Abstract
Among all the genetic factors associated with MS susceptibility, MHC class II molecules have the strongest association. Although a direct role of DR alleles in MS have been confirmed, it has been difficult to understand the role of DQ alleles in disease pathogenesis, due to strong linkage disequilibrium with certain DR alleles. Population studies have indicated that DQ alleles may play a modulatory role in progression of MS. Using HLA- class II transgenic (Tg) mice we investigated gene complementation between DR and DQ genes in the disease process. Previously, using single Tg mice (expressing HLA-DR or DQ gene), we showed that PLP91–110 peptide induced EAE only in DR3.Aβ° mice, suggesting that DR3 (DRB1*0301) is a disease susceptibility gene in the context of PLP. We also showed that DQ6 protect development of EAE in DQ6/DR3 double Tg mice by production of anti-inflammatory IFN-γ. In this study we investigated the ability of DQ8 to modulate disease in DR3/DQ8 double Tg mice. Introduction of DQ8 onto DR3 Tg mice led to higher disease incidence and increased disease severity on immunization with PLP91–110 indicating that DQ8 had an exacerbating effect on development of EAE. Increased susceptibility in DR3/DQ8 Tg mice was due to increased production of pro-inflammatory cytokine IL-17 by DQ8-restricted T-cells. HLA-DR3/DQ8 mice with EAE also demonstrated increased inflammation and demyelination in CNS as compared to single DR3 Tg mice. Thus double Tg mouse provides a novel model to study epistatic interactions between HLA class II molecules in inflammatory and demyelinating disease.
Keywords: EAE/MS, MHC, transgenic mice, neuroimmunology, cytokine, epistasis
Introduction
Multiple sclerosis (MS) is chronic inflammatory disease of central nervous system (CNS) characterized by the infiltration of immune cells such as T cells, B cells and macrophages, resulting in demyelination of axons (1). The pathogenic CD4+ T cells in CNS are mainly restricted to components of self-myelin such as proteolipid protein (PLP), myelin basic protein (MBP) and myelin oligodendrocytic glycoprotein (MOG). PLP, a major component of CNS myelin, has emerged as an important target antigen in experimental autoimmune encephalomyelitis (EAE) and MS. PLP peptide reactive T cells have been identified in MS patients (2–4).
Since myelin-antigen specific CD4+ T cells are restricted by MHC class II molecules, it is not surprising that strongest genetic factors influencing development of MS have been reported with HLA locus on chromosome 6p21. Population studies have reported that individuals with certain HLA class II haplotype such as HLA-DR2/DQ6, DR3/DQ2, and DR4/DQ8 have increased frequency of MS. The class II linkage in MS differs in various populations with highest association with HLA-DR2 (DRB1*1501)/DQ6 (DQB1*0602) (5–8), followed by DR3/DQ2 and DR4/DQ8 genes (8–13). Elegent studies by Dyment et al (14) have shown that beside DRB1*15, DRB1*17 (DR3) allele show a clear association with MS susceptibility. Similar finding on association of DR3 with MS has been shown in non Nothern European, Candian, Mexican and Sardinian MS patients (11, 12, 15, 16). Recent studies have shown that disease outcome might be decided by a complex interaction among different class-I and class-II present in a haplotype, suggesting that haplotype might be basic immunogenetic unit of susceptibility or resistance (14, 17–20). The strong linkage disequilibrium among HLA-DR, -DQ and other genes within HLA region, make it difficult to identify role of individual genes in the immunopathogenesis of MS. In order to understand role of class-II molecules in inflammatory autoimmune diseases, Tg mice expressing human HLA-DR or –DQ genes on mouse endogenous class II negative background were generated. Using these class II Tg mice, we and others have previously shown that PLP91–110 peptide can induce MS-like neurological disease in HLA-DR3 (21) and HLA-DR4 Tg mice (22), while HLA-DR2 Tg mice were susceptible to MOG and MBP induced EAE (23, 24). Neither PLP nor MOG antigens were able to induce any disease in Tg mice expressing human DQ6 or DQ8 gene(25). Thus current data from HLA class II transgenic mice suggests that HLA-DR genes such as HLA-DR2, DR3 and DR4 are responsible for predisposition and susceptibility to demyelinating disease. Population studies in MS indicate that other DR, DQ as well as HLA class I alleles on disease susceptible haplotype influence frequency, progression and severity of disease in human patients. While HLA-DRB1*01, -DRB1*11, -DRB1*14, -DQB1*0601 and -DQB1*0603 protect MS (14, 17, 18, 26, 27), DQB1*0602 and DQB1*0302 alleles can increase disease susceptibility (7, 26, 28–30). Thus it is hypothesized that the epistatic interaction between HLA molecules on disease susceptible haplotype plays an important role in final disease outcome in MS. We undertook this study to investigate the role of HLA-DQ8 (DQB1*0302) gene in disease susceptible HLA-DR3 Tg mice. We generated double Tg mice expressing DQB1*0302 (DQ8) on disease susceptible HLA-DR3 background to determine whether presence of DQ8 allele can modulate development of EAE. Since these HLA class II Tg mice express human class II in the absence of endogenous mouse class II molecule, all the T cell responses are restricted to human class II molecules. Previously, using double Tg mice, we reported that DQ6 (DQB1*0601) can protect DR3 (DRβ1*0301). Aβ° mice from development of EAE (31). In the present study, we report that presence of DQ8 on DR3 background led to increased disease severity and CNS pathology as compared to DR3 single Tg mice. The synergistic effect of DQ8 on EAE might be due to increased production of IL-17 by DQ8 restricted CD4 T cells in DR3DQ8 double Tg mice.
Material and Methods
Transgenic (Tg) mice
The HLA-DQ8 (DQA1*0103, DQB1*0302), HLA-DR3 (DRB1*0301), and HLA-DR3/DQ8 Tg mice were produced, as previously described (28, 32, 33). Briefly HLA class II transgenes were introduced into (B6 × SWR)F1 fertilized eggs. Positive offspring were backcrossed to B10.M mice for several generations. HLA transgenic mice were then mated to class II-deficient (Aβ°) mice and intercrossed to generate the HLA transgenic lines. To generate double transgenic mice, single transgenic DR3.Aβ° mice were mated with DQ8.Aβ° Tg lines to produce HLA-DR3/DQ8 Tg lines. Transgene negative littermates were used as controls. All mice were bred and maintained in the pathogen free Immunogenetics Mouse Colony of Mayo Clinic according to National Institutes of Health and institutional guidelines. All experiments were approved by the Institutional Animal Care and Use Committee (IACUC), at Mayo Clinic, Rochester.
Flow cytometry
Expression of HLA-DR and HLA-DQ molecules on PBLs, lymph node cells (LNCs), and splenocytes were analyzed by flow cytometry using monoclonal antibodies (mAbs) L227 and IVD12, specific for HLA-DR and HLA-DQ (34), respectively, as described previously (32). Surface expression of CD4 (GK1.5), CD8 (53.6.72), B cells (RA3-6B2), DCs (HL3), monocytes/macrophages (M1/70), NK cells (PK136, CD25 (PC61), CD44 (IM7) and CD45RB (16A) were analyzed using fluorescent conjugated mAb from BD Biosciences (San Jose, USA). The T-cell receptor (TCR) Vβ usage of CD4+ T cells was determined on PBLs with mAbs specific for: Vβ2 (B20.6.5), Vβ4 (KT4-10), Vβ5.1,2 (MR9.4), Vβ 5.1 (MR9.8), Vβ6 (44.22.1), Vβ7 (TR.310), Vβ8.1,2 (KJ16–133), Vβ8.2 (F23.2), Vβ9 (MR10–2), Vβ11 (RR–153), Vβ14 (14.2), and Vβ17 (KJ23a), as described previously (21).
