Rapidly identifying an infectious disease outbreak is critical, both for effective initiation of public health intervention measures and timely alerting of government agencies and the general public. Surveillance capacity for such detection can be costly, and many countries lack the public health infrastructure to identify outbreaks at their earliest stages. Furthermore, there may be economic incentives for countries to not fully disclose the nature and extent of an outbreak.1 The Internet, however, is revolutionizing how epidemic intelligence is gathered, and it offers solutions to some of these challenges. Freely available Web-based sources of information may allow us to detect disease outbreaks earlier with reduced cost and increased reporting transparency.
Health surveillance using the Internet
A vast amount of real-time information about infectious disease outbreaks is found in various forms of Web-based data streams.2 These range from official public health reporting to informal news coverage to individual accounts in chat rooms and blogs.3–5 Because Web-based data sources exist outside traditional reporting channels, they are invaluable to public health agencies that depend on timely information flow across national and subnational borders. These information sources, which can be identified through Internet-based tools, are often capable of detecting the first evidence of an outbreak, especially in areas with a limited capacity for public health surveillance. For example, the World Health Organization’s Global Outbreak Alert and Response Network relies on these data for day-to-day surveillance activities.3,4 Revised international health rules have authorized the World Health Organization to act on this information to issue recommendations to prevent the spread of diseases.6
Canadians were leaders in introducing Web-based surveillance technologies to the world. In the 1990s, Health Canada created the Global Public Health Intelligence Network.7 Its software application retrieves articles that provide relevant information pertaining to the possibility of a public health emergency. Every 15 minutes, the network obtains information from news feed aggregators based on established search queries. Although automation is a key component, the Global Public Health Intelligence Network also employs trained analysts who provide essential linguistic, interpretive and analytical expertise. These data are disseminated to various public health agencies, including the World Health Organization, which then perform public health vetting of the informal report. The value of this network was demonstrated when the system identified the outbreak of severe acute respiratory syndrome in Guangdong Province, China, as early as November 20027, more than 2 months before the World Health Organization publically published details on cases of the new respiratory illness.
A parallel pioneering effort in Internet-based surveillance was started by the International Society for Infectious Diseases’ Program for Monitoring Emerging Diseases.8 Rather than rely on automated news scanning, this program draws from its membership to find, comment and disseminate reports on emerging disease threats through a freely available and open mailing list. This program is now one of the largest publicly available emerging disease and outbreak reporting systems in the world.
There are a number of online resources that deliver similar real-time intelligence on emerging infectious diseases to diverse audiences, from public health officials to international travelers on user-friendly, open-access websites. These systems combine freely available data sources and open-source software technology to create online surveillance systems. For example, HealthMap is a freely accessible, automated real-time system that monitors, organizes, integrates, filters, visualizes and disseminates online information about emerging diseases.9 The site pulls from over 20 000 sources every hour, many of which come from news aggregators such as Google News.
Search-term surveillance
Syndromic surveillance has emerged over the last decade as a new strategy for early detection of outbreaks. In this form of surveillance, efforts are focused on monitoring symptoms or other evidence of a disease, which may be identified before the diagnosis is confirmed and formally recognized.10 Unlike traditional surveillance efforts, an outbreak investigation would be triggered when certain health-related outcomes exceeded expected baseline levels. Examples of this type of surveillance include examining increases in visits to emergency departments, the volume of calls to health advice lines and the sales of prescription or over-the-counter medications. A new frontier in syndromic surveillance has emerged that uses Web-based clickstream- and keyword-searching aggregated across Internet users. This application has the opportunity to provide important insights into public health trends for a fraction of the cost. Eysenbach originally demonstrated the potential value and cost-effectiveness of such a strategy for surveillance of influenza in Canada.11 Similarly, recent efforts that used data from Google12 and Yahoo13 have shown that search query data can be harnessed as a form of collective intelligence where patterns of population-level searching mirror and may even predict disease outbreaks. Google Flu Trends, for example, now provides both public health professionals and the general population with a real-time geographically specific view of influenza search activity in the United States.
