Abstract
Our previous allelotyping studies of 59 sporadic colorectal cancers revealed that loss of heterozygosity is most frequent for regions of chromosomes 14 and 18. Yet subsequent BAC microarray comparative genomic hybridization studies of the same tumor DNAs showed no corresponding pattern of copy number alteration for chromosome 14. To clarify this apparent discrepancy, we utilized hybridization to SNP microarrays; this revealed frequent uniparentalism for chromosome 14 and for chromosome 18. Based on the BAC array results combined with fluorescent in situ hybridization data, it was evident that uniparental disomy was occurring in many colorectal cancers as well as in additional chromosomes, and often coordinately involved chromosomes 14 and 18. Further studies examined the possibility that uniparentalism was directed towards the selection for imprinted genes, but no association with imprinting was observed.
Keywords: uniparental disomy, imprinting, genomic instability, colorectal cancer
1. Introduction
Genomic instability in the form of aneuploidy was first recognized to be a factor in cancer by Theodor Boveri nearly a century ago [1, 2]. Several additional forms of instability have been subsequently observed in sporadic colorectal and other cancers, including large chromosomal changes in the form of reciprocal and non-reciprocal translocations, deletions, etc., along with small events as revealed by microarrays, inter-(simple sequence repeat) PCR, microsatellite instability, and direct DNA sequencing [3–5]. Further contributing to overall genomic change are epigenetic disruptions, such as hypermethylation that results in the silencing of key tumor suppressors and hypomethylation at repeat sequences that encourage faulty recombination and loss of imprinting (LOI) [6–9]. Additional genomic changes involve micro RNAs, although at present the cancer-related consequences are unclear [10].
We have previously characterized a set of 59 colorectal carcinomas for loss of heterozygosity (LOH) profiles, using 348 microsatellite markers spaced about every 10Mb spanning the entire genome [11]. This study revealed the highest level of LOH for regions of chromosomes 14 and 18, with lesser involvement of many other chromosomes. Further analysis with BAC-microarray comparative genomic hybridization showed most of these tumors showed no copy number aberration for chromosome 14 [12], generating an apparent conflict. A potential explanation was that uniparental disomy was present in the tumors. And if uniparental disomy was indeed occurring, a key question would be if imprinted genes were being selected for or against, or alternatively if uniparentalism was arising during the course of progression, rendering homozygous those somatic mutations that had occurred previously.
Uniparentalism reflects the loss of an allele and reduplication of the retained allele; uniparental disomy is the case where both copies of a chromosome are from one parent. For tumor progression, this can arise from inappropriate segregation during mitosis initially creating a cell with two copies of a chromosome from one parent and one from the other parent, with the one from the other parent later lost [13]. Imprinted genes show monoallelic transcription, more often from the unmethylated allele, with the methylated allele silent. Loss of imprinting has been often observed in cancer, and has been proposed to plan an etiologic role [14–17].
By applying single nucleotide polymorphism (SNP) microarrays, we have found both whole chromosome uniparentalism and segmental uniparentalism in five of the thirteen sporadic colon cancer samples analyzed. In our analyses, four tumor samples showed coordinate whole chromosome uniparentalism for chromosome 14 and chromosome 18. We have further analyzed the methylation status of imprinted genes present on chromosomes displaying uniparentalism, in order to determine if there exists a selection bias towards an imprinted status. In our analysis of chromosomes that showed uniparentalism, we found the retention of either the methylated or unmethylated allele, indicating that a selection bias toward either allele does not exist.
2. Materials and methods
2.1. Patient specimens
Tumor and adjacent normal colon tissues were obtained from 13 sporadic colorectal cancer patients who underwent surgical resections at Roswell Park Cancer Institute from 1991–1998, including eleven who in previous analyses had shown losses of heterozygosity on chromosome 14 [11,12,18]. Descriptions of tumors and two previously measured genomic instabilities are listed in Table 1, and clinicopathologic features of these tumors have been reported [19].
Table 1.