Peptide
Twenty-amino acid-long synthetic peptide PLP91–110 (YTTGAVRQIFGDYKTTICGK) was synthesized at the peptide core facility of Mayo Clinic, Rochester, MN.
Immunization and T cell proliferation assay
Mice were immunized subcutaneously with PLP91–110 (100 μg) peptide, emulsified in CFA (1:1) containing 100 μg of Mycobacterium tuberculosis H37Ra (Difco, Detroit, MI). Some immunized mice were sacrificed 10 days after immunization, draining lymph nodes removed and challenged in vitro with antigen (28). The results are presented as stimulation indices (CPM of test sample/CPM of the control). For in vitro inhibition experiments, mAbs specific for CD4 (GK1.5), CD8 (TIB 105), HLA-DQ (IVD12), and HLA-DR (L227) were added to LNCs challenged in vitro with human PLP91–110 (20 μg/ml). All of the neutralizing antibodies were generated in-house using the Mayo antibody core facility.
Disease induction
For disease induction 12–14 weeks old Tg mice were immunized subcutaneously in both flanks with 100 μg of PLP91–110 emulsified in CFA containing Mycobacterium tuberculosis H37Ra (400 μg/mice). Pertussis toxin (Sigma Chemicals, St. louis, Mo, USA; 100ng) was injected i.v. at day 0 and 2, post immunization. Mice were observed daily for clinical symptoms and disease severity was scored as follows: 0, normal; 1, loss of tail tone; 2, hind limb weakness; 3, hind limb paralysis; 4, hind limb paralysis and forelimb paralysis or weakness; 5, moribundity/death. Mice of both sexes were used.
Cytokine production
Draining LNs were collected 10 days post immunization and stimulated with PLP91–110 peptide as mentioned before in T cell proliferation section. Supernatants were collected from culture 48 hrs after peptide stimulation. The concentration of cytokines (IFN-γ, IL-2, IL-4, IL-6, IL-10, IL-12, IL-17 and TNF-α) in the supernatant was measured by sandwich ELISA using pairs of relevant anti-cytokine monoclonal antibodies according to manufacturer’s protocol (Pharmingen, San Deigo, California, USA).
Real time PCR
Levels of IL-17, IL-21, IL-23 and IL-27 mRNA in vitro were analyzed using Real time PCR. RNA was extracted from cells using RNAeasy columns (Qiagen) and cDNA was prepared using RNase H-reverse transcriptase (Invitrogen). cDNA was analyzed by real-time quantitative PCR in triplicates by using SYBR® GreenER™ qPCR reagent system (Invitrogen). The expression level of each gene was quantified using the threshold cycle (Ct) method normalized for the house keeping gene β-actin.
Neutralization of IFNγ and IL-17 (anti-cytokine) treatment of EAE
HLA-DQ8 or DR3.DQ8 transgenic mice were injected intraperitoneally either with 250 μg of anti–IFN-γ (clone H22, mouse IgG), or 200 μg of anti–IL-17 (clone TC11-18H10, BD Biosciences) or isotype control (mouse IgG). Anti-IFN-γ was given at day -1, and 10, post immunization (both anti–IFNγ and isotype control antibodies were a kind gift from Dr. R. Schreiber) while anti-IL-17 was administered at 4, 8, 12 and 16 days post-immunization as published previously (35).
In situ apoptosis detection
Apoptotic cells were detected by TUNEL using a kit (IN situ Cell Death Detection Kit, Flourescein, Roches Applied Science, Penzberg, Germany) according to the manufacturer’s directions. Apoptosis (as evidenced by intense nuclear TUNEL staining) was evaluated using a Zeiss Axiovert 510M confocal laser-scanning microscope (Carl Zeiss International, Germany).
Pathology
Mice were perfused via intracardiac puncture with 50 ml of Trump’s fixative. Spinal cords and brains were removed and post-fixed for 24–48 hours in Trump’s fixative in preparation for morphologic analysis. All grading was performed without knowledge of the experimental group.
Spinal cords were cut into one- millimeter coronal blocks and every third block post fixed in osmium and embedded in glycol methacrylate. Two- micron sections were stained with a modified erichrome/cresyl violet stain. Morphological analysis was performed on 12 to 15 sections per spinal cord. Briefly, each quadrant from every coronal section from each spinal cord was graded for the presence or absence of inflammation and demyelination. The score was expressed as the percentage of spinal cord quadrants examined with the pathologic abnormality. A maximum score of 100 indicated that there was a particular pathologic abnormality in every quadrant of all spinal cord sections. Brain pathology was assessed following perfusion. Two coronal cuts were made in the intact brain (one section through the optic chiasm and a second section through the infundibulum). This resulted in three blocks that were embedded in paraffin. This allowed for analysis of the cortex, corpus callosum, hippocampus, brainstem, striatum, and cerebellum. The resulting slides were stained with hematoxylin and eosin. Each area of brain was graded on a 4-point scale. 0 = no pathology; 1 = no tissue destruction but minimal inflammation; 2 = early tissue destruction, demyelination and moderate inflammation; 3 = moderate tissue destruction (neuronal loss, demyelination, parenchymal damage, cell death, neurophagia, neuronal vacuolation); 4 = necrosis (complete loss of all tissue elements with associated cellular debris). Meningeal inflammation was graded as follows: 0 = no inflammation; 1 = one cell layer of inflammation; 2 = two cell layers of inflammation; 3 = three cell layers of inflammation; 4 = four or more cell layers of inflammation. The area with maximal extent of tissue damage was used for assessment of each brain region.
Statistical analysis
The statistical significance of the differences in functional and histological scores between groups was assessed by a one-way analysis of variance on ranks (Kruskall–Wallis test) when comparing more tan two groups, and by the Mann–Whitney rank-sum test when comparing only two groups. The differences in proliferation or in cytokine levels between groups was assessed by a one-way analysis of variance with multiple comparisons of the means when more than two groups were analyzed, or by Student’s t-test when only two groups were analyzed if their data were normally distributed.
Results
Characterization of HLA-DR3.DQ8 transgenic mice
All Tg lines developed normally and showed no gross phenotypic abnormalities. Both HLA-DR and DQ were expressed on 35–50% of cell population in PBLs, and splenocytes (Figure 1A and B). HLA-DR or DQ expression was also detected on 14–21% of LN cell population in DR3, DQ8 or DR3DQ8 mice (Figure 1C). Expression of HLA-DQ and -DR was observed on B cells, macrophages and DCs (data not shown). No endogenous class II expression was seen in class II knockout Aβ° mice (Figure 1A, B and C). Thus both HLA-DR and –DQ molecule were expressed at similar levels in DR3.Aβ°, DQ8.Aβ°, and DR3.DQ8.Aβ° Tg mice.
Figure 1. Class II expression in single and double trangenic mice.
Normal expression of HLA-DR, and/or HLA-DQ was observed in PBLs (A) spleen (B) and LNCs (C) of single and double Tg mice. PBLs, Splenocytes and LNCs were isolated from MHC-Class II deficient control mice (Aβ°), DQ8.Aβ°, DR3.Aβ° or DR3.DQ8.Aβ° Tg mice and analyzed for cell surface markers (HLA-DR/HLA-DQ alone or together with B cells/DCs markers) by flow cytometry. Numbers in histograms indicate the percentage of cells positive for the HLA-DR/HLA-DQ marker. B cells showed maximum class II expression in all the tissue samples analyzed. Data represent one of three experiments performed at different time points.
Cellular analysis of DR3.DQ8.Aβ° Tg mice showed standard T cell repertoire with a normal number of CD4 and CD8 cells in spleen, PBLs and LNCs (data not shown). The CD4 and CD8 T cells also showed a diverse T cell V β repertoire (data not shown). Thus class II expression and cellular profile of double transgenic DR3.DQ8.Aβ° mice was similar to DR3.Aβ° and DQ8.Aβ° single Tg mice.
