Abstract
Exit from mitosis in Saccharomyces cerevisiae is triggered by activation of the phosphatase Cdc14. Throughout interphase and early mitosis, Cdc14 is sequestered in the nucleolus by its inhibitor Cfi1/Net1. In anaphase, the Cdc Fourteen Early Anaphase Release (FEAR) network and the Mitotic Exit Network (MEN) coordinately trigger the release of Cdc14 from the nucleolus. Here we show that the FEAR network component Cdc5 physically associates with two other members of the pathway, the Separase Esp1 and the Esp1-binding protein Slk19. Furthermore, we find that Cdc5 physically interacts with Cdc14 and that this association is mediated by Cdc5’s Polo-box domain, a phospho-serine/phosphothreonine binding domain. Finally, we present evidence that the Cdc5–Cdc14 association is direct, further supporting the central role of Cdc5 in Cdc14 localization.
Keywords: Cdc5, Cdc14, Esp1, Slk19, FEAR network, mitotic exit
Introduction
Cyclin-Dependent Kinases (CDKs) drive entry into the cell cycle, progression through S phase, and entry into mitosis. To exit from mitosis, however, cells must attenuate CDK activity and reverse many of the phosphorylation events carried out by CDKs.1 In the budding yeast Saccharomyces cerevisiae, the protein phosphatase Cdc14 triggers exit from mitosis by dephosphorylating mitotic CDK targets and by promoting CDK inactivation.1–5 Control of Cdc14 activity is therefore the primary mechanism by which cells regulate mitotic exit.
During G1, S, G2 and early M phase, Cdc14 is sequestered in the nucleolus by its inhibitor Cfi1/Net1.2,6–8 Upon entry into anaphase, two signaling pathways, the Cdc Fourteen Early Anaphase Release (FEAR) network and the Mitotic Exit Network (MEN) trigger the release of Cdc14 from the nucleolus, thereby allowing the phosphatase to access its targets.2,8–11 The FEAR network-dependent release of Cdc14 is important, but not essential, for mitotic exit and occurs early during anaphase. Furthermore, the FEAR network-induced release of Cdc14 is transient and, in the absence of a functional MEN, Cdc14 returns to the nucleolus.10–12 Why the FEAR network cannot sustain the release of Cdc14 from the nucleolus in the absence of MEN activity is unclear. The MEN-dependent release of Cdc14 occurs during late anaphase and is essential for exit from mitosis, as MEN mutants arrest in late anaphase with high mitotic CDK activity.1
Cdc14 released by the FEAR network has multiple functions, including the activation of the MEN,10,13 the segregation of the rDNA locus,14,15 the translocation of chromosomal passenger proteins to the mitotic spindle,16 the regulation of anaphase spindle dynamics 17,18 and the positioning of the nucleus.19 These functions of Cdc14 are important for timely progression through anaphase and for the generation of viable daughter cells, as FEAR network mutants exhibit a 15–20 minute anaphase delay and 20% of FEAR network mutant cells lose viability during anaphase.10,14
The FEAR network is composed of Esp1, the protease that triggers sister chromatid separation,10,20 the Esp1-associated protein Slk19,10,20 the Polo-like kinase Cdc5,10–12 Clb1/2-CDKs (mitotic-CDKs),21 protein phosphatase type 2A (PP2A) associated with its targeting subunit Cdc55,22–24 the replication fork block protein Fob1, and a protein of unknown function, Spo12.10,25 How the FEAR network promotes Cdc14 activation is partly understood. Activation of the FEAR network is triggered by the ubiquitin-mediated proteolysis of the Esp1 inhibitor, Pds1, at the metaphase-anaphase transition. The ubiquitylation of Pds1 is catalyzed by an E3 ubiquitin ligase complex, the Anaphase Promoting Complex/Cyclosome, in association with a specificity factor, Cdc20.26–29 Following Pds1 destruction, Esp1 is thought to downregulate PP2A-Cdc55 activity,23 leading to the stable phosphorylation of Cfi1/Net1 by Clb-CDKs and subsequent dissociation of Cdc14-Cfi1/Net1 complexes.21 Esp1 and Slk19 also contribute to Spo12 activation in anaphase (B. Tomson, personal communication), which may further destabilize Cdc14-Cfi1/Net1 complexes.1
Cdc5 is necessary and its overexpression is sufficient to induce Cdc14 release from the nucleolus in vivo.30 However, how Cdc5 brings about this event is not well understood. We find that Cdc5 physically associates with Cdc14, as well as with the FEAR network components Slk19 and Esp1. Moreover, we show that Cdc5’s Polo-box domain, a phospho-serine/phospho-threonine binding domain, is sufficient to interact with Cdc14 and present evidence that it can do so directly. Our results therefore suggest that a direct interaction between Cdc5 and Cdc14 is involved in mediating Cdc5’s regulation of the phosphatase.
Results
Cdc5 physically interacts with Esp1 and Slk19
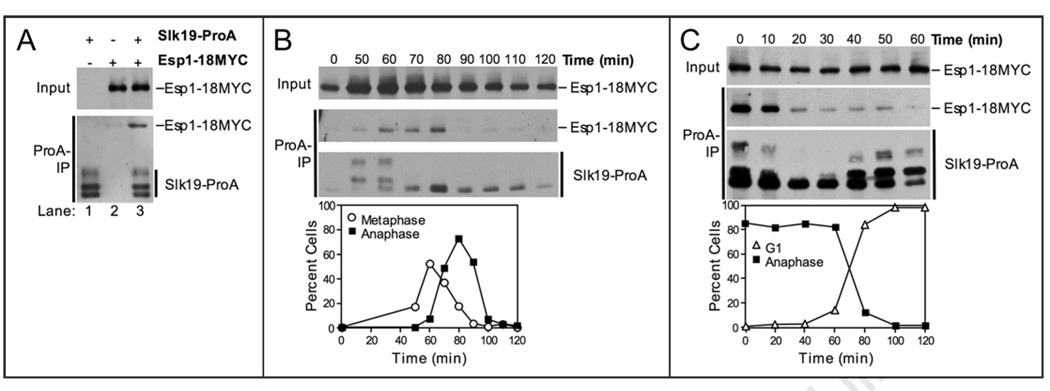
Cdc5 is thought to function downstream of, or in parallel to, the Esp1-Slk19 branch in the FEAR network. Before we examined whether Cdc5 can interact with these FEAR network components, we further characterized the Esp1-Slk19 association.20 For this experiment, Slk19 and Esp1 were C-terminally fused to Protein A (ProA) and 18MYC, respectively. Consistent with previous studies,20 we found that Slk19-ProA coimmunoprecipitated Esp1-18MYC (Fig. 1A). Multiple fragments of Slk19-ProA can be detected because the C-terminal cleavage product of Slk19, Slk19 (78–821), undergoes phosphorylation dependent mobility shifts.20,31
Figure 1.
