Abstract
The signal transducer and activator of transcription (STAT) 3, a transcriptional factor downstream of several cytokines, is activated by Janus kinase families and plays a pivotal role in cardiac hypertrophy through gp130. To determine the physiological significance of STAT3 in vivo, transgenic mice with cardiac-specific overexpression of the Stat3 gene (STAT3-TG) were generated. STAT3-TG manifested myocardial hypertrophy at 12 wk of age with increased expression of the atrial natriuretic factor (ANF), β-myosin heavy chain (MHC), and cardiotrophin (CT)-1 genes. The animals were injected i.p. with 15 mg/kg doxorubicin (Dox), an antineoplastic drug with restricted use because of its cardiotoxicity. The survival rates after 10 days were 25% (5/20) for control littermates (WT), but 80% (16/20) for STAT3-TG (P < 0.01). WT showed increased expression of β-MHC and ANF mRNAs in the hearts 1 day after Dox treatment; this expression peaked at 3 days, suggesting that the WT suffered from congestive heart failure. Although the expression of these mRNAs was elevated in STAT3-TG hearts before Dox treatment, no additional increase was observed after the treatment. Dox administration significantly reduced the expression of the cardiac α-actin and Stat3 genes in WT hearts but not in STAT3-TG. These results provide direct evidence that STAT3 transduces not only a hypertrophic signal but also a protective signal against Dox-induced cardiomyopathy by inhibiting reduction of cardiac contractile genes and inducing cardiac protective factors.
Signal transducer and activator of transcription (STAT) proteins, which consist of six members, are known to play important roles in cytokine signaling pathways (1). STATs are activated by the Janus kinase (JAK) family after cytokine binding to their specific receptors (2–6). Previous studies revealed that STAT3 was important for the growth, differentiation, and cell death or survival in various kinds of cells (7–11). Recently, we have demonstrated that STAT3 is expressed in cardiac myocytes, immediately activated by the IL-6 family of cytokines, and plays a crucial role in generating the hypertrophic signal through gp130 (12, 13). In addition, activation of gp130 has been shown to induce bcl-xL expression by means of STAT1 in cardiac myocytes, resulting in a cytoprotective effect (14). Although recent reports provide evidence of STAT phosphorylation in a pressure-overload model (15) and an acute myocardial infarction model of rat hearts (16), the physiological function of STAT in the heart has not been clarified yet.
Doxorubicin (Dox), an antitumor drug, is effective for a variety of soft and solid human malignancies, but its long-term use is limited because of the development of cardiomyopathy. Several proposals regarding the pathogenesis of Dox-induced cardiac toxicity have been made, including a generation of oxygen-free radicals (17, 18), from which damage is inhibited by free-radical scavengers, and an intercalation of the drug to DNA (19), which is not affected by free-radical scavengers. Transgenic mice with cardiac-specific overexpression of the metallothionein gene, which functions as a scavenger of free radicals, were reported to be resistant to myocardial morphological and functional damages induced by Dox (20). However, the molecular mechanisms for this protection against Dox-induced cardiomyopathy have not been fully identified yet.
A recent study with mice that harbor a ventricular-restricted knockout of the gp130 demonstrated a critical role for a gp130-dependent myocyte survival pathway in the transition to heart failure (21). Accordingly, identifying the downstream pathways by which gp130-dependent ligands can promote cardiac myocyte survival has become of critical interest. To determine the physiological significance of STAT3 in the heart, transgenic mice with cardiac-specific overexpression of the Stat3 gene (STAT3-TG) were generated and examined with special attention being paid to any reduction in cardiotoxicity induced by Dox. Our results demonstrate that overexpression of STAT3 in the heart provides protection against Dox-induced cardiomyopathy, thus resulting in an improved survival rate by preventing progression of heart failure.
Materials and Methods
Generation of Transgenic Mice with Cardiac-Specific Overexpression of the Stat3 Gene.