Presence of HLA-DQ8 molecule increases disease incidence in EAE-susceptible HLA-DR3 Tg mice
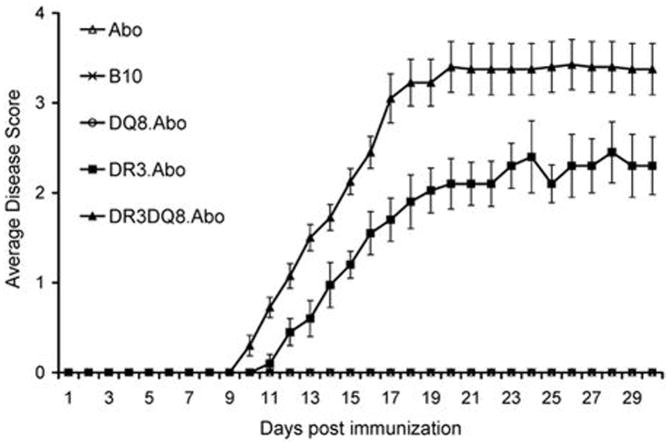
Next we investigated the disease susceptibility of DR3DQ8 double transgenic mice. The susceptibility and clinical features of single Tg, double Tg and control mice to PLP91–110-induced EAE is presented in Table I and Figure 2. Administration of PLP91–110 to DR3.Aβ° Tg mice led to development of chronic progressive disease in 66% (23/35) of Tg mice and disease was characterized by weight loss (data not shown) as well as ascending paralysis (limp tail followed by hind limb weakness and leading to complete hind limb paralysis). DR3 Tg mice with EAE showed a disease onset of 13±1.4 days and mean disease severity score of 2.3±0.4, whereas no disease was seen in DQ8 Tg mice or transgene negative littermates or control Aβ° mice. However, administration of PLP91–110 in DR3DQ8 double Tg mice led to earlier disease onset (9.3±0.9 Vs 13±1.4, p<0.001) and with increased disease frequency (32/35) as compared to single Tg DR3.Aβ° mice (91% Vs 66%, p<0.05). DR3DQ8.Aβ° mice also showed increased disease severity compared to DR3.Aβ° mice (mean clinical score 3.4±0.2 Vs 2.3±0.3, p<0.05). Disease severity in DR3DQ8.Aβ° mice was characterized by weight loss (data not shown) accompanied with paralysis of hind limb (15/32), as well as paralysis of fore limb (10/32) and 5 out of 32 mice also became moribund. Thus the above data indicated that DQ8 plays a modulatory role in DR3DQ8 double transgenic mice by causing more severe EAE in disease susceptible DR3 Tg mice.
Table I.
PLP91–110 induced EAE in HLA Tg micea
| Mouse strain | Disease incidence (%) | Mean onset of disease ± SD | Number of mice with maximum severity score |
||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| B10 | 0/15 (0) | - | - | - | - | - | - |
| Aβ° | 0/15 (0) | - | - | - | - | - | - |
| DQ8.Aβ° | 0/20 (0) | - | - | - | - | - | - |
| DR3.Aβ° | 23/35 (66%) b | 13±1.4 | - | 1 | 18 | - | 2 |
| DR3.DQ8.Aβ° | 32/35 (91%) | 9.3±0.9 | - | - | 15 | 10 | 7 |
Mice were immunized with 100μg of PLP peptide/400μg Mtb in CFA and Ptx was administered at 0 and 48 h post immunization. Mice were scored daily for disease as mentioned in material and methods. The data is from three experiments combined.
p<0.01 Mann-Whitney rank sum test, DR3 compared to other mice.
Figure 2. PLP91–110 induced EAE in single and double Tg mice.
DR3.DQ8.Aβ° mice show higher disease incidence as compared to DR3.Aβ° mice, while no disease was observed in DQ8.Aβ° mice. Five mice per group were immunized with 100μg of PLP peptide/400μg Mtb in CFA and animals also received 100ng of Pertussis toxin at days 0, and 2, post immunization. Mice were scored daily for disease (as stated in material and methods) and the daily mean disease score for each group is plotted. Error bars represent the standard error of mean. The data is from four independent experiments combined.
DR3.DQ8.Aβ° mice produce increased levels of IL-17 and IFN-γ
We previously showed that both DR3 as well as DQ8 molecule recognize PLP91–110 peptide (21). Although DQ8 Tg mice are resistant to PLP91–110 induced EAE, double Tg DR3DQ8 mice showed more severe EAE. Therefore we analyzed T cell proliferative response as well as levels of different inflammatory cytokines in PLP91–110 immunized single and double Tg mice in an attempt to identify the mechanism for this increased severity. LNCs from PLP91–110 immunized DR3.Aβ°, DQ8.Aβ° and DR3.DQ8.Aβ° Tg mice were stimulated in vitro with PLP peptide in the presence or absence of blocking antibodies to HLA-DR (L-227) or HLA-DQ (IVD-12) and the T cell proliferation was measured using standard thymidine incorporation assay. T cells from DR3DQ8.Aβ° Tg mice showed higher T cell response to PLP91–110 peptide as compared to T cell response observed in single Tg DR3 or DQ8 mice (Figure 3A). As expected HLA-DR antibody inhibited T cell proliferation of DR3.Aβ° (Figure 3A) but had no effect on T cell proliferation from DQ8.Aβ° mice, and similarly anti-DQ antibody IVD-12 inhibited T cell response only in DQ8 mice (Figure 3A) but not in DR3 specific T cell cultures. Although, presence of anti-DR antibody inhibited T cell proliferation completely in DR3 mice, only partial inhibition was observed in T cell cultures from double Tg DR3.DQ8.Aβ° mice. Similarly anti-DQ antibody inhibited T cell proliferation completely in DQ8 but partially in DR3.DQ8.Aβ° mice. Thus both DR as well DQ specific T cells were recognizing PLP peptide in double Tg mice.
Figure 3. Ex vivo T cell response and cytokine levels in PLP91–110 immunized single and double Tg mice.
A) DR3.DQ8.Aβ° mice showed 1.5 fold higher T cell proliferative response to PLP91–110 compared to the T cell response observed in DR3.Aβ° Tg mice, while T cell response from DQ8.Aβ° mice showed slightly lower response as comparable to those from DR3.Aβ°. For measurement of antigen specific T cell responses, draining LNCs from Tg mice immunized with PLP91–110 were cultured with or without (control) PLP peptide for 48 h. The proliferative response was assessed by pulsing the cultures with [3H]thymidine for the last 16 h. The data are presented as the mean CPM ± SD and are average of three independent experiments. B) T cells from PLP91–110 immunized DR3.DQ8.Aβ° mice produced high levels of IFN-γ, IL-6, IL-12, IL-17, and IL-23 as compared to DR3.Aβ° mice, while levels of TNF-α, IL-2 and IL-10 were similar between two strains. Levels of Th17 cytokines were also higher in T cell cultures from DQ8 Tg mice as compared to DR3 Tg mice. Cytokines level in culture supernatants were determined by standard sandwich ELISA as described in material and methods. C) T cells from PLP91–110 peptide immunized DR3.DQ8.Aβ° and DQ8.Aβ° mice showed higher expression of IL-17, IL-21 and IL-23 as compared to DR3.Aβ° mice. Expression of Th17 related cytokines in different transgenic mice were quantified by real-time PCR. Expression of β-actin was measured as an internal control. The expression of different cytokines in LNCs stimulated with PLP91–110 relative to that in medium control was calculated by the ΔΔCt method. Data are presented as means ± SD of at least four different mice. *- p ≤0.01 as compared to DR3.Aβ°.