Surveillance of the Listeria outbreak
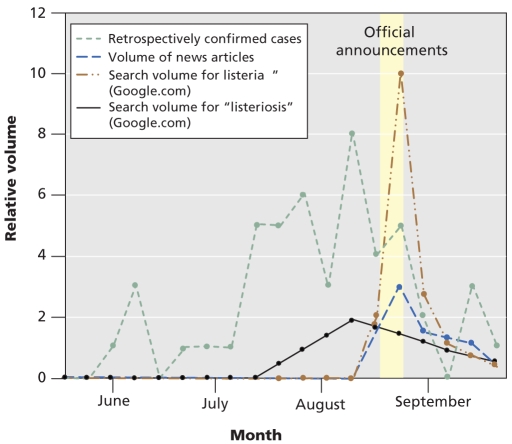
Analysis of the recent listeriosis outbreak in Canada that resulted from contaminated deli meat provides some interesting insights into the potential power of these tools. We investigated data sources other than traditional reporting that may have been available at the time of the outbreak. The public was officially informed by federal officials that 1 death and 16 cases were linked to a listeriosis outbreak on August 20, 2008.14 HealthMap collected 89 original articles that provided detailed information about the outbreak, the earliest of which appeared on August 17, although the majority of reports followed the federal announcement. However, search-term surveillance using the word “listeriorisis” showed a spike beginning in mid to late July, nearly a month before the declaration of the public outbreak. Interestingly, peak searching for “listeriosis” correlated more with the retrospective epidemic curve (Pearson correlation = 0.62, p = 0.005) than with the publicity of the outbreak (Pearson correlation = 0.55, p = 0.014) as measured by news volume. In comparison, a massive increase in searching for the word “Listeria” coincided perfectly with news media attention (Figure 1). Therefore, it appears that there was a clear Internet signal related to “Listeria” that preceded the official federal announcement.
Figure 1.
Epidemic curve for the course of the listeriosis outbreak in Canada in 2008.
A potential explanation for these findings is that the term “listeriosis” is more technical and that the data reflect queries by food inspection or industry officials investigating the possibility of the outbreak. Or it could have reflected queries by family and friends of people diagnosed early or people concerned about the initial voluntary recalls. A question that arises from this analysis is whether knowledge of this information, either by public health officials or members of the public, could have prompted an earlier response that may have reduced exposure to the contaminated products.
Limitations and future advancements
These data and aggregating Web-based technologies provide valuable information, but there are important limitations (Box 1). Although the utility of news media scanning is better established than surveillance of Internet search terms, there is limited evidence of the ability of these systems to detect emerging threats before signals from more traditional systems.4,15,16 Clearly, these data sources require in-depth evaluation, especially with respect to false positives and gaps in coverage.17 Lack of specificity, in particular, may be a primary limitation of these technologies (i.e., spikes in search terms or news stories potentially related to a disease outbreak may not necessarily mean that an outbreak exists). This may be less of an obstacle if the analysis is supported by trained public health officials who can investigate signals as they develop. However, these inefficiencies create the possibility of overload of signals that require verification and suggest that further work be conducted to determine how much of a change from baseline warrants further investigation.
Box 1.
Advantages and disadvantages of Internet-based surveillance
Advantages
Possibility of earlier detection of disease outbreaks than with use of traditional reporting mechanisms
Does not require voluntary reporting on the part of governments or local officials
The systems can provide information outside traditional communication channels
Information is freely available
Systems are relatively inexpensive to operate
Systems can be automated and the information can be disseminated in near real time
Potentially allows the public to have greater access to health surveillance information
Disadvantages
Information is often unstructured and difficult to interpret and requires advanced computational techniques to effectively implement
The sensitivity is unclear, and the percentage of outbreaks that can be identified by these strategies needs to be identified
The specificity is unclear, and a high false-positive rate could create workload issues because of the need for verification
Availability of information to the public may create challenges in risk communication
Privacy concerns for strategies that have the potential to identify individual internet activity
Public awareness of such signals, if they are openly accessible, could create problems in terms of risk communication for public health officials. The operating characteristics of these technologies need to be more precisely defined, as do their ability to detect disease before conventional systems and their application to a wide spectrum of diseases. Privacy implications are also need to be considered and balanced with the public health need to drill down to the highest possible geographic resolution.18 Given that search data contains associated internet provider information, which can be identified to the level of the household, appropriate decisions need to be made as to the level of appropriate spatial aggregation.18
Another potentially major obstacle to the use of these technologies is the requirement for Internet access. This is especially true in developing countries where surveillance is important. However, the dissemination of hand-held devices and mobile phones that connect to the Internet and have the ability to use short message service, or SMS, can help fill in technology gaps in resource-poor settings.19 In the future, we expect that the diagnostic accuracy of these instruments will be improved through an iterative process and that search term surveillance will be expanded to other diseases.
Internet scanning represents an important advancement in health surveillance, and search term surveillance is a provocative new tool that has much potential. However, both technologies merit further evaluation. The new application of these technologies could provide earlier access to information on potential disease outbreaks and promote greater transparency in disease reporting. Most importantly, these technologies may provide important benefits to outbreak control at local, national and international levels, ultimately reducing the health consequences of these outbreaks.
Outbreak surveillance systems
Global Public Health Intelligence
www.phac-aspc.gc.ca/media/nr-rp/2004/2004_gphin-rmispbk-eng.php (currently paid subscription)
International Society for Infectious Diseases’ Program for Monitoring Emerging Diseases
www.promedmail.org (free subscription)
HealthMap
www.healthmap.org (freely available)
Google Flu Trends
Key points
Internet surveillance tools can assist in the early identification of disease outbreaks and raise public awareness about emerging disease threats.