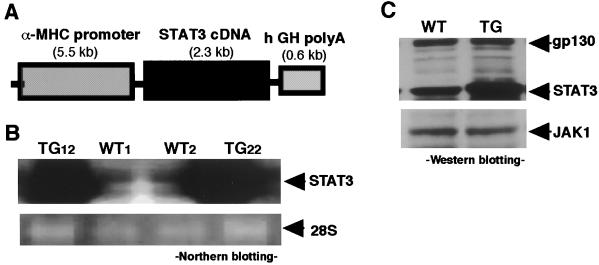
A 5.5-kilobase (kb) fragment of murine α-myosin heavy chain (MHC) gene promoter [generously provided by Jeffrey Robbins (Children's Hospital Research Foundation, Cincinnati)] and a 2.3-kb murine Stat3 cDNA [generously provided by S. Akira (Osaka University, Osaka)] were subcloned into pBsIISK(+) plasmids. The plasmids were then digested with BamHI to generate an 8.4-kb DNA fragment consistent with the α-MHC gene promoter, Stat3 cDNA, and poly(A) of the human growth hormone (see Fig. 1A). Transgenic mice (C57BL/6 strain) were generated as described elsewhere (22). The care of all animals used in the present study was in accordance with Osaka University Animal Care guidelines.
Figure 1.
Transgene construct and STAT3 expression in the hearts. (A) Schematic representation of the transgene, a 2.3-kb murine Stat3 cDNA was ligated downstream of a 5.5-kb fragment of the murine α-myosin heavy chain (MHC) gene promoter and upstream of a 0.6-kb human growth hormone (hGH) poly(A). (B) Northern blot analysis was performed using a murine Stat3 cDNA as a probe. Hearts from transgene-positive animals (TG12 and TG22) showed abundant Stat3 mRNA expression. Equal RNA loadings were documented by 28S RNA (Lower). (C) gp130, JAK1, and STAT3 expressions in the hearts. The lysates from the hearts containing equal amounts of the protein (10 μg) were separated by SDS/PAGE and immunoblotted with rabbit anti-gp130, JAK1, or STAT3 Ab as described in Materials and Methods. WT, Control littermate; TG, STAT3-TG.
To identify the exogenous Stat3 gene, genomic DNA was extracted from the tail tissues of 3-wk-old mouse pups. Screening of STAT3-TG was performed by PCR analysis with a set of primers, 5′-GATGCGACCAACATCCTGGTG-3′ and 5′-GGACATCGGCAGGTCAATGGT-3′, which correspond to the sequences obtained from exons 21 and 23 of the murine Stat3 gene (2). PCR conditions were 96°C for 4 min, followed by 30 cycles of 96°C for 1 min, 58°C for 1 min, and 72°C for 2 min. The length of amplified DNA was 201 bp.
Histological Examination.
The hearts were flushed with PBS and fixed with 4% paraformaldehyde at 4°C for 24 hr and then embedded in paraffin. Three sections from each left ventricle perpendicular to the major axis of the heart were sampled and stained with hematoxylin/eosin. For comparison of the ventricular thickness, the hearts were sectioned at the papillary muscle level.
Northern Blot Analysis.
Total cellular RNA was isolated from the hearts with the guanidinium thiocyanate-phenol-chloroform method (23). Ten micrograms of total RNA were size-fractionated by formaldehyde-agarose gel electrophoresis and blotted onto a nylon membrane (Hybond N+; Amersham) in the presence of 20× SSC [300 mM sodium chloride and 300 mM sodium citrate (pH 7.0)]. A Stat3 cDNA probe was obtained by means of reverse transcription (RT)-PCR by using specific oligonucleotide primers as described above, and a cardiotrophin (CT)-1 cDNA probe was obtained by using the specific oligonucleotide primers 5′-GAGACAGTGCTGGCCGCG CTG-3′ and 5′-AGAGGAGAGCAGAAGAGAGAGA-3′, which are based on the nucleotide sequence of murine CT-1 cDNA (24). Cardiac α-actin cDNA was kindly provided by H. Ito (Tokyo Medical and Dental University, Tokyo) and used as a probe as described previously (25). The membranes were labeled with [32P]dCTP by using a PRIME IT labeling kit (Promega), and hybridization and washing were performed as described previously (13). The membranes were then exposed to x-ray film for 24 hr at −70°C.
Semiquantitative RT-PCR.
