each KIR probe bound Myc as evidenced by the gel shift for KIR3DL1, KIR2DL1, KIR2DS3, and KIR2DL5 (Figure 1C lanes 3-7). The fact that the KIR2DL2 probe did not bind Myc implies that the C-to-T change relative to the consensus abrogates binding at that site for KIR2DL2, -2DS2, -2DL3, and -3DL2. This lack of binding prompted us to look more closely at this particular set of KIR promoters. On further evaluation, we found that, because of a 406-bp deletion and a 317-bp insertion in the intergenic region compared with the other KIR genes, a perfect Myc consensus site exists within an Alu repeat region 1190 bp upstream of the transcriptional start site for KIR2DL2, -2DS2, -2DL3, and -3DL2 (Figure 1A,E).
To confirm that this site is capable of binding c-Myc, we designed a new EMSA probe (KIR2DL2−1190) spanning the Myc site within the Alu repeat (Figure 1B). We observed c-Myc binding to the KIR2DL2−1190 probe as expected (Figure 1D lane 9). Blocking antibodies against c-Myc and its binding partner, Max, were tested to ensure specificity of the assay. The addition of anti–c-Myc or anti-Max blocking antibodies resulted in a significant decrease in the KIR2DL2−1190 probe shift (Figure 1D lanes 10,11). Control blocking antibodies against the transcription factors CREB and YY1 did not interfere with the probe shift (lanes 12 and 13).
c-Myc drives transcription from the KIR3DL1 promoter through a direct interaction with the Myc site in the distal promoter element
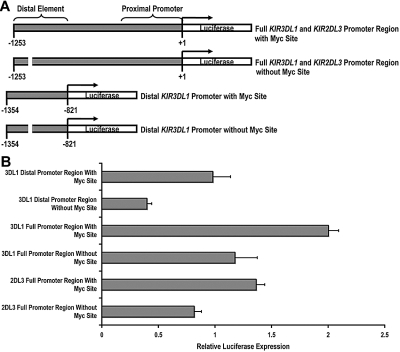
Having established that c-Myc is able to bind to the KIR distal promoter element, we next wanted to determine whether c-Myc could directly promote KIR transcription. We cloned a 1253-bp fragment of the KIR3DL1 promoter, which we refer to as the “full” KIR3DL1 promoter region and a 433-bp fragment, which we refer to as the “distal” KIR3DL1 promoter region. We then used a bridging PCR strategy to specifically eliminate the distal Myc-binding site from each fragment (Figure 2A). The same strategy was used to clone the KIR2DL3 full promoter region. Each intact and mutant fragment was tested using a dual luciferase assay for transcriptional activity in the NKL-cell line. The transcriptional activities of both the distal promoter region alone and the full KIR3DL1 promoter region were significantly decreased by the elimination of the Myc-binding site, demonstrating the direct contribution of c-Myc in enhancing KIR3DL1 promoter activity. The full KIR2DL3 promoter luciferase activity was similarly reduced with the elimination of the Myc site (Figure 2B).
Figure 2.
c-Myc acts in a direct and specific manner at the distal promoter element to influence KIR transcription. (A) Schematics of the full-length and distal promoter fragments with and without the Myc site that were cloned into the pGL3 basic reporter vector. (B) Each pGL3 vector containing full and distal promoter sequences was electroporated into the NKL-cell line, and luciferase expression was determined 6 hours after transfection. Results represent the mean luciferase levels normalized to Renilla signals from 3 independent experiments.