Disease susceptible DR3.Aβ° Tg mice produced moderate to high levels of IFN-γ, TNF-α, IL-2, IL-6 and IL-12 cytokines (Figure 3B), showing classical Th1 phenotype. Although MNCs from DQ8 mice did not produce IFN-γ, they produced significantly higher levels of IL-17 (p<0.01), recently discovered pro-inflammatory cytokine, and associated with development of autoimmune diseases. Double Tg DR3DQ8 mice also produced higher levels of IL-17 as well as IFN-γ, besides producing moderate to high levels of TNF-α, IL-2, IL-6 and IL-12 cytokines. IL-4 levels were below detection limits in all samples from single and double Tg mice. DR3.Aβ° Tg mice also produced moderate amounts of IL-17, IL-21 and IL-23 (Figure 3C), however levels were significantly less (p<0.01) as compared to DQ8 or DR3DQ8 mice. Both DR3 as well as DR3DQ8 mice produced comparable levels of IL-27. Thus, disease susceptible DR3.DQ8 mice produced higher levels of IL-17, IL-21, IL-23 and IFN-γ as compared to DR3 mice.
Increased levels of IFN-γ is produced by DR3 specific T cells, while IL-17 is produced by DQ8 specific T cells
Thus cytokine data suggested that the exacerbating effect of DQ8 molecule on disease severity in double Tg DR3.DQ8.Aβ° mice might be either due to high levels of IL-17 or IFN-γ, which can be produced by either DQ and/or DR specific T cells. Therefore, we performed Elispot assay (a standard assay for analyzing antigen specific cytokine levels), to analyze the source of T cells producing IL-17 and IFN-γ. LNCs from PLP91–110 immunized DR3.Aβ°, DQ8.Aβ° and DR3.DQ8.Aβ° Tg mice were stimulated in vitro with PLP peptide and levels of IL-17 and IFN-γ was analyzed in the presence or absence of blocking antibodies to HLA-DR (L-227) or HLA-DQ (IVD-12) using cytokine specific Elispot assay. Analysis of IFN-γ spots showed that T cells from DR3DQ8 mice produced slightly higher levels of IFN-γ (275±30 Vs 200±25, p<0.01) as compared to T cells from DR3 Tg mice (Figure 4A). Antibody blocking experiments confirmed that most of the IFN-γ was produced by DR specific T cells as it was significantly inhibited by anti-DR antibody in DR3DQ8 as well as DR3-specific cultures, while anti-DQ antibody had no effect on IFN-γ spots (Figure 4A). Further, T cells from DQ8 as well as DR3DQ8 showed significantly higher IL-17 spots as compared to those from DR3 Tg mice [240±27 (DQ8) Vs 62±20 (DR3), p<0.001 and 320±37 (DR3DQ8) Vs 62±20 (DR3), p<0.001]. Although, blocking with anti-DR antibody suppressed IFN-γ producing T cells in DR3.DQ8.Aβ° specific cultures, it had minimal effect on IL-17 producing cells (Figure 4B). In contrast, anti-DQ blocking antibody suppressed most of the IL-17 spots in DR3DQ8 specific cultures but had no effect on IFN-γ producing T cells. Presence of anti-DQ antibody in cultures suppressed (>80%) IL-17 spots in both DQ8 specific as well as DR3DQ8 specific cultures, while anti-DR antibody suppressed only 15–20% of IL-17 spots in DR3DQ8 specific T cell cultures. Anti-DQ antibody had no effect on IL-17 spots observed in cultures from DR3 mice. Thus higher levels of IL-17 in double Tg DR3DQ8.Aβ° mice was produced by PLP restricted DQ8-specific CD4 T cells, while most of the IFN-γ was produced by DR-specific CD4 T cells.
Figure 4. Source and specificity of IFN-γ and IL-17 in DR3, DQ8 and DR3DQ8 Tg mice.
A) IFN-γ was produced by PLP91–110 specific CD4+ T cells as anti-DR antibody suppressed production of IFN-γ in both DR3 as well as DR3DQ8 Tg mice by >90%. B) Increased levels of IL-17 in double Tg DR3DQ8 mice were produced mostly by DQ restricted PLP91–110 specific CD4+ T cells as blocking monoclonal antibody specific for DQ (IVD12) inhibited >80% of PLP91–110 specific IL-17 spots. For blocking experiments LNCs were isolated from PLP91–110 immunized DR3, DQ8 and DR3DQ8 Tg mice and co-cultured in special Elispot plates with PLP91–110 in presence or absence of blocking monoclonal antibodies specific for HLA-DQ (IVD-12) or HLA-DR (L-227). After 48 h in culture, IFN-γ and IL-17 spots were measured as described in methods. The results are representative for three independent experiments for all groups.
Neutralization of IL-17 but not IFN-γ in DR3DQ8.Aβ° Tg mice abolished the synergistic effect of DQ8 molecule
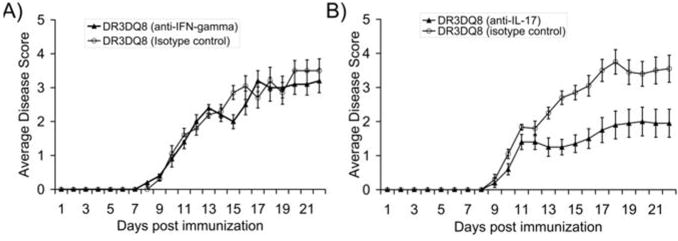
Data from previous experiments suggested that either high levels of IL-17 produced by PLP-specific DQ8-restricted T cells or IFN-γ produced by DR3-specific T cells might be responsible for the synergistic effect of DQ8, leading to increased disease incidence and severity. Therefore, we performed in vivo neutralization of either IL-17 or IFN-γ, with a rationale that blocking of cytokines responsible for increased disease severity would lead to decrease in disease incidence as well as severity. Single and double Tg mice were immunized with PLP91–110 and treated with either neutralizing IFN-γ antibody (clone H-22) or neutralizing IL-17 antibody (clone TC11-18H10) at time points mentioned in material and methods. Five animals in each group were also treated with respective control isotype antibody. Treatment with anti-IFN-γ had no effect on disease incidence or severity in double transgenic DR3DQ8.Aβ° mice (Table 2 and Figure 5A). However, treatment with neutralizing IL-17 antibody led to decrease incidence and severity in double Tg DR3DQ8.Aβ° mice as compared to mice treated with isotype control antibody (Table 2 and Figure 5B). Thus the increased disease severity observed in double Tg DR3DQ8.Aβ° mice was due to high levels of IL-17 produced by DQ8 specific T cells. Blocking of IFN-γ or IL-17 in DQ8.Aβ° mice by neutralizing antibody had no effect on disease phenotype.
Figure 5. Effect of IFN-γ and IL-17 neutralization on double Tg DR3.DQ8.Abo mice.
A) Anti-IFN-γ treatment of PLP91–110 immunized DR3.DQ8.Aβ° mice had no effect on disease incidence or severity as the disease onset and average disease score were similar between anti-IFN-γ treated and isotype control treated mice. B) However, blocking IL-17 decreased disease incidence as well severity in DR3.DQ8.Aβ° mice receiving neutralizing IL-17 antibody as compared to those receiving isotype control antibody. For disease induction, DR3DQ8.Aβ° Tg mice were immunized with 100 μg of PLP91–110 emulsified in CFA containing 400 μg of mycobacterium. Pertussis toxin (100ng) was administered at 0, and 48 h post immunization. For neutralization of IFN-γ, five mice per group were also given 250 μg of anti-IFN-γ Ab or isotype control antibody at day -1 and day 10 postimmunization. IL-17 was neutralized in-vivo by administering 200 μg of anti-IL-17 antibody or isotype control antibody at day 4, 8, 12 and day 16 postimmunization. Mice were scored daily for disease (as stated in material and methods) and the daily mean disease score for each group is plotted. Error bars represent the standard error of mean. The data are pooled from two independent experiments.