Total RNA (0.1 μg) was subjected to first-strand synthesis by using the oligo(dT) (Pharmacia Biotech) and Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL) at 37°C for 1 hr. The first-strand DNA was amplified by PCR (94°C for 4 min, followed by the indicated number of cycles at 94°C for 45 s, 60°C for 1 min, and 72°C for 2 min) with the following sets of primers: α-MHC (302 bp), 5′-CTGCTGGAGAGGTTATTCCTCG-3′ and 5′-GGAAGAGTGAGCGGCGCATCAAGG-3′; β-MHC (205 bp), 5′-TGCAAAGGCTCCAGGTCTGAGGGC-3′ and 5′-GCCAACACCAACCTGTCCAAGTTC-3′; and atrial natriuretic factor (ANF) (400 bp), 5′-CTCTGAGAGACGGCAGTGCT-3′ and 5′-TATGCAGAGTGGGAGAGGCA-3′ (26). PCR cycles were 27, 30, and 24, respectively. Specific primers for β-tubulin (5′-TCACTGTGCCTGAACTTACC-3′ and 5′-GGAACATAGCCGTAAACTGC-3′) were also used to qualitate and quantitate RNA in each sample. Equal volumes of the amplified samples were loaded onto a 1.5% agarose gel and stained with ethidium bromide.
Western Blot Analysis.
The hearts were lysed with sample buffer [62.5 mM Tris (pH 7.4)/2% SDS/5% 2-mercaptoethanol/10% glycerol] and boiled for 10 min. Thereafter, the samples were centrifuged at 15,000 rpm, and the supernatants were used to measure protein content. The samples were then separated by SDS/PAGE and transferred electrophoretically onto a polyvinylidene difluoride membrane (Immobilon; Millipore) with a transfer buffer (25 mM Tris/190 mM glycine/20% methanol) and immunoblotted with anti-gp130, JAK1, or STAT3 Ab (Santa Cruz Biotechnology) as described previously (12). Enhanced chemiluminescence detection reagents (Amersham) were employed to visualize peroxidase reaction products.
Statistics.
Results are shown as mean ± SE. Statistical analysis was performed by unpaired t test. P values of <0.05 were considered significant.
Results
Generation of STAT3-TG.
Fig. 1A shows a schematic representation of the transgene. The α-MHC gene promoter effects a pattern of transgene expression similar to that produced by the endogenous α-MHC gene. PCR analysis of murine genomic DNA identified two founders containing the exogenous Stat3 gene (TG12 and TG22). Fig. 1B shows the Northern blot results for the hearts from two lines probed with murine Stat3 cDNA, which manifested equally augmented Stat3 mRNA expression in the hearts. The expressions of the gp130, JAK1, and STAT3 proteins in the hearts were compared (Fig. 1C). An 8-fold increase in the STAT3 protein was observed in STAT3-TG hearts compared with that in control littermates (WT), whereas no differences were observed in gp130 and JAK1 expressions. Moreover, the amounts of JAK2, STAT1, and extracellular signal-regulated kinases were also equal in the hearts from WT and STAT3-TG (data not shown). Neonatal mortality in STAT3-TG did not differ from that in WT, and no adult death was observed. The TG12 line was used in the subsequent experiments.
Myocardial Hypertrophy in STAT3-TG Hearts.
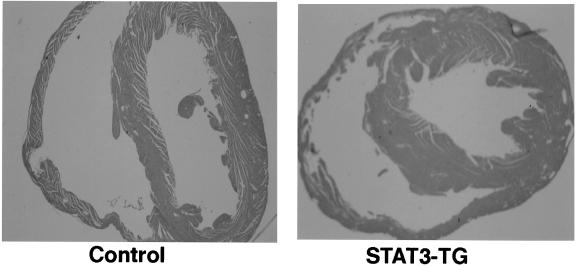
As shown in Fig. 2, pathological analysis of the hearts from 3-mo-old mice demonstrated that the thickness of the left ventricle had increased by 21.3 ± 1.8% (n = 5) in STAT3-TG hearts. Microscopic examination of the longitudinal section of 500 cardiac myocytes revealed that the cell width had increased by 28.8 ± 2.7% in STAT3-TG compared with that in WT (P < 0.05). There was no evidence of fibrosis or infiltration of inflammatory cells in STAT3-TG hearts.
Figure 2.
Histological analysis of the hearts. The sections obtained from the hearts of 3-mo-old control mice and STAT3-TG were stained with hematoxylin/eosin. (Magnification: ×20.)