Transcription from the distal promoter element occurs in NK-cell precursors
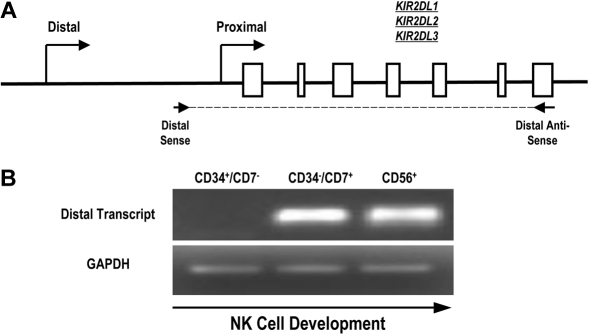
To determine whether the distal promoter element is active during normal human NK-cell development, we designed primers to specifically detect KIR transcripts originating upstream of the classic proximal promoter. Because of the extensive homology of the KIR genes and promoter regions, we were unable to select primers specific for single KIR genes. Therefore, we designed primers for RT-PCR that amplified a region initiating upstream of the proximal promoter and extending to the stop codon of KIR2DL1, KIR2DL2, and KIR2DL3 (Figure 3A). Mononuclear cells from umbilical cord blood of healthy donors were depleted of CD3+ and CD14+ cells to eliminate thymocytes and monocytes and sorted to obtain populations of uncommitted lymphoid progenitor cells (CD34+CD7−), committed NK-cell precursors (CD34−CD7+), and fully committed NK cells (CD56+). We have previously shown that the CD34−CD7+ cord blood fraction is highly enriched for NK-cell progenitors.27 Transcripts were absent from the uncommitted CD34+CD7− population but were present in the CD34−CD7+ NK-cell precursor population (Figure 3B), which is consistent with a previous analysis of KIR expression during NK-cell development.28 The early timing of KIR transcription from the distal promoter element suggests that the distal promoter element is active at the initiation of KIR expression during human NK-cell development.
Figure 3.
Full-length KIR transcripts originate from the distal promoter region during NK-cell development. (A) PCR primers were designed to amplify full-length KIR2DL1, -2DL2, and -2DL3 transcripts originating from the distal promoter region. The amplified region begins upstream of the classical transcriptional start site (distal sense) and extends to the stop codon (distal antisense). Open boxes represent KIR gene exons. (B) Mononuclear cells from umbilical cord blood were sorted for specific NK-cell precursor populations based on the expression of the developmental markers CD34, CD7, and CD56. Sorted cells were analyzed for the presence of distal transcripts by RT-PCR using the primers shown in panel A. Control PCRs were carried out using GAPDH primers to confirm the presence of cDNA.
IL-15 drives KIR expression and induces c-Myc binding to the distal KIR promoter element in peripheral blood NK cells
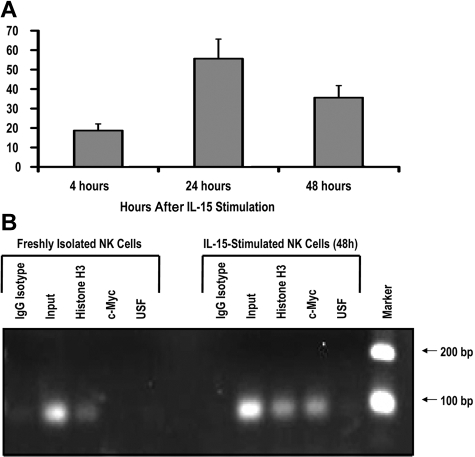
To test whether c-Myc is induced by IL-15 signaling in human NK cells, we isolated CD56+ NK cells from the peripheral blood of healthy donors and stimulated these cells with 10 ng/mL exogenous recombinant human IL-15. After 24 hours of IL-15 stimulation, c-myc transcript levels were increased approximately 50-fold and began to decline by 48 hours (Figure 4A), suggesting that IL-15 is a rapid, potent stimulator of c-myc transcription in NK cells. IL-2 stimulation resulted in a similar increase in c-myc transcript levels (data not shown).
Figure 4.
IL-15 increases c-Myc transcription and c-Myc binding to the KIR distal promoter element. (A) KIR-negative NK cells were isolated from adult peripheral blood and cultured in the presence of 10 ng/mL IL-15 for 4, 24, or 48 hours. Cells were harvested at each time point, and c-myc transcript levels were determined by quantitative RT-PCR (n = 3). Samples are normalized to unstimulated peripheral blood NK-cell controls. (B) ChIP analysis of c-Myc binding to freshly isolated KIR-negative NK cells or NK cells stimulated with 10 ng/mL IL-15 for 48 hours. Input sample represents the total input DNA contained in the chromatin aliquot used for each ChIP. Rabbit antibodies used for the IP were purified rabbit IgG (as a negative control), histone H3 (as a positive control), anti-Myc, and anti-USF. Results from a 35-cycle PCR are shown.