High levels of IL-17 led to increased T cell migration into CNS of DR3.DQ8.Aβ° mice
In recent years IL-17 has emerged as an important inflammatory cytokine in development of auto-immune diseases and it can exert its pro-inflammatory role through number of pathways. We investigated whether antigen specific T cells from DR3DQ8 mice can cross blood brain barrier more efficiently then those from single Tg DR3 mice. Brain and spinal cord infiltrating cells were isolated at different time points from mice immunized with PLP91–110 peptide. At all time points DR3DQ8 mice with EAE showed an increased cellular infiltration in the CNS as compared to DR3 single transgenic mice with EAE (Fig. 6A). CNS infiltrating cells from DR3 as well as DR3DQ8 produced 5–6 fold higher levels of IFN-γ (Fig 6B) and IL-17 (Fig 6C) without any stimulation in ex vivo cultures suggesting highly activated cells. Ex vivo stimulation of CNS infiltrating cells with PLP91–110 peptide lead to 2–3 fold increase in levels of IFN-γ (Fig 6B) and IL-17 (Fig 6C), indicating the presence of antigen specific T cells in CNS. Levels of both cytokines were higher in DR3DQ8 mice as compared to DR3 mice (3000±440 Vs 1150±200, p<0.01). When stained for T cell activation markers, we found that CD4+ T cells showed activated phenotype, as they were CD44high and CD45RBlow (data not shown). Thus above findings indicated that PLP91–110 specific, HLA-DR/DQ restricted CD4+T cells were present in the CNS of both DR3 as well as DR3DQ8 Tg mice with EAE.
Figure 6. CNS infiltrating inflammatory cells.
A) DR3DQ8 mice with EAE showed higher number of CNS infiltrating cells as compared to DR3 mice with EAE. EAE was induced in HLA-Tg mice and CNS (brain and spinal cord) infiltrating mononuclear cells were isolated using percol gradient. B) High levels of IL-17 as well as IFN-γ was detected on culturing CNS infiltrating cells from DR3 mice as well as from DR3DQ8 mice, however levels of both cytokines were significantly higher in DR3DQ8 Tg mice as compared to DR3 Tg mice. Stimulation with PLP91–110 led to 2–3 fold increase in levels of IL-17 and IFN-γ. For cytokine analysis, CNS infiltrating cells from Tg mice (with EAE) were stimulated with or without PLP91–110 for 48 h in vitro culture and cytokines levels were determined as described previously.
DR3DQ8 double transgenic mice show severe CNS pathology as compared to DR3 Tg mice
To determine if presence of DQ8 on disease susceptible DR3 mice also had an effect on CNS pathology, we analyzed brain and spinal cord tissue from mice with EAE. Pathological analysis of CNS tissue showed that double Tg DR3DQ8.Aβ° mice had increased inflammation and demyelination as compared to DR3.Aβ° mice (Fig 7 A-D). While single Tg DR3.Aβ° showed primarily inflammation localized to the meninges of the spinal cord and brain, double Tg DR3DQ8.Aβ° mice showed more widespread brain pathology with severe inflammation and demyelination in all parts of the brain tissue including cerebellum, brain stem, cortex, corpus callosum, stratium and meninges (Fig 7A and 7C). Plastic embedded sections from DR3DQ8.Aβ° mice but not from DR3.Aβ° also showed typical parenchymal white matter loss, the classical pathology observed in MS. Similar pattern of pathology was also observed in the spinal cord with increased inflammation and demyelination in DR3DQ8.Aβ° mice as compared to DR3.Aβ° mice (Fig 7B). Quantitative analysis of spinal cord tissues showed that on average 44±6% of the spinal cord quadrants from DR3DQ8.Aβ° showed inflammation as compared to only 23±7% in single Tg DR3.Aβ° mice (p<0.01, Fig. 7D). While DR3.Aβ° showed demyelination only in 20±5% of spinal cord quadrants, 40±6% of the quadrants from double Tg mice showed loss of myelin (p<0.001, Fig. 7B and 7D). Thus double Tg mice showed severe CNS pathology which was similar to pathology observed in MS.
Figure 7. CNS pathology in DR3.Aβ° and DR3DQ8.Aβ° Tg mice with EAE.
Double Tg DR3.DQ8.Aβ° mice with EAE show severe inflammation and demyelination in CNS as compared to single Tg DR3.Aβ° mice with EAE. Representative photomicrograph of inflammatory lesion in brain (A) and spinal cord (B) of DR3 and DR3DQ8 Tg mice immunized with PLP antigen. A) Brain samples were embedded in paraffin and stained with H&E. Brain pathology was characterized with widespread inflammation and demyelination in DR3.DQ8.Aβ° as compared to mild meningeal inflammation DR3.Aβ° mice with EAE. B) The photographs of 2-μm-thick spinal cord sections show increased inflammation and demyelination in DR3.DQ8.Aβ° mice as compared to DR3.Aβ° mice. These figures are representative of one of four experiments. Quantitative analysis of brain C) and spinal cord D) pathology also showed that double Tg DR3.DQ8.Aβ° mice with EAE have a higher pathology score as compared to DR3.Aβ° mice. C) Pathology scores in brain (each bar represents the histologic score for each mouse) and D) percent of spinal cord quadrants showing inflammation and demyelination (mean ± SD) as described in materials and methods. Tg mice and control mice were immunized with 100 μg of PLP91–110 peptide emulsified in CFA containing 400 μg of mycobacterium. Pertussis toxin (100ng) was administered at 0, and 48 h post immunization and Tg mice were sacrificed on day 25 post-immunization.
Increased Apoptosis of Inflammatory cells in CNS of single Tg DR3.Aβo mice
Next we investigated, why double Tg mice expressing both DR3 and DQ8 show more pathology. It has been suggested previously that apoptosis of CNS infiltrating inflammatory cells play an important part in recovery from EAE. We hypothesized that less pathology in DR3 mice was the result of CNS infiltrating inflammatory cells undergoing increased apoptosis. To investigate this hypothesis, TUNEL staining was performed on brain sections. Inflammatory cells in the CNS of DR3 mice with EAE showed higher percentage of tunnel positive cells as compared to DR3DQ8 double Tg mice with EAE (Figure 8). The TUNEL-stained cells were observed in the inflamed meninges. No TUNEL staining was observed in control mice without EAE. Thus inflammatory cells in the CNS of double Tg mice had a longer survival than in DR3 Tg mice with EAE.
Figure 8. Apoptosis of inflammatory cells inside CNS.
DR3 Tg mice with EAE showed significantly higher number of tunnel positive cells in CNS as compared to DR3DQ8 double Tg mice. No staining was observed in DQ8 Tg mice. Representative photomicrograph showing TUNNEL positive cells within an inflammatory infiltrate brain section of HLA-DR3 and DR3.DQ8 mice with EAE. Disease was induced in HLA Tg mice and mice were sacrificed on day 25. Brain tissue was isolated, snap frozen in liquid nitrogen and embedded in Tissue-Tek® OCT compound. 10 μm thick section were stained for tunnel staining as mentioned in material and methods. The stained sections were analyzed by confocal laser scanning microscope (Carl Zeiss).
Discussion
MS, like other putative autoimmune diseases has a strong genetic component associated with certain HLA-DR/DQ haplotypes such as DR2/DQ6, DR3/DQ2 and DR4/DQ8. Population studies in MS as well as EAE studies done in HLA class II transgenic mice have confirmed that HLA-DR alleles such as DR2, DR3 and DR4 are major susceptibility genes. The role of HLA-DQ allele in disease pathogenesis has not been well understood due to strong linkage disequilibrium between DR and DQ genes; prevailing data suggests that HLA-DQ allele might play a modulatory role. Using double HLA class II transgenic mice expressing both HLA-DR3 as well as HLA-DQ8, we show that whereas HLA-DR3 is the main disease predisposing gene in EAE in context of PLP91–110, HLA-DQ8 molecule plays a modulatory role. Although, DQ8 (DQB1*0302) transgenic mice were resistant to PLP91–110 induced EAE, presence of DQ8 gene on disease susceptible DR3 (DRB1*0301) Tg mice led to increased incidence and disease as well as severe CNS pathology suggesting a exacerbating role for DQ8 molecule in inflammation and demyelination. This increased severity observed in DR3DQ8 double Tg mice might be due to high levels of pro-inflammatory cytokine IL-17 produced by DQ8 restricted PLP91–110 specific- T cells.