The Esp1-Slk19 interaction is cell cycle regulated. (A) Western blots showing that Esp1-18MYC coimmunoprecipitates with Slk19-ProA. The top panel (Input) shows the amount of Esp1-18MYC in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Esp1-18MYC coimmunoprecipitated with Slk19-ProA (lane 3). ProA-tagged proteins were detected in anti-MYC immunoblots because ProA binds to primary and secondary antibodies. The following strains were used (from left to right): A8196, A3072, A8444. (B) Cells carrying Slk19-ProA and Esp1-18MYC fusions (A8444) were arrested in G1 in YEPD with alpha factor (5 µg/ml) at room temperature. After 2.5 hours, cells were washed with 10 volumes YEP and released into pheromone-free YEPD media at room temperature. After 100 minutes, alpha factor (10 µg/ml) was re-added to prevent cells from entering a new cell cycle. Samples were taken at the indicated times to determine the ability of Esp1 and Slk19 to interact (top) and the percentage of metaphase (open circles) and anaphase (closed squares) spindles (bottom). The Western blots show the amount of Esp1-18MYC in whole cell extracts (Input, top), the amount of Esp1-18MYC coimmunoprecipitated with Slk19-ProA (ProA-IP, middle), and the amount of Slk19-ProA immunoprecipitated (ProA-IP, bottom) at the indicated times after release from the G1 arrest. (C) cdc15-2 cells carrying Slk19-ProA and Esp1-18MYC (A11605) fusions were arrested in G1 in YEPD with alpha factor (5 µg/ml) for 2.5 hours at room temperature. Cells were washed with 10 volumes YEP and released into pheromone-free YEPD media at 37°C for 2.5 hours to arrest cells in anaphase. Cells were then released into YEPD media at room temperature in the presence of alpha factor (5 µg/ml) for cells to exit from mitosis and arrest in the following G1 stage. Samples were taken at the indicated times to determine the ability of Esp1 and Slk19 to interact (top) and the percentage of anaphase closed squares) and G1 (open triangles) spindles (bottom). The Western blots show the amount of Esp1-18MYC in whole cell extracts (Input, top), the amount of Esp1-18MYC coimmunoprecipitated with Slk19-ProA (ProA-IP, middle), and the amount of Slk19-ProA immunoprecipitated (ProA-IP, bottom) at the indicated times after release from the late anaphase arrest.
To address whether the association between Slk19 and Esp1 was occurring throughout the cell cycle, we examined complex formation between these two proteins as cells progressed through the cell cycle in a synchronous manner. Consistent with previous reports, we found that Esp1 levels increased upon release from a G1 arrest32 and that full-length Slk19 was absent in G1,31 (Fig. 1B). Full length Slk19-ProA accumulated as cells progressed through S phase and cleavage of Slk19 occurred by 70 minutes, when the majority of cells entered anaphase (Fig. 1B, bottom).
The interaction between Slk19-ProA and Esp-18MYC was maximal from 60–80 minutes after release from G1, when the majority of cells were in M-phase (Fig. 1B). During late stages of anaphase and exit from mitosis, the interaction between Slk19 and Esp1 decreased (Fig. 1B, 80–100 min). To more precisely determine when the interaction between Esp1 and Slk19 diminished, we examined the ability of Slk19-ProA to coimmunoprecipitate Esp1-18MYC in cells released from an anaphase arrest (a cdc15-2 arrest in which the MEN is inactive). The interaction between Esp1 and Slk19 decreased shortly after cells were shifted to the permissive temperature indicating that the MEN and hence Cdc14 activation was important for dissociation of the complex (Fig. 1C). We conclude that the interaction between Esp1 and Slk19 is cell cycle regulated, being maximal during M-phase and minimal during mitotic exit.
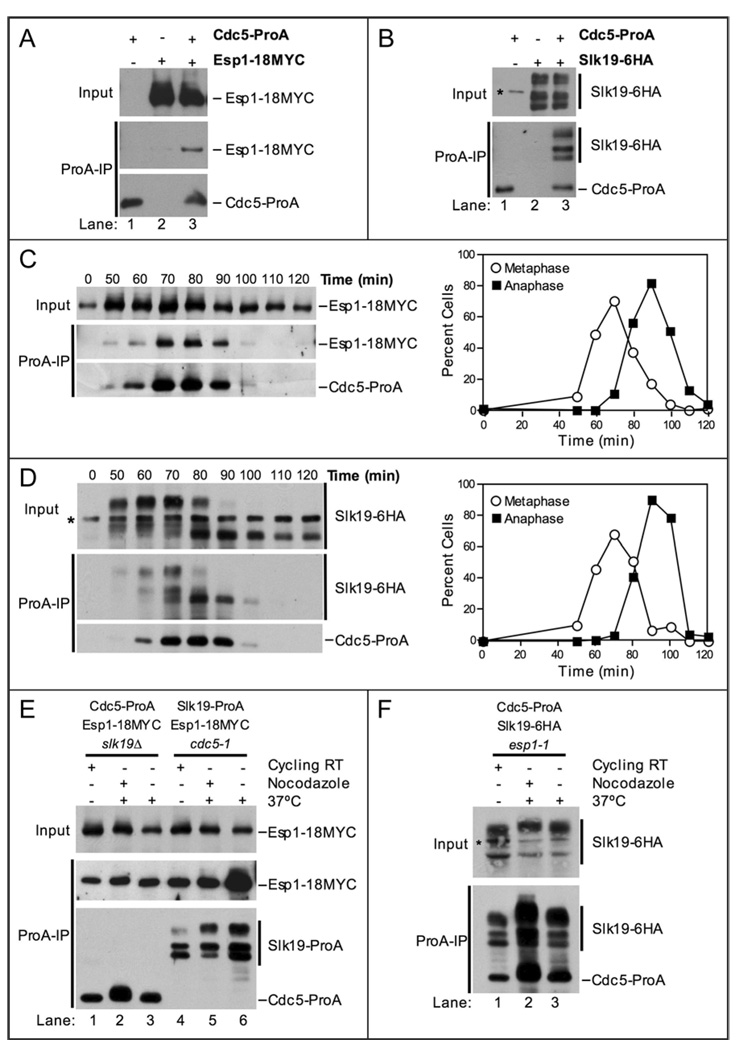
To determined whether Cdc5 associates with either Slk19 or Esp1, we tagged Cdc5 and Slk19 at their C-termini with Protein A (ProA) and 6HA, respectively, and Esp1 with 18MYC. Immunoprecipitation of Cdc5-ProA resulted in the coimmunoprecipitation of Esp1-18MYC (Fig. 2A). Similarly, Slk19-6HA coimmunoprecipitated with Cdc5-ProA (Fig. 2B). All three primary fragments of Slk19-6HA coimmunoprecipitated with Cdc5-ProA (Fig. 2B). The three primary fragments of Slk19 are the full-length form (the slowest migrating form), the hyper-phosphorylated C-terminal cleavage product (the second fastest migrating form), and the hypo-phosphorylated C-terminal cleavage product (the fastest migrating form) (Fig. 2B). We conclude that Esp1, Slk19 and Cdc5 are able to form complexes with each other in cycling cells.
Figure 2.