Colorectal tumors and corresponding normal tissues analyzed on GeneChip SNP Microarrays.
| Sample | S/NS | FAL | GII | Histology |
|---|---|---|---|---|
| 6374 | NS | 0.207 | 3.9 | Tumour |
| 6375 | NS | Normal | ||
| 6376 | NS | 0.202 | 3.9 | Tumour |
| 6377 | NS | Normal | ||
| 3199 | S | 0.247 | 6.5 | Tumour |
| 3200 | S | Normal | ||
| 3118 | NS | 0.043 | 3.9 | Tumour |
| 3119 | NS | Normal | ||
| 3043 | S | 0.075 | 6.5 | Tumour |
| 3044 | S | Normal | ||
| 3125 | S | 0.354 | 6.5 | Tumour |
| 3126 | S | Normal | ||
| 3180 | S | 0.43 | 13 | Tumour |
| 3181 | S | Normal | ||
| 3031 | S | 0.202 | 3.9 | Tumour |
| 3032 | S | Normal | ||
| 12026 | NS | 0.341 | 7.8 | Tumour |
| 12028 | NS | Normal | ||
| 3147 | NS | 0.179 | 1.3 | Tumour |
| 3148 | NS | Normal | ||
| 3936 | NS | 0.032 | 6.5 | Tumour |
| 3937 | NS | Normal | ||
| 6386 | S | 0.078 | 5.2 | Tumour |
| 6387 | S | Normal | ||
| 6388 | S | 0.052 | 2.6 | Tumour |
| 6389 | S | Normal |
S=smoker, NS=nonsmoker
FAL: fractional allelic loss rate
GII: genomic instability index, the percentage of inter-(simple sequence repeat) PCR bands altered as compared with corresopnding normal tissue.
2.2. DNA Preparation and Array comparative genomic hybridization (aCGH)
DNA was extracted from the tumors and normal tissues, as previously described [11,18,19]; small pieces of tissue (approximately 8 mm3) were digested with 1 μg/ml proteinase K for 3 hr at 65°C, followed by 0.5 μg/ml RNase treatment for 1 hr and then phenol: chloroform: isoamyl alcohol extraction and ethanol precipitation [18, 19]. BAC microarray comparative genomic hybridization was performed as described [12].
2.3. Single Nucleotide Polymorphism (SNP) arrays
The Affymetrix GeneChip Mapping 10K 2.0 Array, containing 11,500 SNP markers at an average distance of 210kb was used. The construction of the 10K array has been described elsewhere [20]. Hybridization of the samples to the GeneChip Mapping 10K 2.0 Array and subsequent data generation were carried out in the Functional Genomics Center at University of Rochester Medical Center. Relative allele signal values (RAS) were calculated based on hybridization signal intensities, and genotypes were assigned as AA and BB for homozygous genotypes and AB for heterozygous genotypes at each locus [21]. To compare the genotypes graphically, values were assigned to the homozygous and heterozygous genotypes for each chromosome of the tumor and paired-normal tissue and plotted.
2.4. Bisulfite Treatment of DNA
500ng genomic DNA was denatured and deaminated by treating at 55°C for 4 hours with a solution of saturated sodium bisulfite and 10mM hydroquinone, pH5. DNA was desalted using Wizard DNA Cleanup Columns (Promega) ethanol precipitated and dissolved in 50ul PCR quality water [22,23].
2.5. Methylation Specific PCR (MSP)
MSP for the Meg3 and SNRPN genes on chromosomes 14 and 15 was performed using 1ul bisulfite treated DNA in a 30 ul reaction mixture containing, 0.2mM dNTPs, 2mM MgCl2 (SNRPN) [or 3mM MgCl2 with (Meg3)], 0.4mM primers (Supplementary Table 1) and 2.5u of Jumpstart Taq (Sigma). PCR products were visualized by agarose gel electrophoresis. The expected sizes for the methylated and unmethylated reaction products were 174bp and 100bp for SNRPN and 160bp and 120bp for Meg3.
2.6. Bisulfite Sequencing
Bisulfite treated DNA was PCR amplified and cloned using the TOPO-TA pCR2 kit according to manufacturers’ instructions. Primer sequence and PCR conditions are included in supplementary matter. A minimum of 10 transformants was sequenced using the ABI PRISM 3130XL Genetic Analyzer.
2.7. Methylation Analysis by Mass Array
Bisulfite treated DNA was PCR amplified with primers specific to the differentially methylated region of the imprinted gene being assayed. Primers were modified to contain a T7 promoter sequence on the reverse primer as per manufactures requirements. The PCR products were gel purified using the QIAquick gel purification kit and analyzed by the Sequenom MassARRAY Compact that utilizes the EpiTyper platform for the detection and quantitative analysis of DNA methylation. Primer sequence and PCR conditions are included in the supplementary information.