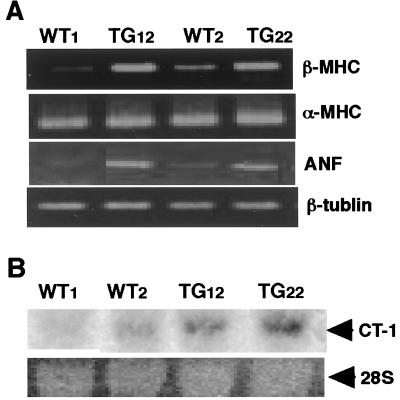
Ventricular hypertrophy induced by pressure or volume overload reportedly accompanies the induction of several specific genes, such as β-MHC and ANF (27). As shown in Fig. 3A, the expression of β-MHC and ANF mRNAs increased in STAT3-TG hearts (TG12 and TG22), whereas α-MHC and β-tubulin mRNA expression did not change. In addition, the expression of CT-1 mRNA was augmented in STAT3-TG hearts (Fig. 3B).
Figure 3.
Induction of the hypertrophy genes program in STAT3 transgenic mice. (A) Semiquantitative RT-PCR analysis. Equal volumes of the amplified PCR products were loaded onto a 1.5% agarose gel. (B) Northern blot analysis using a CT-1 cDNA as a probe. WT, Control littermate; TG, STAT3-TG.
Effect of Dox Treatment on STAT3-TG.
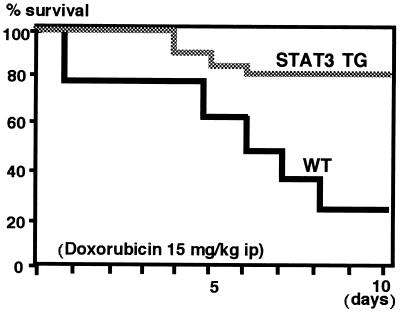
STAT3 is reported to transduce protective signals against various stimuli in several types of cells (10, 11), and its activity is especially critical for the gp130-mediated antiapoptotic signal in murine pro-B BaF/B03 cells (10). Therefore, STAT3 may provide a protective effect against cardiac toxicity induced by Dox. Three-month-old mice were treated i.p. with Dox at a single dose of 15 mg/kg. As shown in Fig. 4, only 25% (5/20) of the WT mice survived compared with 80% (16/20) of the STAT3-TG after 10 days of Dox treatment (P < 0.01). The dead mice were found to have massive bilateral pleural effusion and ascites, which led to a diagnosis of congestive heart failure.
Figure 4.
Survival rates for the mice treated with Dox. Three-month-old STAT3-TG and WT mice weighing 21–23 g (n = 20) were injected i.p. with Dox at a single dose of 15 mg/kg and bred under normal condition. Kaplan–Meier survival curves represent significantly better survival (P < 0.01) in STAT3-TG than in WT.
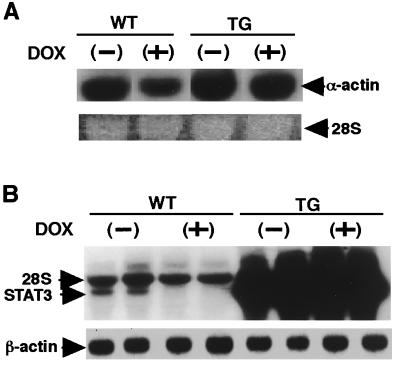
Dox is known to selectively reduce the levels of mRNA for the sarcomeric genes, cardiac α-actin, troponin I, and myosin light chain 2 (25). To investigate the molecular mechanisms of these ameliorative effects of STAT3 on the heart, the expression of these sarcomeric genes was examined in the hearts after 2 days of Dox treatment. Cardiac α-actin mRNA expression was significantly reduced in WT hearts after Dox treatment, and this finding was compatible with previously published data (25). However, STAT3-TG hearts showed continuously augmented cardiac α-actin gene expression (Fig. 5A). In addition, endogenous STAT3 mRNA expression was dramatically reduced in WT hearts after 2 days of Dox treatment (Fig. 5B) and remained elevated throughout the experimental periods in STAT3-TG hearts because its expression was controlled by the α-MHC gene promoter. The transcript of the α-MHC gene itself was not affected by Dox treatment (data not shown).
Figure 5.
Alteration of cardiac gene expressions after Dox treatment. WT and STAT3-TG were treated with single dose of 15 mg/kg Dox, and the hearts were excised after 2 days. Northern blot analysis was performed using a cardiac α-actin cDNA (A) or a murine Stat3 cDNA (B) as a probe. Equal RNA loadings were documented by 28S RNA and β-actin controls. WT, Control littermate; TG, STAT3-TG.