Cdc5 physically interacts with Esp1 and Slk19. (A) Western blots showing that Esp1-18MYC coimmunoprecipitates with Cdc5-ProA from whole cell extracts. The top panel (Input) shows the amount of Esp1-18MYC in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Esp1-18MYC coimmunoprecipitated with Cdc5-ProA (lane 3). The following strains were used (from left to right): A5366, A3072, A8450. (B) Western blots showing that Slk19-6HA coimmunoprecipitates with Cdc5-ProA from whole cell extracts. The top panel (Input) shows the amount of Slk19-6HA in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Slk19-6HA coimmunoprecipitated with Cdc5-ProA (lane 3). The slowest migrating form of Slk19-6HA is the full-length protein, while the fastest migrating form is the C-terminal cleavage product, Slk19 (78–821)-6HA. The C-terminal cleavage product migrates as a doublet due to phosphorylation-dependent mobility shift (middle band). * denotes a protein that cross-reacts with the anti-HA.11 antibody. The following strains were used (from left to right): A5366, A5070, A8661. (C) Cells carrying Cdc5-ProA, Esp1-18MYC and Cdc14-3HA fusions (A8450) were grown and analyzed as described in Figure 1B. Samples were taken at the indicated times to determine the ability of Esp1 and Cdc5 to interact (left) and the percentage of metaphase (open circles) and anaphase (closed squares) spindles (right). The Western blots show the amount of Esp1-18MYC in whole cell extracts (Input, top), the amount of Esp1-18MYC coimmunoprecipitated with Cdc5-ProA (ProA-IP, middle), and the amount of Cdc5-ProA immunoprecipitated (ProA-IP, bottom) at the indicated times after release from the G1 arrest. (D) Cells carrying Cdc5-ProA and Slk19-6HA fusions (A8662) were grown and analyzed as described in Figure 1B. Samples were taken at the indicated times to determine the ability of Slk19 and Cdc5 to interact (left) and the percentage of metaphase (open circles) and anaphase (closed squares) spindles (right). The Western blots show the amount of Slk19-6HA in whole cell extracts (Input, top), the amount of Slk19-6HA coimmunoprecipitated with Cdc5-ProA (ProA-IP, middle), and the amount of Cdc5-ProA immunoprecipitated (ProA-IP, bottom) at the indicated times after release from the G1 arrest. * denotes a protein that cross-reacts with the anti-HA.11 antibody. (E and F) Exponentially grown cells of the indicated genotype were split into 3 cultures: one culture was left untreated (Cycling RT), a second culture was treated with nocodazole (15 µg/ml) for 2 hours at room temperature (RT) and then shifted to 37°C for 1 hour (Nocodazole), and a third culture was shifted to 37°C for 2 hours (37°C). The following strains were used A10823 (lanes 1–3), A10846 (lanes 4–6) in (E) and A10848 in (F). The Western blots in (E) show that Esp1-18MYC coimmunoprecipitates with Cdc5-ProA in the absence of SLK19 (lanes 1–3) and that Esp1-18MYC coimmunoprecipitates with Slk19-ProA in the absence of Cdc5 activity (lanes 4–6). The Western blots show the amount of Esp1-18MYC in whole cell extracts (Input, top), the amount of Esp1-18MYC coimmunoprecipitated with Cdc5-ProA or Slk19-ProA (ProA-IP, middle), and the amount of Cdc5-ProA or Slk19-ProA immunoprecipitated (ProA-IP, bottom) under each condition. The Western blots in (F) show that Slk19-6HA coimmunoprecipitates with Cdc5-ProA in the absence of Esp1 activity. The top panel (Input) shows the amount of Slk19-6HA in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Slk19-6HA coimmunoprecipitated with Cdc5-ProA. * denotes a protein that cross-reacts with the anti-HA.11 antibody.
To address whether the Cdc5-Esp1 and Cdc5-Slk19 associations were cell cycle regulated, we examined these interactions using synchronized cell populations. Cdc5 is degraded by the APC/C-Cdh1 during exit from mitosis and G1.30,33–37 Cdc5-ProA was therefore absent during the pheromone-induced G1 arrest (Fig. 2C). The protein began to accumulate during G2 and reached maximal levels during metaphase and early anaphase (Fig. 2C). The ability of Cdc5-ProA to coimmunoprecipitate Esp1-18MYC correlated with the abundance of Cdc5-ProA (Fig. 2C). Similar results were obtained when we examined the interaction between Cdc5-ProA and Slk19-6HA (Fig. 2D). We conclude that the Cdc5-Esp1 and Cdc5-Slk19 interactions, as detected by coimmunoprecipitation analysis, are largely determined by the abundance of Cdc5.
The physical interactions between FEAR network components are independent of FEAR network function
Next, we examined whether the interactions between Esp1 and Slk19 required CDC5, whether the binding between Slk19 and Cdc5 needed ESP1 and whether SLK19 was important for the association between Esp1 and Cdc5. SLK19 was inactivated by deleting the gene. To inactivate the essential genes CDC5 and ESP1, we employed the temperature sensitive cdc5-1 and esp1-1 alleles, respectively. At the restrictive temperature of 37°C, the esp1-1 allele prevents anaphase onset but does not block mitotic exit,27,30,38 whereas the cdc5-1 allele does not affect anaphase onset but blocks mitotic exit.10,30,35,37 To eliminate effects brought about by arresting cells in different cell cycle stages, we evaluated each interaction under 3 conditions: in cultures grown at room temperature, in cells arrested in metaphase using the microtubule depolymerizing drug nocodazole prior to a temperature shift to 37°C, and in cultures incubated at 37°C.
Deletion of SLK19 did not affect the association between Cdc5-ProA and Esp1-18MYC under any conditions tested (Fig. 2E). Similarly, inactivation of CDC5 did not affect the interaction between Slk19-ProA and Esp1-18Myc (Fig. 2E). The association between Cdc5-ProA and Slk19-6HA was not affected by the esp1-1 mutation either (Fig. 2F). These findings indicate that the physical associations among FEAR network components are independent FEAR network function. However, we cannot exclude the possibility that the inactivated Cdc5-1 and Esp1-1 proteins, despite being non-functional, contribute to the observed associations because we do not know if the proteins are degraded at 37°C.
Cdc5 physically associates with Cdc14
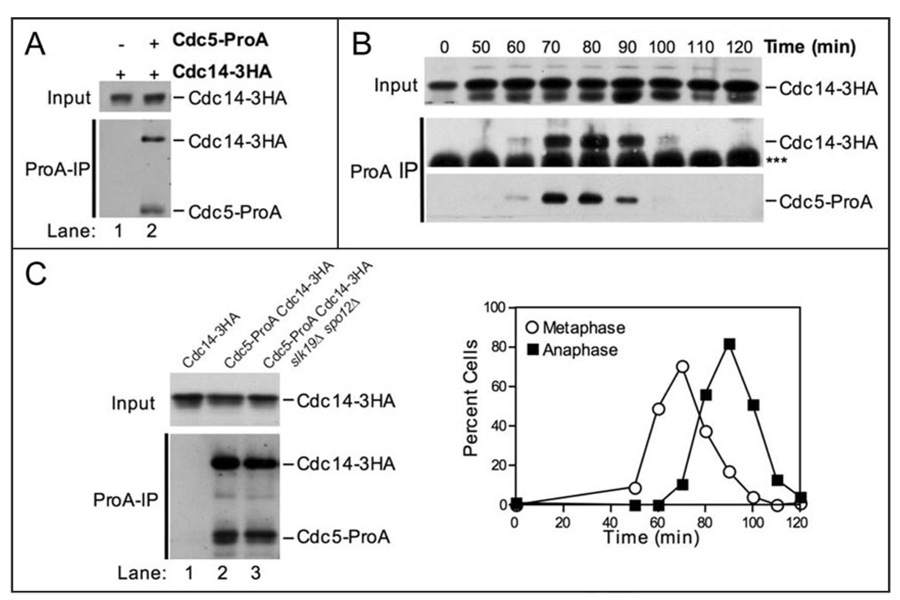
Cdc5 is capable of disassembling Cdc14-Cfi1/Net1 complexes in vitro and is required maintain Cdc14 out of the nucleolus in vivo.30,39 We therefore tested whether Cdc5-ProA can stably associate with Cdc14. As shown in Figure 3A, Cdc5-ProA coimmunoprecipitated Cdc14-3HA from cycling cells. Like the Cdc5-Esp1 and Cdc5-Slk19 interactions, ability of Cdc5 to interact with Cdc14 throughout the cell cycle was determined by Cdc5 levels (Fig. 3B). This association was also independent of FEAR network function, as Cdc5 coimmunoprecipitated Cdc14 in the absence of both SLK19 and SPO12 (Fig. 3C). We conclude that Cdc5’s interaction with Esp1, Slk19 and Cdc14 in extracts is largely determined by Cdc5 levels and is independent of FEAR network function.
Figure 3.
Cdc5 physically interacts with Cdc14. (A) Western blots showing that Cdc14-3HA coimmunoprecipitates with Cdc5-ProA from whole cell extracts. The top panel (Input) shows the amount of Cdc14-3HA in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Cdc14-3HA coimmunoprecipitated with Cdc5-ProA (lane 2). The following strains were used (from left to right): A1411, A7216. (B) Immunoprecipitates of Cdc5-ProA from the experiment shown in Figure 2C were probed with an HA.11 antibody to determine the amount of Cdc14-3HA coimmunoprecipitated. The Western blots show the amount of Cdc14-3HA in whole cell extracts (Input, top), the amount of Cdc14-3HA coimmunoprecipitated with Cdc5-ProA (ProA-IP, middle), and the amount of Cdc5-ProA immunoprecipitated (ProA-IP, bottom) at the indicated times after release from the G1 arrest. The bottom panel is a lighter exposure of the panel shown in Figure 2C. *** denotes the IgG heavy chain. The graph showing the percentage of cells in metaphase and anaphase is the same as that shown in Figure 2C. (C) Western blots showing that Cdc14-3HA coimmunoprecipitates with Cdc5-ProA in the absence of SLK19 and SPO12. The top panel (Input) shows the amount of Cdc14-3HA in whole cell extracts. The bottom panel (ProA-IP) shows the amount of Cdc14-3HA coimmunoprecipitated with Cdc5-ProA. The following strains were used (from left to right): A1411, A7216, A12424.