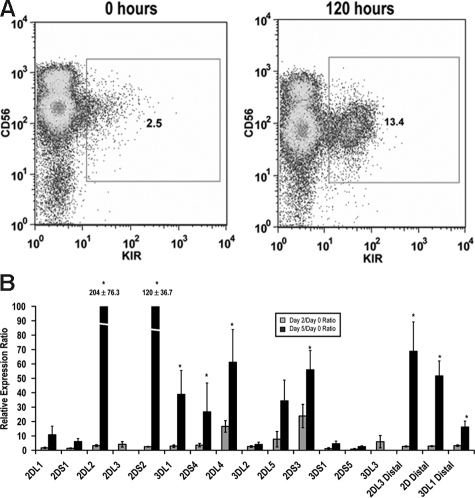
Although it is known that stimulation with either IL-15 or IL-2 can enhance KIR expression,29,30 a direct link between stimulation and transcription factor binding within the KIR promoter is lacking. Therefore, we isolated KIR−CD56+ NK cells from peripheral blood and stimulated these cells ex vivo with IL-15. KIR expression was rapidly induced in KIR− cells as measured by both surface staining with monoclonal antibodies and by quantitative RT-PCR (Figure 5). Importantly, distal transcripts for KIR2DL3, KIRs 2DL1/2DL2/2DL3/2DS1/2DS2 (2D distal), and KIR3DL1 were also strongly induced by IL-15 stimulation (Figure 5B).
Figure 5.
IL-15 induces KIR expression in KIR-negative NK cells. KIR-negative NK cells were isolated from adult peripheral blood and stimulated with 10 ng/mL IL-15 for 48 to 120 hours. (A) FACS analysis was performed at baseline before culture and 120 hours after IL-15 stimulation staining with a cocktail of APC-conjugated NCAM16.2 and PE-conjugated DX9, EB6, GL183, and FES172 monoclonal antibodies (n = 9). (B) Proximal coding KIR transcript and distal KIR transcript levels were measured after 48 and 120 hours by quantitative RT-PCR. Values are presented as a ratio of gene expression after 2 and 5 days compared with gene expression at the initial purification (n = 9). Error bars represent the SEM. *P < .05.
To test the hypothesis that IL-15 stimulation induces c-Myc binding at the distal promoter element, we performed a ChIP assay with freshly isolated peripheral blood NK cells and cells that were stimulated ex vivo with IL-15 for 48 hours. The primers used in the ChIP assay perfectly match the KIR3DL1 and KIR2DL1 distal promoters, and sequencing of PCR products showed that KIR3DL1 was the predominant product. Thus, most of the enrichment in this assay is coming from the KIR3DL1 distal promoter. c-Myc binding to the KIR distal promoter element was approximately 48-fold higher in NK cells stimulated with IL-15 compared with unstimulated controls (Figure 4B). Therefore, the induction of c-myc transcription by IL-15 correlates with binding of the transcription factor to the KIR distal promoter element.