A disease enhancing role for DQ8 is in agreement with human linkage studies. Population studies have shown that DQ8 (DQB1*0302) allele is associated with more severe forms of MS (9, 13, 36, 37). Olerup et al (36) as well as Marrosu et al (37) showed that DQ8/DR4 haplotype was associated with MS in Sardinia and since then a number of other studies have confirmed association of DQ8 with MS in different ethnic population. Using a high-resolution HLA genotyping analysis, Zivadinov et al (13) showed that beside DR2/DQ6, presence of DQ8 allele was associated with more severe damage on inflammatory and neurodegenerative MRI measures. We previously showed that DR3DQ8 double Tg mice develop severe disease on immunization with whole myelin extract as compared to DR3 single transgenic mice (28). Similarly, MOG induced more severe form of EAE in DR2/DQ8 double transgenic mice as compared to DR2 single transgenic mice (24). In this study, we show that presence of DQ8 on DR3 background increase disease incidence and severity in PLP-induced EAE, indicating a modulating role of DQ8. Since, DQ8 can worsen disease severity in both DR2/DQ8 as well as DR3/DQ8 double Tg mice, we hypothesize that presence of DQ8 with any of the disease susceptible –DR allele such as DR2, DR3 or DR4 might lead to more severe demyelinating disease.
How certain HLA-class II molecules such as DQ8 modulates disease when present with disease susceptible HLA-DR allele is not understood. There are several explanations for this modulatory effect of DQ8 molecule. The first possibility is that there is reduced number of CD4CD25+FoxP3+ Treg cells in double Tg DR3DQ8 mice as compared to DR3 single Tg mice. However, we observed similar number of Tregs in all three strains of mice (data not shown), ruling out the role of Tregs in modulating disease. Another possibility is that DR3DQ8 Tg mice show a stronger T cell response to encephalitogenic epitope of PLP, which leads to production of increased levels of pro-inflammatory cytokine resulting in increased inflammation and severe disease. Analysis of T cell responses indicated that although DQ8 showed a moderate T cell response to PLP91–110, T cells from immunized double Tg DR3DQ8 mice showed 1.5 fold stronger T cell responses as compared to T cell response observed in DR3 Tg mice. Because, it is well established that both Th1 as well as Th17 play an important role in immunopathogenesis of EAE, we next analyzed the cytokine(s) responsible for this severe disease and CNS pathology. Disease susceptible DR3 mice produce only moderate amounts of IFN-γ and IL-17, whereas DR3DQ8 double Tg mice produced significantly higher levels of IFN-γ (1180±150 Vs. 650±90, p<0.05) and IL-17 (1200±150 Vs. 215±40, p<0.001). DR3DQ8 mice also produced higher levels of IL-6 and IL-23, cytokines associated with IL-17 and Th17 pathway. However levels of TNF-α were similar between DR3 and DR3DQ8 double Tg mice. Disease resistant DQ8 mice also produced high levels of IL-17 as compared to DR3 Tg mice but no IFN-γ. The failure of DQ8 Tg mice to produce IFN-γ in response to PLP91–110 might be a reason for the allele being disease resistant despite producing high levels of IL-17. Thus both pro-inflammatory IFN-γ and IL-17 may be required for increased inflammation and demyelination of CNS.
To confirm the source of IL-17 and IFN-γ in DR3 DQ8 mice, we did Elispot assay in presence of anti-DQ or anti-DR blocking antibodies. Blocking with anti-DR antibody suppressed IFN-γ levels in both DR3 as well as DR3DQ8 mice indicating that DR-specific T cells are the major source of the IFN-γ in double Tg mice. However, most of IL-17 in DR3DQ8 mice was produced by DQ specific T cells as anti-DQ but not anti-DR antibody suppressed IL-17 levels.
Thus DR3DQ8 mice have higher levels of IFN-γ (produced by DR specific T cells) as well as high levels of IL-17 (produced by DQ specific T cells). Either IFN-γ or IL-17 or both of these pro-inflammatory cytokines might be responsible for severe disease observed in DR3DQ8 mice. To confirm role of these cytokines, we did in vivo neutralization of IFN-γ and IL-17 using blocking antibodies. Of interest neutralization of only IL-17 led to decrease in disease incidence as well as severity in DR3DQ8 mice, where as anti-IFN-γ had no effect on disease incidence and severity. Thus IL-17 produced by DQ8 specific T cells appears to be responsible for increased disease incidence and severity in DR3DQ8 mice. We also observed that T cells from DQ8 as well as DR3DQ8 mice produced high levels of IL-23. Our results that IL-17 neutralization decreased disease incidence as well as severity in DR3DQ8 support the hypothesis that IL-23/IL-17 axis plays an important role in immuno-pathogenesis of EAE in DR3DQ8 mice. Although neutralization of IFN-γ had no effect on disease course in our animal model, we can not completely rule out role of Th1 cells in pathogenesis of EAE. Recent data also support the idea that both Th1 and Th17 cells are capable of inducing autoimmunity. Generation of IL-17 and IFN-γ knock out on DR3DQ8 background in future will help us in defining clear role of Th1 and Th17 response in our animal model.
IL-17 (IL-17A) is a member of IL-17 family (IL-17A-F) and stimulates various types of cells, such as epithelial cells, endothelial cells and fibroblasts to produce proinflammatory cytokines and chemokines (38, 39). IL-17 can modulate disease through multiple pathways such as CNS recruitment of inflammatory cells, induction of pro-inflammatory mediators inside the CNS or direct injury to CNS tissue. DR3DQ8 mice with higher disease incidence and severity also had higher frequency of CNS infiltrating cells suggesting that IL-17 might help to recruit cells in to the CNS. Recently, Carlson et al (40) showed that Th17 induced chemokine pathway is essential for blood brain barrier breakdown, and CNS infiltration of inflammatory cells. These CNS infiltrating cells also produced high levels of IL-17 and IFN-γ suggesting an important role in CNS pathology. Because, high IL-17 in DR3DQ8 mice was associated with increased inflammation and demyelination in both spinal cord as well as brain, it is possible that IL-17 might be responsible for severe pathology observed in these mice. Earlier, Lees et al (41), showed that IL-17 deficient mice develop less severe EAE on adoptive transfer of encephalitogenic T cells. Beside IL-17 and IFN-γ, other inflammatory cytokines, chemokines and chemical mediators can also play an important role in disease pathogenesis (1). It is possible that severe CNS pathology in DR3DQ8 double Tg mice may be due to less apoptosis of inflammatory cells in the CNS of these mice compared to DR3 single Tg mice. Our TUNNEL staining data support this hypothesis as there were more Tunnel positive cells in CNS tissue section from DR3 mice as compared to DR3DQ8 mice. Apoptotic elimination of encephalitogenic CD4+ T cells is a well-documented early step in recovery from EAE (42, 43).