3. Results
3.1. Detection of uniparentalism
We have characterized a set of 59 sporadic colon carcinomas and adjoining normal tissue specimens for several different genomic instabilities associated with the disease. Previously all 59 tumors had been allelotyped and regions of LOH defined, using 348 microsatellite markers spaced approximately every 10 Mb [12]. A sub-set of 32 tumors were further analyzed utilizing BAC-array comparative genomic hybridization [11]. We observed an apparent discrepancy between the two data sets: some regions with LOH did not display loss in copy number via aCGH. This discrepancy was most evident for chromosome 14, where extensive LOH was confirmed using fourteen additional microsatellite markers (data not shown).
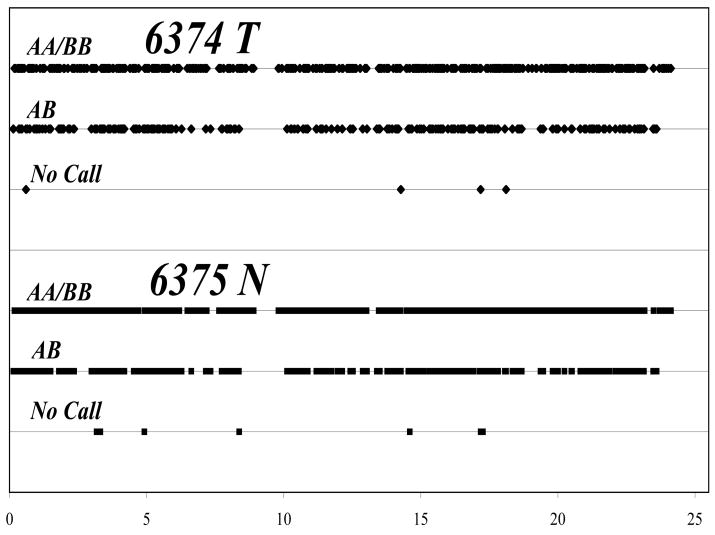
To investigate this apparent discrepancy between absence of copy number changes and losses of heterozygosity, thirteen of the paired tumor-normal sporadic colon cancer samples were analyzed using the Affymetrix GeneChip Mapping SNP array, comprised of more than 11,500 SNPs that map across the genome. Genotypes for each SNP were generated as a result of the clustering of relative allele signal (RAS) values into three types AB, AA and BB. A fourth set was composed of those SNPs that could not be genotyped by the computer analysis, which were designated as “no calls”. To illustrate, figure 1 is a comparison of the genotypes along chromosome 2 for a tumor/normal pair showing no significant difference between the patient’s tumor and normal DNA.
Figure 1.
Representation of chromosome 2 SNP genotypes for tumor and paired-normal DNA. Algorithm-generated SNP genotypes (AA, BB, AB) for each position on the chromosome were assigned separate numerical values for tumor 6374T and normal 6375N respectively to enable graphical comparison.
AA/BB - homozygous allele calls; AB – heterozygous allele calls.
NoCalls are a result of signals that cannot be classified as either homozygous or heterozygous calls.
We identified five tumors that demonstrated regions of conversion of heterozygous genotypes to homozygous genotypes, in comparison with corresponding normal tissue DNA from each patient; regions of conversion included both entire chromosomes and segments along various chromosomes. Loss of heterozygosity is generally associated with a loss of one allele, resulting in copy number reduction. Alternatively, amplification of an allele can shift the relative copy number ratio such that the algorithm reads the signal as loss of heterozygosity. Our previous BAC array CGH studies found that the tumors in our study do not exhibit significant copy number changes in these same regions; conversion of these regions to uniparentalism underlies the copy number neutral losses of heterozygosity.