c-Myc overexpression promotes KIR acquisition in developing NK cells
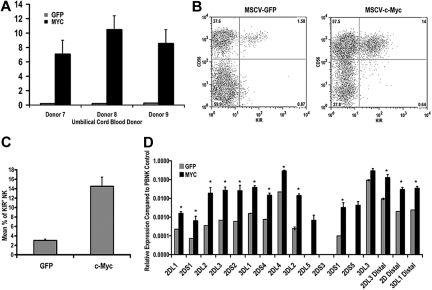
Because the signaling events that occur downstream of IL-15 binding to its receptor complex are multifarious,31 we wanted to look specifically at the ability of c-Myc to promote KIR expression in NK cells. To this end, we transduced CD34+ hematopoietic precursor cells with either eGFP or c-Myc retroviral constructs and differentiated these cells in vitro for 21 days. Overexpression of c-myc transcript after 21 days in culture was confirmed by quantitative RT-PCR (Figure 6A). Cells from c-Myc–transduced and control cultures were analyzed by flow cytometry for KIR expression. c-Myc overexpression enhanced NK-cell maturation, as measured by the percentage of CD56+ cells in culture, as well as the percentage of KIR+ NK cells (14.5% ± 1.95% vs 3.01% ± 0.29%, n = 28, P = .003; Figure 6B). We also performed quantitative RT-PCR using cells from day 21 cultures and observed a statistically significant increase in the expression of variegated KIR transcripts and transcripts originating from the distal promoter element (Figure 6C,D).
Figure 6.
c-Myc overexpression leads to an increase in KIR expression during NK-cell development. CD34+ cells were isolated from umbilical cord blood and transduced with MSCV retroviral constructs containing egfp or c-myc. These cells were cultured on the EL08.1D2 cell line in the presence of exogenous cytokines. After 21 days, cultured cells were harvested for (A) quantitative RT-PCR to determine c-myc transcript levels. All values are normalized to GAPDH (n = 5). Error bars represent the SEM for each group of samples. *P < .05. (B) These cells were also immunophenotyped with APC-conjugated NCAM16.2 and PE-conjugated DX9, EB6, GL183, and FES172 monoclonal antibodies. The FACS plots in panel B are representative examples of cells harvested from day 21 cultures. (C) The percentage of KIR+ NK cells in eGFP- and c-Myc–transduced cultures was determined by FACS analysis (n = 28). (D) RNA was harvested from day 21 cultures and used for quantitative RT-PCR using primers designed to amplify coding KIR transcripts and to detect transcripts originating from the distal promoter element for KIR2DL3, KIR2DL1/2DL2/2DL3/2DS1/2DS2 (2D distal), and KIR3DL1 (n = 8). Expression levels were normalized to an IL-2–activated peripheral blood NK population known to express all KIR genes. Error bars represent the SEM for each group of samples. *P < .05.
c-Myc overexpression leads to de novo KIR acquisition in the NK92 cell line
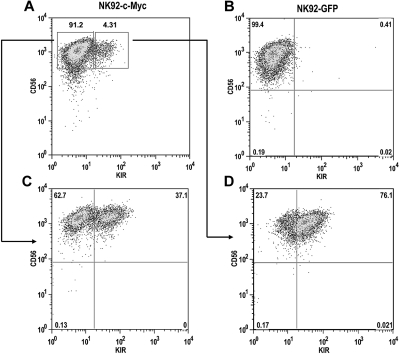
The NK92 cell line does not express most KIR, with the exception of KIR2DL4, resulting from extensive CpG DNA methylation within the KIR promoters.32 To determine whether c-Myc expression can induce de novo KIR expression in this line, we transduced NK92 cells with either eGFP or c-Myc retroviral constructs. After a period of 4 weeks, c-Myc–transduced NK92 cells began to express KIR, whereas GFP-transduced cells remained KIR-negative (Figure 7A,B). KIR-positive and KIR-negative cells from c-Myc–transduced cultures were sorted with more than 98% purity and placed back into culture. After 8 weeks, approximately one-third of the cells in the KIR-negative culture acquired KIR (Figure 7C). Nearly two-thirds of the cells in the KIR-positive culture retained KIR expression, suggesting that c-Myc can stably maintain KIR expression in NK92 cells (Figure 7D).
Figure 7.
c-Myc induces de novo KIR expression in the NK92 cell line. NK92 cells were transduced with either (A) MSCV–c-myc or (B) MSCV-egfp vectors and cultured for a period of 4 weeks. (C) KIR-negative and (D) KIR-positive cells from c-Myc–transduced cultures were then sorted by flow cytometry into separate cultures and phenotyped for CD56 and KIR expression 8 weeks later.