The polo-box domain of Cdc5 is sufficient to bind Cdc14
The C-terminus of Cdc5, like other Polo-like kinases (Plks), contains a phospho-serine/phospho-threonine binding domain called the Polo-box Domain (PBD).40–42 The PBD is thought to bind Plk substrates after the substrates undergo “priming” phosphorylation by another protein kinase.42,43 Phospho-specific binding of the PBD in human Plk1 requires two highly conserved residues, His-538 and Lys-540, that function as molecular pincers, making direct contact with the phosphate group on the substrate.43 A “pincer” mutant Plk1 PBD containing the His-538-Ala/Lys-540-Met substitutions is severely reduced in its ability to bind phosphorylated ligands.43
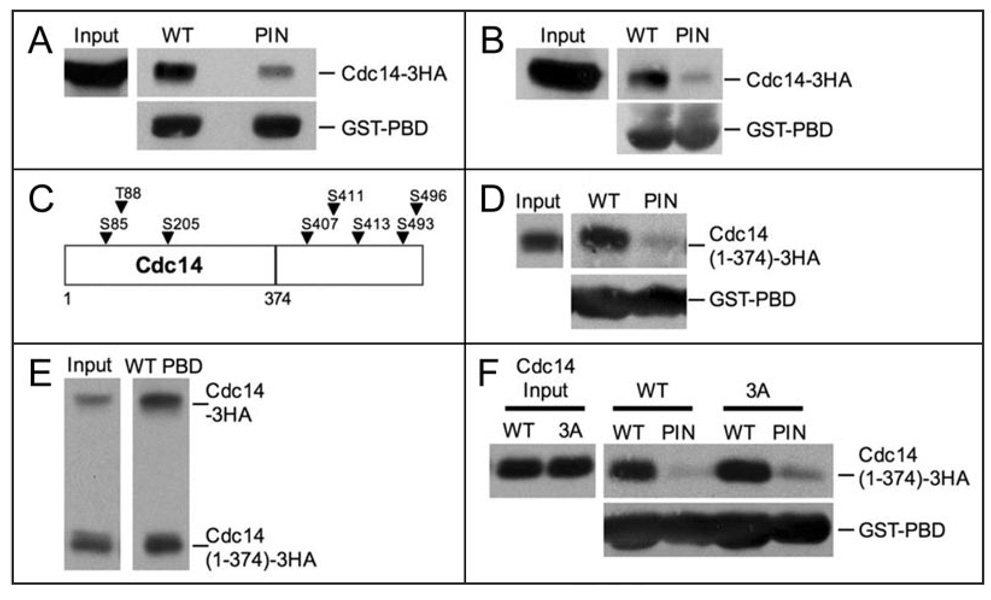
We tested if the PBD of Cdc5 was sufficient to associate with Cdc14 and if this interaction was dependent on the His/Lys phosphate pincer. For these experiments, recombinant wild-type (WT) PBD-GST or pincer (PIN) mutant PBD-GST fusions were purified from E. coli and used to precipitate Cdc14-3HA from yeast extracts. Wild-type PBD-GST was more efficient at binding to Cdc14-3HA than pincer PBD-GST (Fig. 4A). These results show that the Cdc5-PBD is sufficient to associate with Cdc14-3HA. In addition, the interaction between the PBD and Cdc14 may be phospho-dependent since it is reduced by mutation of the PBD His/Lys phosphate pincer.
Figure 4.
The Polo-box domain (PBD) of Cdc5 binds to Cdc14. (A) Whole cell extracts from cells carrying Cdc14-3HA fusion (A1411) were incubated with WT or PIN PBD-GST fusions immobilized on glutathione (GSH) beads. Western blots show the amount of Cdc14-3HA in whole cell extracts (Input, top left), the amount of Cdc14-3HA coprecipitated with WT or PIN PBD-loaded GSH beads (top right), and the amount of WT or PIN PBD-GST fusion immobilized on GSH beads (bottom). (B) Lysates from cells carrying a Cdc14-3HA fusion (A1411) were prepared under denaturing conditions as described in Materials and methods. WT or PIN PBD-GST fusions immobilized on GSH beads were incubated with “denatured” extract. Western blots show the amount of Cdc14-3HA in “denatured” extracts (Input, top left), the amount of Cdc14-3HA coprecipitated with WT or PIN PBD-loaded GSH beads (top right), and the amount of WT or PIN PBD-GST fusion immobilized on the GSH beads (bottom). (C) Schematic diagram of Cdc14 showing the locations of putative PBD binding sites (arrows). The line denotes where the Cdc14(1–374) truncation ends. (D) WT or PIN PBD-GST fusions immobilized on GSH beads were incubated with “denatured” extract from cells carrying a Cdc14(1–374)-3HA fusion (A19448). Western blots show the amount of Cdc14(1–374)-3HA in “denatured” extracts (Input, top left), the amount of Cdc14(1–374)-3HA pulled-down with WT or PIN PBD-loaded GSH beads (top right), and the amount of WT or PIN PBD-GST fusion immobilized on the GSH beads (bottom). (E) WT PBD-GST fusions immobilized on GSH beads were incubated with “denatured” extract from diploid cells carrying a Cdc14-3HA and a Cdc14(1–374)-3HA fusion (A19422). Western blots show the amount of Cdc14-3HA and Cdc14(1–374)-3HA in “denatured” extracts (Input, left), the amount of Cdc14-3HA and Cdc14(1–374)-3HA coprecipitated with WT PBD-loaded GSH beads (right). (F) WT or PIN PBD-GST fusions immobilized on GSH beads were incubated with “denatured” extract from cdc14Δ cells carrying Cdc14(1–374)-3HA (A21384) or Cdc14(1–374, 3A)-3HA fusions (A21386) on centromeric plasmids. Western blots show the amount of Cdc14(1–374)-3HA and Cdc14(1–374, 3A)-3HA in “denatured” extracts (Input, top left), the amount of Cdc14(1–374)-3HA and Cdc14(1–374, 3A)-3HA precipitated with WT or PIN PBD-loaded GSH beads (top right), and the amount of WT or PIN PBD-GST fusion immobilized on the GSH beads.
To examine whether the interaction between the Cdc5 PBD-GST fusion and Cdc14 was direct, we asked whether recombinant PBD-GST could precipitate Cdc14-3HA from “denatured” yeast extracts (see Material and Methods). Wild-type PBD-GST precipitated Cdc14-3HA from extracts previously boiled in 2% SDS followed by a 25 fold dilution in hypertonic buffer (Fig. 4B). The PBD carrying the pincer mutation did not interact with Cdc14-3HA efficiently (Fig. 4B). These experiments suggest that the Cdc5-PBD directly binds to Cdc14 in a phospho-dependent manner.