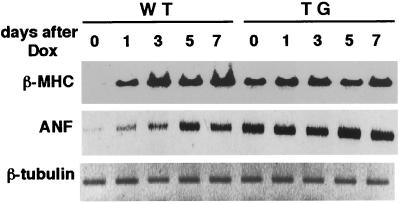
The expression of ANF and β-MHC genes is induced not only in myocardial hypertrophy but also in heart failure (27, 28). As shown in Fig. 6, the transcript of β-MHC gradually increased in WT hearts after 1 day of Dox treatment, peaked at 3 days, and remained at that level throughout the experiment. Although β-MHC mRNA expression was elevated in untreated STAT3-TG hearts, no additional increase was observed after Dox treatment, and the peak expression level was much higher in WT than in STAT3-TG hearts. The expression levels of the α-MHC, myosin light chain 2v, and β-tubulin mRNAs were equal in both hearts at the time examined (data not shown).
Figure 6.
Expression of β-MHC and ANF mRNAs after Dox treatment. Semiquantitative RT-PCR was performed as described in Fig. 3. RNAs were obtained from the hearts at the indicated day after 15 mg/kg Dox treatment. WT, Control littermate; TG, STAT3-TG.
Although the expression pattern of the ANF gene after Dox treatment was similar to that of the β-MHC gene in WT hearts and STAT3-TG hearts showed no significant change, the induced expression level of ANF mRNA was always higher in STAT3-TG hearts than in WT throughout the experimental period (Fig. 6).
Discussion
In this study, we showed that the activation of STAT3 induces hypertrophy in vivo as observed in the forms of adenovirus-mediated Stat3 gene transfer in cardiac myocytes (13). STAT3 is activated by a variety of factors, such as the IL-6 family of cytokines, interferons, epidermal growth factor, growth hormone, and angiotensin II (Ang II), in a manner dependent on the JAK family of tyrosine kinases (2–6). Although the candidates for activating STAT3 during normal cardiac development have not been specifically identified yet, the IL-6 family of cytokines, especially CT-1 and leukemia inhibitory factor (LIF) or Ang II, appear to be the most likely candidates for inducing myocardial hypertrophy in STAT3-TG. CT-1, LIF, and Ang II are known to express in hearts and have the potential to induce hypertrophy. The present study specifically showed the augmented expression of CT-1 mRNA in STAT3-TG hearts. Recently, we reported that dominant-negative STAT3 abrogated LIF-induced increased leucine incorporation in cardiac myocytes (13), which suggests that STAT3 is a key molecule in gp130-dependent hypertrophic signaling. The cis-acting DNA sequences recognized by STAT dimers, the STAT-binding element, locate in the 5′ flanking region of the cardiac α-actin, and β-MHC genes (29, 30). In addition, recent observations have focused on GATA5 as an important transcriptional factor in gp130-dependent β-MHC gene induction (31).
Dox is reported to produce free radicals, cause lipid peroxidation, and disrupt cell membrane functions (17, 18). In addition, Dox binds to DNA and interferes with DNA replication (19). Cardiomyopathy produced by Dox is known to associate with alterations in the expression of genes important for the structural integrity and enzymatic function of cardiac myocytes (25). These phenomena can be expected to lead to defective formation or inadequate maintenance of the contractile apparatus in the hearts. In the present study, a single high dose (15 mg/kg) of Dox was injected on the basis of findings by Myers et al. (18), who had demonstrated severe ultrastructural damage to the myocardium after administration of Dox in mice. Although numerous investigators have reported on possible mechanisms of Dox-induced cardiotoxicity, the etiology of Dox-induced cardiomyopathy has not been clearly elucidated. A recent finding indicates the importance of cardiac myocyte apoptosis in the transition between compensatory cardiac hypertrophy and heart failure (21). Expression of Bcl-xL and Bcl-2 was examined in Dox-treated hearts, but no significant change was observed nor were any apoptotic cardiac myocytes. Therefore, apoptosis was not considered to be important for the cardiac decompensation induced by Dox, while Dox reduced sarcomeric gene expression.