Discussion
We have shown that KIR transcription can originate from an upstream regulatory region in the noncoding sequence of most KIR genes. Despite some polymorphism at this Myc site, transcription is activated by direct Myc binding. The physiologic mechanism is triggered through IL-15, providing an important link between signals required for NK-cell development and KIR acquisition. These mechanisms are operant in NK-cell lines, primary blood NK cells, and NK cells derived from early progenitors. This provides definitive evidence that IL-15 is not only important for development and homeostatic expansion21,23 but also for generation of the NK-cell repertoire.
It is well established that the DNA methylation status of CpG islands within the promoter region proximal to the translational start site is predictive of KIR gene expression. For those KIR alleles that are expressed in a variegated fashion, promoter hypomethylation is strongly correlated with active transcription, whereas those alleles that are hypermethylated are silent.32 DNA methylation may inhibit KIR gene expression by blocking access of transcription factors that are necessary for the initiation of transcription.33 However, the regulatory elements directly responsible for the induction of KIR expression have not previously been determined.
Our study was based on the description of a novel distal promoter element, referred to as Pro 1, first identified upstream of the previously studied Ly49g promoter in murine NK cells.34 The Ly49 family of MHC class I receptors are expressed in a variegated fashion and are the functional analogs of KIR.35 The Pro 1 element is expressed only in immature murine NK cells and has bidirectional activity resulting from the presence of overlapping, divergent promoters. The direction of transcription from Pro 1 is determined by competitive interactions between transcription factors, such as nuclear factor-κB, CCAAT/enhancer-binding protein (C/EBP), and TATA-binding protein binding to forward or reverse TATA and C/EBP elements.36
The mechanism of KIR gene transcription is distinct from that of the Ly49 genes. The KIR distal promoter element does not have bidirectional activity,10 and as shown here, is responsive to direct c-Myc binding rather than nuclear factor-κB and C/EBP as seen in the mouse. The Myc sites identified 1150 bp upstream of the translational start sites of the KIR3DL1/3DS1/2DL1/2DL5/2DS1/2DS3/3DS2 genes are located within an L1 repeat, which is a non-LTR retrotransposon of the long interspersed element family. The Myc sites identified 1190 bp upstream of the translational start sites of the KIR2DL2/2DS2/2DL3/3DL2 are located within a 317-bp Alu insertion (Figure 1E). The presence of L1s and Alus within the genome allows for DNA mispairing and unequal crossing over, which can lead to the deletion or duplication of sequences between the repeats.37 A significant percentage of L1 retrotranspositions are also involved in exon shuffling and the swapping of regulatory sequences via 3′ transductions.38 This is particularly interesting in the context of the evolutionary history of the KIR genes in light of a recent analysis suggesting that unequal crossing over is responsible for expansion and contraction within the KIR locus.39
We show that full-length KIR transcripts originate from the distal promoter element in committed NK-cell precursors, and there appears to be a 1:1 ratio between the levels of transcription from the distal promoter element and total KIR transcript levels. In mature NK cells, there is approximately 5-fold more proximal transcript resulting from the higher activity of the proximal promoter. Thus, the KIR distal promoter element seems to act either independently or synchronously with the proximal promoter to promote forward transcription within individual KIR genes.
We detected transcripts originating from the KIR distal promoter in a NK-cell progenitor-enriched CD34−CD7+ population isolated from umbilical cord blood (Figure 3). Because KIR are not detected on the surface of NK cells before commitment to the NK-cell lineage, as defined by CD56 expression, we suggest that transcription is initiated at low levels from the distal promoter in NK-cell progenitors, but translation does not take place until a later stage of development. The same phenomenon has been described for IL-2Rβ (CD122) expression in human NK-cell development where low levels of CD122 transcript can be detected in progenitors, but protein cannot be detected on the surface of cells until cells are fully committed to the NK-cell lineage.40
Our results are consistent with the current proposed model for human NK-cell development. We have shown that CD56dim KIR-negative cells can acquire KIR on stimulation with exogenous IL-15, implying that IL-15 alone is sufficient for KIR expression. These findings are consistent with a recent study showing that high doses of IL-2 can induce KIR expression on CD56dim KIR-negative NK cells.29,30
Whether CD56bright cells differentiate into CD56dim KIR-negative cells that subsequently acquire KIR or whether CD56bright cells differentiate separately into CD56dim KIR-negative and CD56dim KIR-positive cells has not been formally addressed. However, our results support the former possibility because KIR expression can be induced by cytokine signaling, suggesting that the recently described CD56dim KIR-negative population of NK cells may represent an immature population within the continuum of NK-cell development. The question of whether CD56dim inhibitory receptor-negative cells are mature hyporesponsive cells or whether they represent a developmentally immature subpopulation is of considerable interest in the context of NK-cell education.