The N-terminus of Cdc14 is sufficient to bind to Cdc5’s PBD
Next we wished to identify the region within Cdc14 that binds to the Cdc5 PBD. The optimal phosphopeptide binding motif recognized by the Cdc5 PBD is [Ala-Ala/Thr-Ser-pSer/pThr-Pro] (“p” denotes phosphorylated residues). The minimal phosphopeptide motif is [Ser-pSer/pThr].43 Cdc14 does not contain an optimal Cdc5-PBD binding motif, but harbors at least 8 minimal motifs (Fig. 4C). 5 of the minimal Cdc5-PBD binding motifs are located in the C-terminus of Cdc14, which has previously been shown to be dispensable for Cdc14 function.44 Deletion of the C-terminus of Cdc14 (Cdc14 (1–374)) did not abolish the interaction between the Cdc5 PBD and Cdc14 (Fig. 4D). Binding was not dramatically reduced compared to wild-type when the interaction between the Cdc5-PBD and full length and truncated Cdc14 was measured in the same extract either (Fig. 4E), indicating that the C-terminus of Cdc14 is dispensable for Cdc5-PBD binding.
The N-terminus of Cdc14 contains three Ser/Thr residues that form minimal phospho-peptide binding motifs for the Cdc5 PBD: Ser85, Thr88 and Ser205 (Fig. 4C). Substituting these three sites with alanine in the cdc14(1–374) truncation (cdc14(1–374, 3A) neither affected CDC14 function nor binding to the Cdc5 PBD. When placed on a centromeric- plasmid under the CDC14 promoter, both cdc14(1–374) and cdc14(1–374, 3A) rescued the lethality of a CDC14 deletion (data not shown).44 Finally, the Cdc14(1–374, 3A) protein bound the Cdc5 PBD as efficiently as Cdc14(1–374) (Fig. 4F). Our results suggest that the PBD of Cdc5 binds to the N-terminus of Cdc14-3HA. This binding is dependent on PBD residues important for recognizing phospho-binding motifs (i.e., the His/Lys pincer), but it is not mediated by traditional PBD-binding motifs in Cdc14. We conclude that the Cdc5 PBD makes atypical contacts with Cdc14.
Cdc14 function is in part controlled by its C-terminus
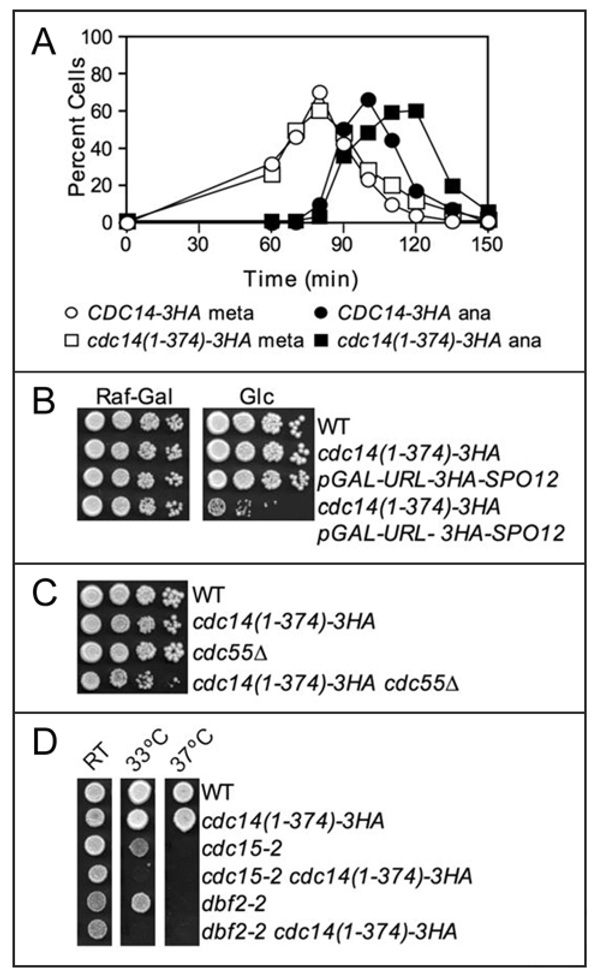
Although the cdc14(1–374) allele did not interfere with Cdc5 binding, we characterized its properties in more detail because we noticed that cells that expressed this construct from a centromeric-plasmid were slightly delayed in anaphase (data not shown). We therefore examined cell cycle progression in yeast strains that expressed this truncation as the sole source of CDC14. Upon release from a pheromone-induced G1 arrest, cdc14(1–374)-3HA cells entered metaphase and anaphase at the same time as wild-type cells (Fig. 5A). However, cdc14(1–374-)-3HA cells exhibited a 10–15 minute delay in exit from mitosis (Fig. 5A). This result suggests that the cdc14(1–374)-3HA allele is hypomorphic for CDC14’s functions in anaphase.
Figure 5.
The cdc14(1–374) allele is hypomorphic. (A) Cells carrying Cdc14-3HA (A1411) or Cdc14(1–374)-3HA fusions (A19448) were grown as described in Figure 1B. The percentage of cells in metaphase (open circles) and anaphase (closed squares) was determined at the indicated time points. (B) FEAR network signaling is essential in cdc14(1–374)-3HA mutants. Cells of the indicated genotype were grown in YEP media containing 2% raffinose-2% galactose, serially diluted 10-fold, and spotted on solid YEP media containing 2% raffinose-2% galactose (Raf-Gal) or 2% glucose (Glc). The plates were incubated at 30°C for 2 days. The following strains were used (from top to bottom): wild-type cells carrying a Cdc14-3HA fusion (A1411), cdc14(1–374)-3HA (A19448) cells, pGAL-URL-3HA-SPO12 cells (A12737), and pGAL-URL-3HA-SPO12 cdc14(1–374)-3HA (A20798) cells. (C) cdc14(1–374)-3HA is not synthetically lethal with cdc55Δ. 10-fold serial dilution of wild-type (A2587), cdc14(1–374)-3HA (A19448), cdc55Δ (A15396), and cdc14(1–374)-3HA cdc55Δ (A20810) cells on solid YEPD media. Cells were grown at 30°C for 2 days. (D) cdc14(1–374)-3HA enhances the temperature sensitivity of cdc15-2 and dbf2-2 mutants. Cells of the indicated genotype were spotted on solid YEPD media and grown for 2 days at room temperature (RT), 33°C, or 37°C. The following strains were used (from top to bottom): A1411, A19448, A1674, A19765, A2505, A20873. A1411, A1674 and A2505 carry a Cdc14-3HA fusion.
To characterize which aspects of Cdc14 function were impaired in the cdc14(1–374) mutant, we examined whether the truncation allele exhibited genetic interactions with FEAR network or MEN mutants. The cdc14(1–374)-3HA allele caused lethality when combined with a deletion of SLK19 or SPO12 or with the temperature-sensitive esp1-1 or cdc5-1 alleles (data shown). To confirm the synthetic lethality of cdc14(1–374)-3HA spo12Δ double mutants, we used a conditional depletion allele SPO12, pGAL-URL-3HA-SPO12.30 URL-3HA-Spo12 is continuously degraded due to the N-end rule pathway 45 and is rapidly destabilized when transcription from GAL1–10 promoter is repressed glucose.30 As shown in Figure 5B, cdc14(1–374)-3HA pGAL-URL-3HA-SPO12 double mutants were not viable on glucose containing media, but could proliferate on galactose containing media. The synthetic lethality of cdc14(1–374)-3HA with FEAR network mutants was specific to activators of this network since cdc14(1–374)-3HA was not synthetically lethal with the deletion CDC55, a FEAR network inhibitor (Fig. 5C). However, we do note that cdc14(1–374)-3HA cdc55Δ double mutants exhibited a mild proliferation defect compared to either single mutant.
The cdc14(1–374)-3HA allele also exhibited genetic interactions with hypomorphic MEN mutations. Deletion of the MEN activator LTE1 or introduction of a temperature sensitive allele of the MEN GTPase Tem1 was lethal in cdc14(1–374)-3HA cells (data not shown). The cdc14(1–374)-3HA allele lowered the restrictive temperature of temperature-sensitive cdc15-2 dbf2-2 mutants (Fig. 5D). Together, these data indicate that the cdc14(1–374)-3HA allele is hypomorphic and enhances the mitotic exit defects of both FEAR network and MEN mutants.