Important determinants for the STAT function are the intensity and duration of the STAT activity and the amounts of STAT proteins ready to be activated in cells. All of these factors eventually determine the range of target genes and the duration and intensity of target gene activation and thereby determine the outcome, such as cell growth and cell survival. c-fos mRNA expression was augmented in STAT3-TG hearts after i.v. injection of LIF (data not shown); that is, the signals transduced by STAT3 were augmented in STAT3-TG hearts in spite of the same stimulation as that applied to WT hearts. The tyrosine-phosphorylated level of STAT3 was always higher in STAT3-TG hearts than in WT, which indicates that the genes were probably continuously activated even after Dox administration. The Stat3 gene itself is activated by a gp130-dependent signal (32), and the transcript of CT-1 is also activated even after Dox treatment in STAT3-TG hearts. These findings and our results suggest the existence of an autoregulatory mechanism in the gp130-dependent signal-transducing system at the level of the ligands, the receptor, and the signal-transducing transcription factor.
Recently, ANF was reported to prevent myocardial damage not only through reduction of plasma volume and pressure overload but also through suppression of the activation of renin-angiotensin and sympathetic nervous systems (33–35). The augmented expression of the ANF and β-MHC mRNAs observed in the WT hearts after Dox treatment suggested that they had died of heart failure. Although no additional increase in ANF gene expression was observed in STAT3-TG hearts after Dox treatment, the expression level remained higher than that observed in WT hearts. These results suggest that STAT3-TG did not suffer from severe congestive heart failure and that the enhanced expression of ANF and CT-1 mRNAs in STAT3-TG hearts may partially explain the protective function of STAT3. These protective factors might then be constitutively induced in STAT3-TG hearts and prevent cell damage caused by Dox toxicity.
The present study has demonstrated that STAT3-TG survive significantly longer after Dox treatment and that the overexpression of STAT3 in the hearts reduces the deteriorative effects of Dox. Inhibition of the reduction in sarcomeric gene expression and induction of cytoprotective factors are though to be possible mechanisms for STAT3-induced myocardial protection. The most important finding, however, is that Stat3 mRNA expression in the heart is selectively and dramatically reduced by Dox treatment. This suggests that STAT3 may play a crucial role in cardiac myocyte survival, thus resulting in the prevention of heart failure.
Acknowledgments
We thank Dr. S. Akira for his critical reading of the manuscript and C. Kusunoki for her secretarial assistance. This study was supported by a Grant-in Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan and grants from the Ministry of Health and Welfare of Japan and the study group of Molecular Cardiology.
Abbreviations
- STAT
signal transducer and activator of transcription
- JAK
Janus kinase
- Dox
doxorubicin
- MHC
myosin heavy chain
- CT
cardiotrophin
- RT
reverse transcription
- ANF
atrial natriuretic factor
- LIF
leukemia inhibitory factor
References
- 1.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Yoshida K, Sudo T, Naruo M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 3.Ruff-Jamison S, Zhong Z, Wen Z, Chen K, Darnell J E, Jr, Cohen S. J Biol Chem. 1994;269:21933–21935. [PubMed] [Google Scholar]
- 4.Yang C H, Murti A, Pfeffer L M. Proc Natl Acad Sci USA. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell G S, Meyer D J, Raz R, Levy D E, Schwartz J, Carter-Su C. J Biol Chem. 1995;270:3974–3979. doi: 10.1074/jbc.270.8.3974. [DOI] [PubMed] [Google Scholar]
- 6.McWhinney C D, Hunt R A, Conrad K M, Dostal D E, Baker K M. J Mol Cell Cardiol. 1997;29:2513–2524. doi: 10.1006/jmcc.1997.0489. [DOI] [PubMed] [Google Scholar]