The conventional explanation for self-tolerance by mature NK cells was that each cell expressed at least one inhibitory receptor that recognizes “self” and prevents autoimmunity.41 However, studies in both mice and humans have provided evidence of phenotypically mature peripheral blood NK cells that lack expression of all known inhibitory receptors.26,42,43 These cells exhibit poor functional responses to stimulation, leading to their designation as “hyporesponsive.”43 Several hypotheses have been put forward to account for the existence of hyporesponsive NK cells. First, these cells may be induced to enter a hyporesponsive state in response to chronic stimulation.41 Second, these cells may persist as developmental “dead ends” because they cannot be functionally educated by inhibitory receptor ligation.41 Third, the hyporesponsive cells may represent a late stage of development. These cells may acquire inhibitory receptors, given the proper stimulation, and subsequently gain functional competency through an educational process.26,44 Because CD56dim KIR-negative cells can acquire KIR when stimulated by IL-15, we favor the hypothesis that CD56dim cells differentiate from CD56bright cells and remain hyporesponsive until inhibitory receptor expression is induced by cytokine signaling.
Thus, distinct events in NK-cell education, which depend on acquisition of KIR followed by acquisition of effector function, may be difficult to separate. Once a sufficient amount of inhibitory receptor expression is achieved, these cells acquire functional competency through an educational process that presumably depends on inhibitory receptor ligation. The development of new methods to track acquisition of NK-cell function will be needed to study this further.
Another informative finding is that c-Myc overexpression leads to surface expression of KIR in the NK92 cell line (Figure 7), which is KIR−, resulting from dense promoter methylation. We hypothesize that c-Myc can bind to the KIR distal promoter independent of its methylation status. This is supported by a recent study showing that the distal promoter is densely methylated in both KIR-positive and KIR-negative cells.45 The overexpression of c-Myc in NK92 cells may force high levels of transcriptional initiation from the distal promoter and generate distal transcripts over time despite methylation of CpG islands within the proximal promoter. Single-stranded distal mRNA transcripts could then complex with DNA demethylase enzymes, leading to sequence-specific DNA demethylation of KIR promoters over time. One recent report demonstrates the ability of single-stranded RNA to bind a protein involved in active DNA demethylation in Arabidopsis. A similar mechanism may exist in mammalian cells.46 This model of DNA demethylation is currently under investigation in our laboratory.
In conclusion, KIR play a central role in both NK-cell development and function. We have found that IL-15 stimulation increases c-Myc expression, which in turn binds at a distal promoter element to enhance KIR transcription in NK cells. Further studies on how distal transcription is modulated by activity at the proximal promoter and how inhibitory KIR signaling affects NK-cell development may provide a basis for new strategies in the design of NK cell–based therapies.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grants P01-CA-111412 and R01-HL-55417 (J.S.M.), in part with federal funds from the National Cancer Institute (NCI), NIH under contract N01-CO-12400 (S.K.A.), and in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research (S.K.A.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.C., R.J.H., S.K.A., and J.S.M. designed research, performed experiments, and wrote manuscript; T.L. contributed to design of plasmid constructs; M.P. designed and performed quantitative RT-PCR analysis; V.M. assisted with NK-cell cultures and flow cytometry analysis; and H.L. performed EMSA analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Correspondence: Jeffrey S. Miller, University of Minnesota Cancer Center, MMC 806, Division of Hematology, Oncology, and Transplantation, Harvard St at East River Rd, Minneapolis, MN 55455; e-mail: mille011@umn.edu.