The C-terminus of Cdc14 is important for controlling its sub-cellular localization
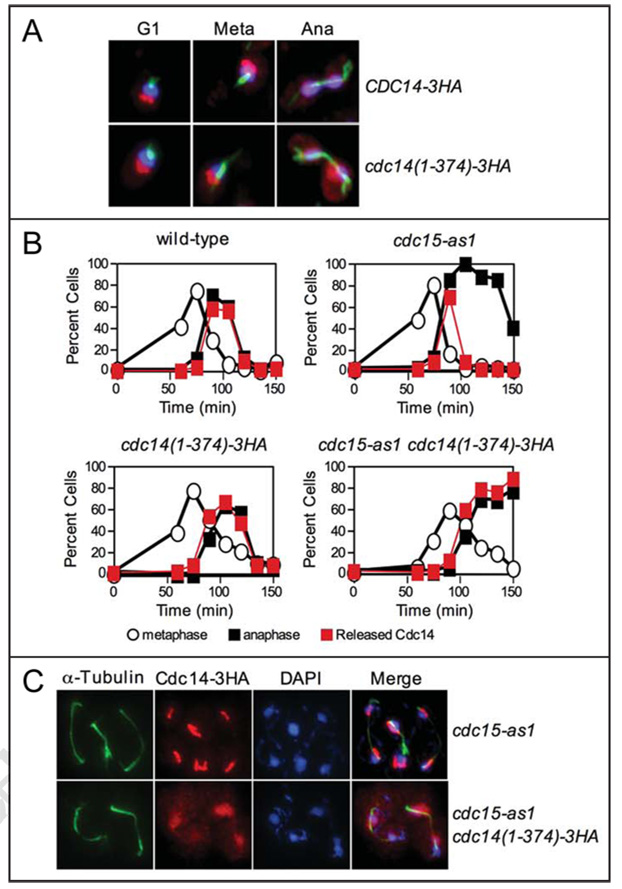
In wild-type cells, the sub-cellular localization of the Cdc14(1–374)-3HA truncation was similar to that of wild-type Cdc14. Cdc14(1–374) localized to the nucleolus during early stages of the cell cycle and was found released from the nucleolus during anaphase (Fig. 6A). However in MEN mutants, Cdc14(1–374) localization was abnormal. To inactivate the MEN, we used an ATP analog sensitive allele of the MEN kinase CDC15 (cdc15-as1).46 Upon release from a pheromone induced G1 arrest in the presence of inhibitor, wild-type Cdc14 was transiently released from the nucleolus during early anaphase and it was found sequestered in the nucleolus in the terminal anaphase arrest of the cdc15-as1 mutant (Fig. 6B and C). The Cdc14(1–374) protein was also released from the nucleolus in early anaphase in cdc15-as1 cells, but unlike the wild-type protein, failed to return to the nucleolus as cdc15-as1 cells entered the late anaphase arrest (Fig. 6B and C). Furthermore, whereas the transient release of wild-type Cdc14 was restricted to the nucleus, Cdc14(1–374) spread throughout the nucleus and cytoplasm (Fig. 6C and data not shown). These results suggest the C-terminus of Cdc14 is required for its return into the nucleolus in the absence of MEN activity.
Figure 6.
The C-terminus of Cdc14 is required for proper localization of Cdc14 during anaphase. (A) The subcellular localization of Cdc14-3HA (A1411) and Cdc14(1–374)-3HA (A19448) fusions in exponentially growing cells. Cdc14 is shown in red, microtubules are shown in green, and DNA is shown in blue. (B) mad1Δ cells carrying a Cdc14-3HA fusion (A2853), mad1Δ cdc15-as1 cells carrying a Cdc14-3HA fusion (A21212), mad1Δ cdc14(1–374)-3HA cells (A20792), and mad1Δ cdc15-as1 cdc14(1–374)-3HA cells (A21215) were grown and arrested in G1 as described in Figure 1B. MAD1 was deleted in these strains to ensure that the spindle assembly checkpoint did not interfere with cell cycle progression. After 2.5 hours, cells were washed with 10 volumes YEP and released into pheromone-free YEPD media containing 10 µM cdc15-as1 inhibitor (PP1 analog 8). After 100 minutes, alpha factor (10 µg/ml) was re-added to prevent cells from entering a new cell cycle. The percentage of cells with metaphase spindles (open circles), anaphase spindles (closed black squares), and Cdc14 released from the nucleolus (closed red squares) was determined at the indicated times. At least 100 cells were counted at each time point. (C) The subcellular localization of Cdc14 in mad1Δ cdc15-as1 carrying a Cdc14-3HA fusion and mad1Δ cdc15-as1 cdc14(1–374)-3HA cells 135 min after release from a G1 arrest. Cdc14 is shown in red, microtubules are shown in green, and DNA is shown in blue.
Discussion
FEAR network components physically interact
We have found that the Polo-like kinase Cdc5 robustly interacts with Esp1, Slk19 and Cdc14; and that Slk19 binds to Esp1. We could not detect interactions between Spo12 and other FEAR network components or between Spo12 and Cdc14, indicating that Spo12 either does not form complexes with these proteins or that these interactions are transient (data not shown). The Esp1 and Slk19 interaction was the only association among FEAR network components that was not solely determined by protein abundance. Binding between these two proteins was maximal during early mitosis and decreased during mitotic exit (Fig. 1B and C). The dissociation of Esp1 from Slk19 during mitotic exit raises the interesting possibility that Cdc14, which is activated during this cell cycle transition, is responsible for disrupting the Esp1-Slk19 complex.
In the coimmunoprecipitation analyses, the interactions between Cdc5 and Esp1, Slk19, or Cdc14 were determined by the abundance of Cdc5. Whether this accurately reflects the associations that occur in vivo is not clear. We found that FEAR network components coimmunoprecipitated when extracts prepared from strains carrying a single tagged FEAR network component were mixed. For example, Cdc5-ProA from one lysate can coprecipitate Esp1-18MYC from another lysate (Rahal R, unpublished observations). It is thus possible that in vivo regulatory mechanisms other than protein abundance also control the interactions among FEAR network components, but that these are absent in cell extracts. In the case Cdc5 and Esp1, the interaction between the two proteins may not solely reflect the proteins’ role(s) in the FEAR network, but may due to other cell cycle functions of these proteins. Cdc5 and Esp1 not only promote Cdc14 release from the nucleolus during early anaphase, but also function together earlier in the cell cycle to bring about the metaphase-anaphase transition. Cdc5 is thought to phosphorylate the cohesin complex subunit Scc1/Mcd1 to promote proper cleavage by Esp1.47 Identifying the residues that mediate the Cdc5-Esp1 interaction and analyzing the consequences of mutating them on Cdc14 release and other functions of Esp1 and Cdc5 will address this question.
A direct interaction between Cdc5 and Cdc14?
Cdc5 is unique among FEAR network and MEN components in that (1) its inactivation abolishes the release of Cdc14 from the nucleolus throughout anaphase10–12 and (2) overexpression of Cdc5 can promote the release of Cdc14 from the nucleolus in S phase as well as in metaphase.30,37 In vitro, Cdc5 can disrupt Cdc14-Cfi1/Net1 complexes 39 and, vivo, Cdc5 is continuously needed throughout anaphase to maintain Cdc14 in the released state.30 These results suggest that Cdc5 directly affects the binding of Cdc14 to its inhibitor. We now show that the PBD of Cdc5 can bind Cdc14 in extracts, confirming previous results by Snead et al.,48 Our observation that the PBD of Cdc5 interacts with Cdc14 in extracts that were previously denatured by boiling SDS indicates that the interaction is likely direct. The finding that depends on the His/Lys phosphate pincer strongly suggests that the Cdc5-Cdc14 interaction is phosphorylation dependent. Because of its dependence on residues important for phospho-Ser/Thr binding, we did not examine the binding between Cdc5 PBD-GST and recombinant Cdc14, which we presume lacks the necessary phosphorylated residues.