Our study show that DQ8 molecule in trans modulate the disease incidence and severity in HLA- DR3.Aβ° Tg mice. The modulatory effect of DQ8 might be due to production of pro-inflammatory cytokine IL-17. IL-17 might exert its effect through multiple pathways such as increased recruitment of inflammatory cells in to CNS and/or increased tissue injury inside CNS. We previously reported that DQ6 molecule produce anti-inflammatory IFN-γ to protect DR3DQ6 double Tg mice from development of EAE (31). Our studies imply that incidence, progression, severity and modulation of EAE are dependent on epistatic interactions in trans between MHC class II molecules. Similarly, interactions between HLA genes on disease susceptible haplotype in humans may determine the course of the demyelinating diseases. Linkage analysis from MS patients indicate that there is complex interaction among MHC haplotypes in MS. MHC genes within a haplotype such as HLA-DR, -DQ, -DP or HLA class I genes can influence disease outcome either through cis or trans interactions. Some protective -DR alleles such as DRB1*11 and DRB1*14 act in cis, while other protective –DR allele DRB1*01 and DRB1*10 act in trans (20). Similarly HLA-class I alleles might influence risk of MS susceptibility by their cis interaction with HLA-DRB1*15 (17). Knowledge of such interactions could aid in designing individualized therapy for MS patients.
Table II.
Neutralization of IFN-γ and IL-17 in HLA Tg mice immunized with PLP91–110a
| Mouse strain | Treatment IFN-γ/IL-17 or Isotype control | Disease incidence (%) | Mean onset of disease ± SD | Number of mice with maximum severity score |
||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||||
| DQ8.Aβ° | IFN-γ | 0/10 (0) | - | - | - | - | - | - |
| DQ8.Aβ° | Isotype | 0/10 (0) | - | - | - | - | - | - |
| DR3.DQ8.Aβ° | IFN-γ | 9/10 (90%) | 9.5±1.2 | - | - | 6 | 2 | 1 |
| DR3.DQ8.Aβ° | Isotype | 9/10 (90%) | 9.2±0.9 | - | - | 5 | 2 | 2 |
| DQ8.Aβ° | IL-17 | 0/10 (0) | - | - | - | - | - | - |
| DQ8.Aβ° | Isotype | 0/10 (0) | - | - | - | - | - | - |
| DR3.DQ8.Aβ° | IL-17 | 6/10 (60%)b | 11±2.0 | - | 3 | 3 | - | - |
| DR3.DQ8.Aβ° | Isotype | 9/10 (90%) | 9.5±1.2 | - | - | 5 | 2 | 2 |
Mice were immunized with 100μg of PLP peptide/400μg Mtb in CFA and Ptx was administered at 0 and 48 h post immunization. Anti-IFN-γ (250μg/mice) or isotype control was administered at days -1 and 10 days post immunization. In IL-17 group mice were treated with anti-IL-17 antibody (200μg/mice) or isotype control antibody at day 4, 8, 12 and 16 postimmunization. Mice were scored daily for disease (as mentioned in material and methods) and daily mean clinical score is presented for each group. The data is from two experiments combined.
p<0.05 Mann-Whitney rank sum test, DR3.DQ8 treated with anti-IL-17 antibody compared to DR3.DQ8 treated with isotype control antibody mice.
Acknowledgments
This work was supported by grants from the National Institute of Health, NS 0521732, NS24180, NS32149 and grants from National Multiple Sclerosis Society (CA1011-03 and RG3172).
We thank Julie Hanson and her staffs for mouse husbandry and Michele Smart for tissue typing of transgenic mice. We also thank Lauri Zoecklein, Louiza Papke and Mable Peirce for excellent technical assistance.
Abbreviations used in this paper
- Tg
transgenic
- MS
multiple sclerosis
- CNS
central nervous system
- PLP
proteolipid protein
- MNCs
mononuclear cells
- LNCs
lymph node cells,
References
- 1.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 2.Greer JM, Csurhes PA, Cameron KD, McCombe PA, Good MF, Pender MP. Increased immunoreactivity to two overlapping peptides of myelin proteolipid protein in multiple sclerosis. Brain. 1997;120(Pt 8):1447–1460. doi: 10.1093/brain/120.8.1447. [DOI] [PubMed] [Google Scholar]
- 3.Martino G, Hartung HP. Immunopathogenesis of multiple sclerosis: the role of T cells. Curr Opin Neurol. 1999;12:309–321. doi: 10.1097/00019052-199906000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt S. Candidate autoantigens in multiple sclerosis. Multiple sclerosis (Houndmills, Basingstoke, England) 1999;5:147–160. doi: 10.1177/135245859900500303. [DOI] [PubMed] [Google Scholar]
- 5.McDonald WI. Multiple sclerosis: epidemiology and HLA associations. Ann N Y Acad Sci. 1984;436:109–117. doi: 10.1111/j.1749-6632.1984.tb14781.x. [DOI] [PubMed] [Google Scholar]
- 6.Oksenberg JR, Begovich AB, Erlich HA, Steinman L. Genetic factors in multiple sclerosis. Jama. 1993;270:2362–2369. [PubMed] [Google Scholar]
- 7.Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, Cree BC, Begovich AB, Villoslada P, Montalban X, Uccelli A, Savettieri G, Lincoln RR, DeLoa C, Haines JL, Pericak-Vance MA, Compston A, Hauser SL, Oksenberg JR. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 8.Dyment DA, Sadovnick AD, Ebers GC. Genetics of multiple sclerosis. Hum Mol Genet. 1997;6:1693–1698. doi: 10.1093/hmg/6.10.1693. [DOI] [PubMed] [Google Scholar]
- 9.Weinshenker BG, Santrach P, Bissonet AS, McDonnell SK, Schaid D, Moore SB, Rodriguez M. Major histocompatibility complex class II alleles and the course and outcome of MS: a population-based study. Neurology. 1998;51:742–747. doi: 10.1212/wnl.51.3.742. [DOI] [PubMed] [Google Scholar]
- 10.Marrosu MG, Muntoni F, Murru MR, Spinicci G, Pischedda MP, Goddi F, Cossu P, Pirastu M. Sardinian multiple sclerosis is associated with HLA-DR4: a serologic and molecular analysis. Neurology. 1988;38:1749–1753. doi: 10.1212/wnl.38.11.1749. [DOI] [PubMed] [Google Scholar]
- 11.Marrosu MG, Murru MR, Costa G, Cucca F, Sotgiu S, Rosati G, Muntoni F. Multiple sclerosis in Sardinia is associated and in linkage disequilibrium with HLA-DR3 and -DR4 alleles. Am J Hum Genet. 1997;61:454–457. doi: 10.1016/S0002-9297(07)64074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarado-de la Barrera C, Zuniga-Ramos J, Ruiz-Morales JA, Estanol B, Granados J, Llorente L. HLA class II genotypes in Mexican Mestizos with familial and nonfamilial multiple sclerosis. Neurology. 2000;55:1897–1900. doi: 10.1212/wnl.55.12.1897. [DOI] [PubMed] [Google Scholar]
- 13.Zivadinov R, Uxa L, Bratina A, Bosco A, Srinivasaraghavan B, Minagar A, Ukmar M, Benedetto S, Zorzon M. HLA-DRB1*1501, -DQB1*0301, -DQB1*0302, -DQB1*0602, and -DQB1*0603 alleles are associated with more severe disease outcome on MRI in patients with multiple sclerosis. Int Rev Neurobiol. 2007;79:521–535. doi: 10.1016/S0074-7742(07)79023-2. [DOI] [PubMed] [Google Scholar]
- 14.Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, Risch N, Ebers GC. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 15.Cullen CG, Middleton D, Savage DA, Hawkins S. HLA-DR and DQ DNA genotyping in multiple sclerosis patients in Northern Ireland. Hum Immunol. 1991;30:1–6. doi: 10.1016/0198-8859(91)90062-e. [DOI] [PubMed] [Google Scholar]
- 16.Masterman T, Ligers A, Olsson T, Andersson M, Olerup O, Hillert J. HLA-DR15 is associated with lower age at onset in multiple sclerosis. Annals of neurology. 2000;48:211–219. [PubMed] [Google Scholar]