References
- 1.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 2.Valiante N, Uhberg M, Shilling H, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 3.Terme M, Ullrich E, Delahaye NF, Chaput N, Zitvogel L. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nat Immunol. 2008;9:486–494. doi: 10.1038/ni1580. [DOI] [PubMed] [Google Scholar]
- 4.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 5.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 6.Van Bergen J, Stewart CA, van den Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. 2005;35:2191–2199. doi: 10.1002/eji.200526201. [DOI] [PubMed] [Google Scholar]
- 7.Trompeter HI, Gomez-Lozano N, Santourlidis S, et al. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. 2005;174:4135–4143. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]
- 8.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 9.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stulberg MJ, Wright PW, Dang H, et al. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 11.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouchard C, Dittrich O, Kiermaier A, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freytag SO, Geddes TJ. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- 14.Walker W, Zhou ZQ, Ota S, Wynshaw-Boris A, Hurlin PJ. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J Cell Biol. 2005;169:405–413. doi: 10.1083/jcb.200411013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 16.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orian A, van Steensel B, Delrow J, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi T, Gasser S, Trumpp A, MacDonald HR. c-Myc acts downstream of IL-15 in the regulation of memory CD8 T-cell homeostasis. Blood. 2006;107:3992–3999. doi: 10.1182/blood-2005-09-3851. [DOI] [PubMed] [Google Scholar]
- 21.Mrozek E, Andereson P, Caligiuri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 22.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:385–387. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper MA, Bush JE, Fehniger TA. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 24.Maasho K, Marusina A, Reynolds NM, Coligan JE, Borrego F. Efficient gene transfer into the human natural killer cell line NKL, using the Amaxa nucleofection system. J Immunol Methods. 2004;284:133–140. doi: 10.1016/j.jim.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Oostendorp RA, Harvey KN, Kusadasi N, et al. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]
- 26.Cooley S, Xiao F, Pitt M, et al. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JS, Alley KA, McGlave P. Differentiation of natural killer (NK) cells from human primitive marrow progenitors in a stroma-based long-term culture system: identification of a CD34+7+ NK progenitor. Blood. 1994;80:2594–2601. [PubMed] [Google Scholar]
- 28.McCullar V, Oostendorp R, Panoskaltsis-Mortari A, et al. Mouse fetal and embryonic liver cells differentiate human umbilical cord blood progenitors into CD56-negative natural killer cell precursors in the absence of interleukin-15. Exp Hematol. 2008;36:598–608. doi: 10.1016/j.exphem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Rham C, Ferrari-Lacraz S, Jendly S, et al. The proinflammatory cytokines IL-2, IL-15, and IL-21 modulate the repertoire of mature human natural killer cell receptors. Arthritis Res Ther. 2007;9:R125. doi: 10.1186/ar2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romagnani C, Juelke K, Falco M, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178:4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 31.Budagian V, Bulanova E, Paus R. Bulfone-Paus, S IL-15/IL-15 receptor biology: a guided tour through an expanding universe. Cytokine Growth Factor Rev. 2006;17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Chan HW, Kurago ZB, Stewart CA, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 34.Saleh A, Makrigiannis AP, Hodge DL, Anderson SK. Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J Immunol. 2002;168:5163–5169. doi: 10.4049/jimmunol.168.10.5163. [DOI] [PubMed] [Google Scholar]
- 35.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 36.Saleh A, Davies GE, Pascal V, et al. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 38.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 39.Martin MP, Bashirova A, Traherne J, Trowsdale J. Carrington, M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 40.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 41.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;7:747–754. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 42.Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez NC, Treiner E, Vance RE, et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–160. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 45.Santourlidis S, Graffmann N, Christ J, Uhrberg M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J immunol. 2008;180:419–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 46.Zheng X, Pontes O, Zhu J, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Science. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]