Interestingly, we found that the binding of the Cdc5 PBD to Cdc14 is likely mediated by an as-yet-uncharacterized PBD binding motif. Mutation of all sequence motifs previously implicated PBD binding did not reduce binding of Cdc14 to the Cdc5 PBD. This observation is not surprising because many of the proteins that interact with the PBD of Plk1 contain a phospho-Ser/Thr-Pro motif instead of the canonical Ser-phospho-Ser/Thr motif.49 Several such motifs exist in Cdc14. It will therefore be interesting to examine the consequences of mutating these sites on Cdc5 PBD binding in vitro and the consequences on Cdc14 function in vivo.
The C-terminus of Cdc14 is important for Cdc14 function and localization
In our examination of the PBD binding sites within Cdc14, we utilized an allele of CDC14 that lacks the last 177 amino acids of the phosphatase. This allele was capable of supporting cell proliferation when present as the sole source of CDC14 in otherwise wild-type cells, but several lines of evidence indicate that the allele is hypomorphic. First, cells carrying this allele as the sole source of CDC14 exhibit a 10–15 minute delay in exit from mitosis. Second, even though the protein is released from the nucleolus in cdc-5-as1 mutants, the protein cannot promote exit from mitosis. Finally, the mutation impairs proliferation of FEAR network and MEN mutants to the extent that double mutants are inviable or more severely growth impaired. Thus, the C-terminus encodes functions of Cdc14 that are particularly important when the FEAR network and the MEN are compromised. We also note that the observation that Cdc14(1–374) is released in cdc15-as1 mutants, but fails to promote exit from mitosis raises the interesting possibility that the MEN is required to promote Cdc14 activity, not only through releasing it from its inhibitor, but also to activate the phosphatase after it has been released. This putative activation step is not essential for wild-type Cdc14 to promote exit from mitosis because deletion of CFI1/NET1 suppresses the lethality of MEN mutants, but may be important when cells rely on the hypomorphic truncation allele to promote exit from mitosis.
The C-terminus of Cdc14 also appears important for accurate localization of the phosphatase. In wild-type cells progressing through the cell cycle in a synchronous manner, Cdc14(1–374) localization was largely normal. However, in a cdc15-as1 mutant, the truncated protein failed to return into the nucleolus after release during early anaphase. The sustained release of Cdc14(1–374) was not due to defects in the organization of the nucleolus, as Nop1 localization was unaffected under these conditions (R. R., unpublished observations). The observation that the Cdc14(1–374) leaves the nucleolus and returns normally in wild-type cells argues against a general structural defect that impairs Cdc14’s ability to bind to Cfi1/Net1. In vitro, p-nitrophenyl phosphate (PNPP) dephosphorylation by Cdc14 or Cdc14(1–374) was equally inhibited by Cfi1/Net1,7 further excluding this possibility. It is not yet clear if the FEAR network or other novel mechanisms are responsible for causing the sustained release of the truncated protein in cdc15-as1 cells because FEAR network mutants are synthetically lethal with this allele. We attempted to test a conditional depletion allele of Cdc5 by placing Cdc5 under the GAL1–10 promoter, but pGAL-CDC5 was synthetically lethal with cdc14(1–374)-3HA. In the future, it will be important to construct conditional alleles of other FEAR network regulators to determine whether the sustained release of Cdc14(1–374) in MEN mutants is FEAR network dependent.
What could be the basis for the failure of the Cdc14(1–374) truncation to return into the nucleolus in MEN mutants? It is possible that the C-terminus of Cdc14 is required for regulatory signals to induce Cdc14’s return into the nucleolus in the absence of MEN activity. Alternatively, high Cdc14 phosphatase activity may be required for Cdc14 to cause its own return into the nucleolus after FEAR network signaling ceases. Although the phosphatase activity of Cdc14(1–374) towards protein substrates is similar to full-length Cdc14, its activity towards PNPP reduced 10 fold.44 Our in vivo data certainly indicate that the truncated Cdc14 protein is not fully functional. Interestingly, when both full length and truncated Cdc14 are expressed simultaneously, Cdc14(1–374) does return into the nucleolus in cells lacking Cdc15 function (R. R. unpublished observations). Although other interpretations are possible, this result is consistent with the idea that wild-type levels of Cdc14 phosphatase activity are needed for the resequestration of Cdc14 into the nucleolus in MEN mutants. Identifying the molecular basis for the abnormal localization of Cdc14(1–374) will help determine how Cdc14 is controlled during anaphase.
Materials and Methods
Strains
All strains are derivatives of W303 (K699). Strain genotypes can be found in Table 1. Esp1-18MYC was described in ref. 39 and Cdc14-3HA in ref. 2. Cdc5-ProA, Slk19-ProA and Slk19-6HA were generated by a PCR-based method.50 All tagged constructs were functional as judged by the fact that cells carrying the tagged proteins as the sole source of the protein did not exhibit a discernable proliferation defect.
Table 1.
Strains used in this study
| Strain number |
Relevant genotype |
|---|---|
| 1411 | MATa CDC14-3HA |
| 1674 | MATa cdc15-2 CDC14-3HA |
| 2505 | MATa dbf2-2 CDC14-3HA |
| 2587 | MATa |
| 2853 | MATa mad1::URA3 CDC14-3HA |
| 3072 | MATa ESP1-18MYC-TRP1 |
| 5070 | MATa slk19::HIS3 ura3::Slk19-6HA |
| 5366 | MATa CDC5-TEV-ProA-KANMX6 |
| 7216 | MATa CDC14-3HA CDC5-TEV-ProA-KANMX6 |
| 8196 | MATa SLK19-TEV-ProA-KANMX6 |
| 8444 | MATa SLK19-TEV-ProA-KANMX6 ESP1-18MYC-TRP1 |
| 8450 | MATa CDC5-TEV-ProA-KANMX6 ESP1-18MYC-TRP1 |
| 8661 | MATalpha slk19::HIS3 ura3::Slk19-6HA CDC5-TEV-ProA-KANMX6 |
| 10823 | MATa slk19::HIS3 CDC5-TEV-ProA-KANMX6 ESP1-18MYC-TRP1 |
| 10846 | MATa cdc5-1 SLK19-TEV-ProA-KANMX6 ESP1-18MYC-TRP1 CDC14-3HA |
| 10848 | MATa esp1-1 slk19::HIS3 ura3::Slk19-6HA CDC5-TEV-ProA-KANMX6 |
| 11605 | MATa cdc15-2 SLK19-TEV-ProA-KANMX6 ESP1-18MYC-TRP1 |
| 12424 | MATa CDC14-3HA CDC5-TEV-ProA-KANMX6 spo12::his3 slk19::kanmx6 |
| 12737 | MATa spo12::pGAL-URL-3HA-SPO12-KANMX6 |
| 15396 | MATa cdc55::KANMX6 |
| 19422 | MATa/alpha CDC14-3HA/cdc14(1–374)-3HA::KANMX6 |
| 19448 | MATa cdc14(1–374)-3HA::KANMX6 |
| 19765 | MATa cdc15-2 cdc14(1–374)-3HA::KANMX6 |
| 20792 | MATa mad1::URA3 cdc14(1–374)-3HA::KANMX6 |
| 20798 | MATa spo12::pGAL-URL-3HA-SPO12-KANMX6 cdc14(1–374)-3HA::KANMX6 |
| 20810 | MATa cdc55::KANMX6 cdc14(1–374)-3HA::KANMX6 |
| 20873 | MATa dbf2-2 cdc14(1–374)-3HA::KANMX6 |
| 21212 | MATa mad1::URA3 cdc15-as1(L99G)::URA3 CDC14-3HA |
| 21215 | MATa mad1::URA3 cdc15-as1(L99G)::URA3 cdc14(1–374)-3HA::KANMX6 |
| 21384 | MATa cdc14::HIS3 Ycplac22-cdc14(1–374)-3HA::KanMX6,-TRP1 |
| 21386 | MATa cdc14::HIS3 Ycplac22-cdc14(1–374, S85A T88A S205A)::KANMX6,-TRP1 |
Coimmunoprecipitation analysis
Cells were grown overnight in YEPD to an OD of 0.8. 50 mls of cells were harvested, washed with 10 mM Tris (pH 7.5), frozen in liquid nitrogen, and stored at −80°C (for 1–3 days). Cell pellets were thawed on ice and resuspended in 200 µl of NP40 lysis buffer (1% NP40, 150 mM NaCl, 50 mM TRIS (pH 7.5), 1 mM dithiothreitol (DTT), 60 mM β-glycerophosphate, 1 mM NaVO3, 2 µM Microcystin-LR (EMD Biosciences), and complete EDTA-free protease inhibitor cocktail (Roche)). Cells were disrupted with ~100 µl glass beads in a FastPrep FP120 (Savant) homogenizer for 3 cycles of 45 sec (6.5 m/s). 1–5 mg of extract (in 120 µl of NP40 buffer) was used for immunoprecipitations. 24 µl of rabbit-IgG coupled dynabeads (Dynal Biotech) was added to each sample and incubated with rotation for 2 hrs at 4°C. Samples were washed five times with NP40 buffer, boiled in SDS-based sample buffer, and run on SDS-PAGE gels for subsequent Western blot analysis.