- 17.Chao MJ, Barnardo MC, Lincoln MR, Ramagopalan SV, Herrera BM, Dyment DA, Montpetit A, Sadovnick AD, Knight JC, Ebers GC. HLA class I alleles tag HLA-DRB1*1501 haplotypes for differential risk in multiple sclerosis susceptibility. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13069–13074. doi: 10.1073/pnas.0801042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca GC, Ramagopalan SV, Herrera BM, Dyment DA, Lincoln MR, Montpetit A, Pugliatti M, Barnardo MC, Risch NJ, Sadovnick AD, Chao M, Sotgiu S, Hudson TJ, Ebers GC. An extremes of outcome strategy provides evidence that multiple sclerosis severity is determined by alleles at the HLA-DRB1 locus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20896–20901. doi: 10.1073/pnas.0707731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramagopalan SV, Deluca GC, Morrison KM, Herrera BM, Dyment DA, Lincoln MR, Orton SM, Chao MJ, Degenhardt A, Pugliatti M, Sadovnick AD, Sotgiu S, Ebers GC. Analysis of 45 candidate genes for disease modifying activity in multiple sclerosis. Journal of neurology. 2008;255:1215–1219. doi: 10.1007/s00415-008-0878-7. [DOI] [PubMed] [Google Scholar]
- 20.Ramagopalan SV, Morris AP, Dyment DA, Herrera BM, DeLuca GC, Lincoln MR, Orton SM, Chao MJ, Sadovnick AD, Ebers GC. The inheritance of resistance alleles in multiple sclerosis. PLoS genetics. 2007;3:1607–1613. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangalam AK, Khare M, Krco C, Rodriguez M, David C. Identification of T cell epitopes on human proteolipid protein and induction of experimental autoimmune encephalomyelitis in HLA class II-transgenic mice. Eur J Immunol. 2004;34:280–290. doi: 10.1002/eji.200324597. [DOI] [PubMed] [Google Scholar]
- 22.Klehmet J, Shive C, Guardia-Wolff R, Petersen I, Spack EG, Boehm BO, Weissert R, Forsthuber TG. T cell epitope spreading to myelin oligodendrocyte glycoprotein in HLA-DR4 transgenic mice during experimental autoimmune encephalomyelitis. Clin Immunol. 2004;111:53–60. doi: 10.1016/j.clim.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Madsen LS, Andersson EC, Jansson L, krogsgaard M, Andersen CB, Engberg J, Strominger JL, Svejgaard A, Hjorth JP, Holmdahl R, Wucherpfennig KW, Fugger L. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet. 1999;23:343–347. doi: 10.1038/15525. [DOI] [PubMed] [Google Scholar]
- 24.Khare M, Mangalam A, Rodriguez M, David CS. HLA DR and DQ interaction in myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in HLA class II transgenic mice. J Neuroimmunol. 2005;169:1–12. doi: 10.1016/j.jneuroim.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Mangalam AK, Rajagopalan G, Taneja V, David CS. HLA class II transgenic mice mimic human inflammatory diseases. Advances in immunology. 2008;97:65–147. doi: 10.1016/S0065-2776(08)00002-3. [DOI] [PubMed] [Google Scholar]
- 26.Amirzargar A, Mytilineos J, Yousefipour A, Farjadian S, Scherer S, Opelz G, Ghaderi A. HLA class II (DRB1, DQA1 and DQB1) associated genetic susceptibility in Iranian multiple sclerosis (MS) patients. Eur J Immunogenet. 1998;25:297–301. doi: 10.1046/j.1365-2370.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 27.Herrera BM, Ebers GC. Progress in deciphering the genetics of multiple sclerosis. Curr Opin Neurol. 2003;16:253–258. doi: 10.1097/01.wco.0000073924.19076.bb. [DOI] [PubMed] [Google Scholar]
- 28.Das P, Drescher KM, Geluk A, Bradley DS, Rodriguez M, David CS. Complementation between specific HLA-DR and HLA-DQ genes in transgenic mice determines susceptibility to experimental autoimmune encephalomyelitis. Hum Immunol. 2000;61:279–289. doi: 10.1016/s0198-8859(99)00135-4. [DOI] [PubMed] [Google Scholar]
- 29.Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, Lincoln RR, Swerdlin A, Mignot E, Lin L, Goodin D, Erlich HA, Schmidt S, Thomson G, Reich DE, Pericak-Vance MA, Haines JL, Hauser SL. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serjeantson SW, Gao X, Hawkins BR, Higgins DA, Yu YL. Novel HLA-DR2-related haplotypes in Hong Kong Chinese implicate the DQB1*0602 allele in susceptibility to multiple sclerosis. Eur J Immunogenet. 1992;19:11–19. doi: 10.1111/j.1744-313x.1992.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 31.Mangalam A, Luckey D, Basal E, Behrens M, Rodriguez M, David C. HLA-DQ6 (DQB1*0601)-restricted T cells protect against experimental autoimmune encephalomyelitis in HLA-DR3.DQ6 double-transgenic mice by generating anti-inflammatory IFN-gamma. J Immunol. 2008;180:7747–7756. doi: 10.4049/jimmunol.180.11.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley DS, Nabozny GH, Cheng S, Zhou P, Griffiths MM, Luthra HS, David CS. HLA-DQB1 polymorphism determines incidence, onset, and severity of collagen-induced arthritis in transgenic mice. Implications in human rheumatoid arthritis. J Clin Invest. 1997;100:2227–2234. doi: 10.1172/JCI119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss G, Vignali DA, Schonrich G, Hammerling GJ. Negative and positive selection by HLA-DR3(DRw17) molecules in transgenic mice. Immunogenetics. 1994;40:104–108. [PubMed] [Google Scholar]
- 34.Lampson LA, Levy R. Two populations of Ia-like molecules on a human B cell line. Journal Of Immunology (Baltimore, Md: 1950) 1980;125:293–299. [PubMed] [Google Scholar]
- 35.Jain R, Tartar DM, Gregg RK, Divekar RD, Bell JJ, Lee HH, Yu P, Ellis JS, Hoeman CM, Franklin CL, Zaghouani H. Innocuous IFN{gamma} induced by adjuvant-free antigen restores normoglycemia in NOD mice through inhibition of IL-17 production. J Exp Med. 2008 doi: 10.1084/jem.20071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olerup O, Hillert J, Fredrikson S, Olsson T, Kam-Hansen S, Moller E, Carlsson B, Wallin J. Primarily chronic progressive and relapsing/remitting multiple sclerosis: two immunogenetically distinct disease entities. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7113–7117. doi: 10.1073/pnas.86.18.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrosu MG, Muntoni F, Murru MR, Costa G, Pischedda MP, Pirastu M, Sotgiu S, Rosati G, Cianchetti C. HLA-DQB1 genotype in Sardinian multiple sclerosis: evidence for a key role of DQB1 *0201 and *0302 alleles. Neurology. 1992;42:883–886. doi: 10.1212/wnl.42.4.883. [DOI] [PubMed] [Google Scholar]
- 38.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 40.Carlson T, Kroenke M, Rao P, Lane TE, Segal B. The Th17-ELR+ CXC chemokine pathway is essential for the development of central nervous system autoimmune disease. J Exp Med. 2008;205:811–823. doi: 10.1084/jem.20072404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lees JR, Iwakura Y, Russell JH. Host T cells are the main producers of IL-17 within the central nervous system during initiation of experimental autoimmune encephalomyelitis induced by adoptive transfer of Th1 cell lines. J Immunol. 2008;180:8066–8072. doi: 10.4049/jimmunol.180.12.8066. [DOI] [PubMed] [Google Scholar]
- 42.Gold R, Hartung HP, Lassmann H. T-cell apoptosis in autoimmune diseases: termination of inflammation in the nervous system and other sites with specialized immune-defense mechanisms. Trends in neurosciences. 1997;20:399–404. doi: 10.1016/S0166-2236(97)01079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pender MP, Rist MJ. Apoptosis of inflammatory cells in immune control of the nervous system: role of glia. Glia. 2001;36:137–144. doi: 10.1002/glia.1103. [DOI] [PubMed] [Google Scholar]