Surveillance based on trends of specific terms entered into search engines offers the potential to assist in earlier detection, but this technique requires further evaluation.
Search engine queries of the term “listeriosis” demonstrated a possible signal of an outbreak before the official announcement was made in Canada.
Footnotes
Contributors: Both of the authors made substantial contributions to the conception and design of the article, drafted the article, revised it critically for important intellectual content and gave final approval of the version to be published.
This article has been peer reviewed.
Competing interests: Kumanan Wilson is supported by a Canada Research Chair in Public Health Policy. John Brownstein is supported by the National Library of Medicine, National Institutes of Health, and by research funding from Google.org. John Brownstein is the co-creator of the HealthMap Project.
REFERENCES
- 1.Woodall J. Official versus unofficial outbreak reporting through the Internet. Int J Med Inform. 1997;47:31–4. doi: 10.1016/s1386-5056(97)00079-8. [DOI] [PubMed] [Google Scholar]
- 2.Brownstein JS, Freifeld CC, Reis BY, et al. Surveillance Sans Frontieres: Internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med. 2008;5:e151. doi: 10.1371/journal.pmed.0050151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grein TW, Kamara KB, Rodier G, et al. Rumors of disease in the global village: outbreak verification. Emerg Infect Dis. 2000;6:97–102. doi: 10.3201/eid0602.000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heymann DL, Rodier GR. Hot spots in a wired world: WHO surveillance of emerging and re-emerging infectious diseases. Lancet Infect Dis. 2001;1:345–53. doi: 10.1016/S1473-3099(01)00148-7. [DOI] [PubMed] [Google Scholar]
- 5.M’Ikanatha N M, Rohn DD, Robertson C, et al. Use of the Internet to enhance infectious disease surveillance and outbreak investigation. Biosecur Bioterror. 2006;4:293–300. doi: 10.1089/bsp.2006.4.293. [DOI] [PubMed] [Google Scholar]
- 6.Wilson K, von Tigerstrom B, McDougall C. Protecting global health security through the International Health Regulations: requirements and challenges. CMAJ. 2008;179:44–8. doi: 10.1503/cmaj.080516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mykhalovskiy E, Weir L. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Can J Public Health. 2006;97:42–4. doi: 10.1007/BF03405213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madoff LC. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis. 2004;39:227–32. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- 9.Freifeld CC, Mandl KD, Reis BY, et al. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J Am Med Inform Assoc. 2008;15:150–7. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandl KD, Overhage JM, Wagner MM, et al. Implementing syndromic surveillance: a practical guide informed by the early experience. J Am Med Inform Assoc. 2004;11:141–50. doi: 10.1197/jamia.M1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eysenbach G. Infodemiology: Tracking flu-related searches on the Web for syndromic surveillance. AMIA Annu Symp Proc. 2006;2006:244–8. [PMC free article] [PubMed] [Google Scholar]
- 12.Ginsberg J, Mohebbi MH, Patel RS, et al. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–4. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 13.Polgreen PM, Chen Y, Pennock DM, et al. Using internet searches for influenza surveillance. Clin Infect Dis. 2008;47:1443–8. doi: 10.1086/593098. [DOI] [PubMed] [Google Scholar]
- 14.CBCnews.ca. Meat recall timeline. Canadian Broadcast Corporation; 2009. [(accessed 2009 Mar. 9)]. Available: www.cbc.ca/health/story/2008/08/26/f-meat-recall-timeline.html. [Google Scholar]
- 15.Zeldenrust ME, Rahamat-Langendoen JC, Postma MJ, et al. The value of ProMED-mail for the Early Warning Committee in the Netherlands: more specific approach recommended. Euro Surveill. 2008;13:8033. [PubMed] [Google Scholar]
- 16.Cowen P, Garland T, Hugh-Jones ME, et al. Evaluation of ProMED-mail as an electronic early warning system for emerging animal diseases: 1996 to 2004. J Am Vet Med Assoc. 2006;229:1090–9. doi: 10.2460/javma.229.7.1090. [DOI] [PubMed] [Google Scholar]
- 17.German RR, Lee LM, Horan JM, et al. Updated guidelines for evaluating public health surveillance systems: recommendations from the Guidelines Working Group. MMWR Recomm Rep. 2001;50:1–35. [PubMed] [Google Scholar]
- 18.Brownstein JS, Cassa CA, Mandl KD. No place to hide — reverse identification of patients from published maps. N Engl J Med. 2006;355:1741–2. doi: 10.1056/NEJMc061891. [DOI] [PubMed] [Google Scholar]
- 19.Chretien JP, Burkom HS, Sedyaningsih ER, et al. Syndromic surveillance: adapting innovations to developing settings. PLoS Med. 2008;5:e72. doi: 10.1371/journal.pmed.0050072. [DOI] [PMC free article] [PubMed] [Google Scholar]