For analysis of complex formation in synchronous cultures, a 600 ml culture (OD600 0.2) was arrested in G1 with alpha factor (5 µg/ml) for 2.5 hours at room temperature in a 4 litre flask. Cells were washed with 4 litres YEP and released into 410 ml of pheromone-free YEPD media. 70 mls of cells were collected for the 0 time point (prior to release); 70 mls of cells were collected for the 50–70 min time points; and 40 mls of cells were collected for the 80–120 min time points. Alpha factor (10 µg/ml) was re-added at 100 minutes to arrest cells in the next G1 phase. Coimmunoprecipitation experiments were performed as described above for cycling cultures.
GST-PBD purification
Wild-type PBD (Cdc5 residues 357–705)-GST and pincer (His-641-Ala/Lys-643-MET) PBD-GST fusion vectors (pGEX4T1) were expressed in the E. coli Rosetta cell line (Novagen). Culture (500 mls) were grown to an OD600 of 0.7 and protein expression was induced by addition of IPTG to a final concentration of 0.4 µm for 3 hours. Bacteria were centrifuged, frozen at −80°C for 30 min, and resuspended in 15 ml TPI buffer (25 mM Tris pH 7.4, 2 mM EDTA, 1 mM DTT, 137 mM NaCl, 2.6 mM KCl, 1% v/v Triton X-100, and Roche protease inhibitor cocktail). Cells were disrupted by three passages through a French pressure cell (Sim-Aminco). Cleared lysates were incubated with 2 ml glutathione-Sepharose 4B beads (GSH beads) (Pharmacia-Biotech) and rotated for 1 hr at 4°C. GSH beads were washed 4 times with: 25 ml TPI for 10 min; 25 ml TPI for 5 min, 15 ml TPI lacking Triton X-100 for 5 min; and 15 ml TPI lacking Triton X-100 for 5 min. The beads were split into two pools: one pool was left in TPI and the other was incubated with 0.1 M reduced glutathione (pH 7.5) overnight at 4°C to elute the PBD-GST fusions.
GST pull-downs
Yeast whole cell extracts were prepared as described above. WT or PIN PBD-GST fusions immobilized on GSH beads were incubated with yeast extract for 1 hour at 4°C. Samples were washed three times with NP40 buffer, boiled in SDS-based sample buffer, and analyzed by Western blot analysis. “Denaturing” extracts were prepared according to a protocol developed by the Tansey lab (tanseylab.cshl.edu/pdf/Denat_IP_Yeast.pdf). Briefly, OD600 units worth of cells were resuspended in 50 µl SHD buffer (2% SDS, 90 mM HEPES pH 7.5, 30 mM DTT, Roche protease inhibitor cocktail) and incubated at 95°C for 10 minutes. The cell lysate was cleared and diluted with 1200 µL TNN (50 mM Tris pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% NP40) buffer. WT or PIN PBD-GST fusions immobilized on GSH beads were incubated with the “denatured” extract for 4 hours at room temperature. Samples were washed three times with TNN buffer, boiled in SDS-based sample buffer and analyzed by Western blot analysis.
Other techniques
Western blot analysis was performed described by ref. 26. Esp1-18Myc was detected using a mouse anti-Myc 9E10 antibody (Covance) at a 1:500 dilution. Cdc14-3HA and Slk19-6HA were detected using mouse anti-HA.11 antibody (Covance) at a 1:500 dilution. Sheep anti-mouse antibody conjugated to horseradish peroxidase (HRP) was used at a 1:2000 dilution. GST-PBD fusions were detected using a mouse anti-GST B14 antibody conjugated to HRP at a 1:100000 dilution (Santa Cruz Biotechnology). Indirect immunofluorescence for tubulin and Cdc14-3HA was performed as described in.2 Cells were imaged on Zeiss Axioplan 2 microscope using a Hamamatsu camera controller. Openlab 3.02 software was used for processing immunofluorescence images. At least 100 cells were counted at each time point.
Acknowledgments
We are grateful to Drew Lowery and Mike Yaffe for providing the Cdc5 PBD-GST constructs and members of the Amon Lab for their critical reading of the manuscript. This work was supported by a National Institute of Health grant GM 56800. A.A. is also investigator of the Howard Hughes Medical Institute.
References
- 1.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 2.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 3.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 4.Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 5.Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 6.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 7.Traverso EE, Baskerville C, Liu Y, Shou W, James P, Deshaies RJ, et al. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J Biol Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- 8.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, et al. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 9.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 10.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida S, Asakawa K, Toh-e A. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr Biol. 2002;12:944–950. doi: 10.1016/s0960-9822(02)00870-9. [DOI] [PubMed] [Google Scholar]
- 12.Pereira G, Manson C, Grindlay J, Schiebel E. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 14.D’Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 16.Pereira G, Schiebel E. Separase Regulates INCENP-Aurora B Anaphase Spindle Function Through Cdc14. Science. 2003 doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci USA. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross KE, Cohen-Fix O. A role for the FEAR pathway in nuclear positioning during anaphase. Dev Cell. 2004;6:729–735. doi: 10.1016/s1534-5807(04)00128-5. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, et al. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- 22.Yellman CM, Burke DJ. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Ng TY. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:80–89. doi: 10.1091/mbc.E04-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stegmeier F, Huang J, Rahal R, Zmolik J, Moazed D, Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr Biol. 2004 doi: 10.1016/j.cub.2004.03.009. In press. [DOI] [PubMed] [Google Scholar]
- 26.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 27.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 28.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 29.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 30.Visintin C, Tomson BN, Rahal R, Paulson J, Cohen M, Taunton J, et al. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev. 2008;22:79–90. doi: 10.1101/gad.1601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen S, Segal M, Clarke DJ, Reed SI. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 34.Cheng L, Hunke L, Hardy CF. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol Cell Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. Embo J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visintin R, Stegmeier F, Amon A. The role of the polo kinase cdc5 in controlling cdc14 localization. Mol Biol Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 39.Shou W, Azzam R, Chen SL, Huddleston MJ, Baskerville C, Charbonneau H, et al. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol Biol. 2002;3:3. doi: 10.1186/1471-2199-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KS, Song S, Erikson RL. The polo-box-dependent induction of ectopic septal structures by a mammalian polo kinase, plk, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14360–14365. doi: 10.1073/pnas.96.25.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 43.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 44.Taylor GS, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272:24054–24063. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- 45.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 46.D’Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, et al. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 48.Snead JL, Sullivan M, Lowery DM, Cohen MS, Zhang C, Randle DH, et al. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem Biol. 2007;14:1261–1272. doi: 10.1016/j.chembiol.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, et al. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. Embo J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]