- 7.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y Y, Bradshaw R A. J Biol Chem. 1996;271:13023–13032. doi: 10.1074/jbc.271.22.13023. [DOI] [PubMed] [Google Scholar]
- 9.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Immunity. 1994;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 11.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 12.Kunisada K, Hirota H, Fujio Y, Matsui H, Tani Y, Yamauchi-Takihara K, Kishimoto T. Circulation. 1996;94:2626–2632. doi: 10.1161/01.cir.94.10.2626. [DOI] [PubMed] [Google Scholar]
- 13.Kunisada K, Tone E, Fujio Y, Matsui H, Yamauchi-Takihara K, Kishimoto T. Circulation. 1998;98:346–352. doi: 10.1161/01.cir.98.4.346. [DOI] [PubMed] [Google Scholar]
- 14.Fujio Y, Kunisada K, Hirota H, Yamauchi-Takihara K, Kishimoto T. J Clin Invest. 1998;99:2898–2905. doi: 10.1172/JCI119484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan J, Fukuda K, Kodama H, Makino S, Takahashi T, Sano M, Hori S, Ogawa S. Circ Res. 1997;81:611–617. doi: 10.1161/01.res.81.4.611. [DOI] [PubMed] [Google Scholar]
- 16.Negoro S, Kunisada K, Tone E, Funamoto M, Yamauchi-Takihara K. Circulation. 1999;100:I–491. [Google Scholar]
- 17.Lee V, Randhawa A K, Singal P K. Am J Physiol. 1991;261:H989–H995. doi: 10.1152/ajpheart.1991.261.4.H989. [DOI] [PubMed] [Google Scholar]
- 18.Myers C E, McGuire W P, Liss R H, Ifrim I, Grotzinger K, Young R C. Science. 1977;197:165–167. doi: 10.1126/science.877547. [DOI] [PubMed] [Google Scholar]
- 19.Tewey K M, Rowe T C, Yang R H, Halligan B D, Liu L F. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y J, Chen Y, Anding Y, Voss-McCowan M, Epstein P N. J Clin Invest. 1997;100:1501–1506. doi: 10.1172/JCI119672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota H, Chen J, Betz U A K, Rajewsky K, Gu Y, Ross J, Jr, Muller W, Chien K R. Cell. 1999;97:189–198. doi: 10.1016/s0092-8674(00)80729-1. [DOI] [PubMed] [Google Scholar]
- 22.Ikawa M, Yamada S, Nakanishi T, Okabe M. FEBS Lett. 1998;23:83–87. doi: 10.1016/s0014-5793(98)00593-6. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Pennica D, Wood W I. Proc Natl Acad Sci USA. 1995;92:1142–1146. doi: 10.1073/pnas.92.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito H, Torti F M. Proc Natl Acad Sci USA. 1990;87:4275–4279. doi: 10.1073/pnas.87.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller-Hance W C, LaCorbiere M, Fuller S J, Evans S M, Lyons G, Schmidt C, Robbins J, Chien K R. J Biol Chem. 1993;268:25244–25252. [PubMed] [Google Scholar]
- 27.Feldman A M, Weinberg E O, Ray P E, Lorell B H. Circ Res. 1993;73:184–192. doi: 10.1161/01.res.73.1.184. [DOI] [PubMed] [Google Scholar]
- 28.Drexler H, Hanze J, Finckh M, Lu W, Just H, Lang R E. Circulation. 1989;79:620–633. doi: 10.1161/01.cir.79.3.620. [DOI] [PubMed] [Google Scholar]
- 29.Eldridge J, Zehner Z B, Paterson M. Gene. 1985;36:55–63. doi: 10.1016/0378-1119(85)90069-1. [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi-Takihara K, Sole M J, Liew J, Ing D, Liew C C. Proc Natl Acad Sci USA. 1989;86:3504–3508. doi: 10.1073/pnas.86.10.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morimoto T, Hasegawa K, Kaburagi S, Kakita T, Masutani H, Kitsis R N, Matsumori A, Sasayama S. J Biol Chem. 1999;274:12811–12818. doi: 10.1074/jbc.274.18.12811. [DOI] [PubMed] [Google Scholar]
- 32.Ichiba M, Nakajima K, Yamanaka Y, Kiuchi N, Hirano T. J Biol Chem. 1998;273:6132–6138. doi: 10.1074/jbc.273.11.6132. [DOI] [PubMed] [Google Scholar]
- 33.Weidmann P, Saxehofer H, Shaw S G, Ferrier C. J Steroid Biochem. 1989;32:229–241. doi: 10.1016/0022-4731(89)90170-2. [DOI] [PubMed] [Google Scholar]
- 34.Naess P A, Christensen G, Kiil F. Am J Physiol. 1991;261:E240–E245. doi: 10.1152/ajpendo.1991.261.2.E240. [DOI] [PubMed] [Google Scholar]
- 35.Fujisaki H, Ito H, Hiroe M. J Clin Invest. 1995;96:1059–1065. doi: 10.1172/JCI118092. [DOI] [PMC free article] [PubMed] [Google Scholar]