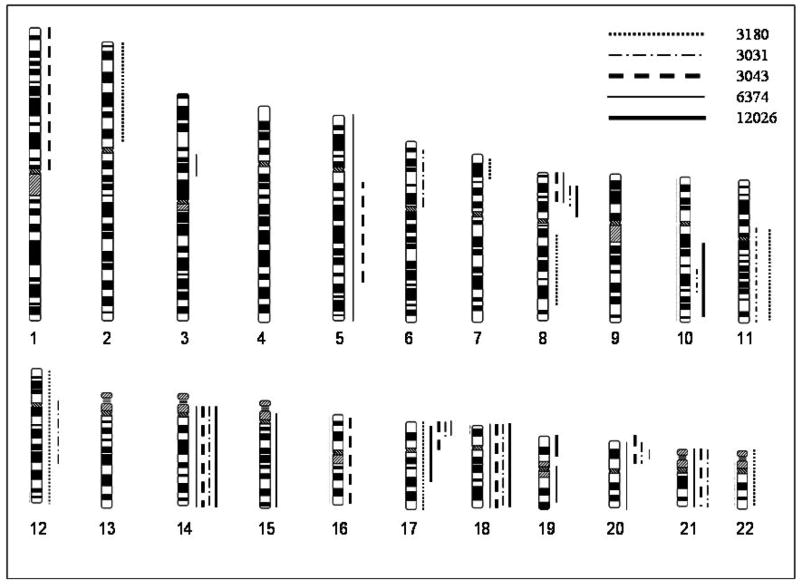
Chromosome 14 and chromosome 18 showed coordinate uniparentalism in four samples (fig 2), which was confirmed by the detection of two copies of each chromosome by FISH analysis (data not shown). Although we have shown how a history of cigarette smoking can affect tumor genomic profiles [24], it did not appear to strongly impact this coordinate uniparentalism; two of the four patients with uniparentalism of chromosomes 14 and 18 were cigarette smokers, and two were not. Figure 3 illustrates for all tumors studied those chromosomes demonstrating a conversion of heterozygous genotypes to homozygous genotypes, either across an entire chromosome or for chromosomal segments. Our previous analyses of 59 sporadic colorectal tumors found the elevated LOH for regions of chromosomes 5, 8, 11, 12, 17, and foremost 14 and 18; these are the same chromosomes that were seen in those cases with either whole chromosome or segmental uniparentalism on the SNP arrays [12]. It is important to note, however, that many of the tumors with LOH for these chromosomes did not exhibit uniparental disomy.
Figure 2. Uniparentalism observed coordinately in chromosomes 14 and 18.

Four tumors, 6374, 3031, 3043, 12026 displayed whole chromosome homozygosity or uniparentalism in chromosomes 14 and 18 compared to their corresponding normal samples 6375, 3032, 3044, 12028.
AA/BB indicate homozygous genotype of SNPs distributed across chromosomes 14 and 18. AB indicates the heterozygous genotype.
Figure 3.
Karyotype displaying whole chromosome and partial uniparentalism of five tumors. Tumors 6374, 3031, 3043, 3180 and 12026 are represented as indicated in the legend.
3.2. Does imprint selection occur for uniparental chromosomes?
Conversion to uniparentalism might reflect a selection process for somatic mutations occurring over the course of tumor progression, with elimination of the remaining normal allele. Alternatively, uniparentalism may reflect a process of selection for or against selection of imprinted alleles. In either case, potential problems with haploinsufficiencies are evaded.
Five chromosomes showing uniparentalism included an imprinted gene in the uniparental region. Analysis of the imprint status at the five different loci in eleven samples was performed using methylation specific PCR (MSP), bisulfite sequencing and Sequenom mass array analyses to evaluate if there is a selection bias towards the methylated or the unmethylated imprinted state in uniparental chromosomes. The imprinted status of a gene may vary between tissues; to confirm the imprinted status in colon tissue we also utilized the normal DNA from ACF-paired normal colon samples as a control. Such ACF (aberrant crypt foci) are believed to represent benign precursors to colon carcinomas. Sequenom mass array data for all the genes analyzed verified a 50% methylation profile, indicating their imprinted status in normal colon tissue (Sup. fig 1). Similarly DNA from normal tissues and tumors that do not show uniparentalism demonstrated a 50% methylation profile, as expected.
Chromosome 1, which includes the imprinted gene Noey2 (ARHI), showed segmental uniparentalism. Bisulfite sequencing and Sequenom mass-array analysis indicated the retention of the unmethylated allele in the tumor. The paired normal sample revealed the presence of both the unmethylated and methylated allele in bisulfite sequencing and a 50% methylation profile via mass-array (Fig 5a and fig 6a).
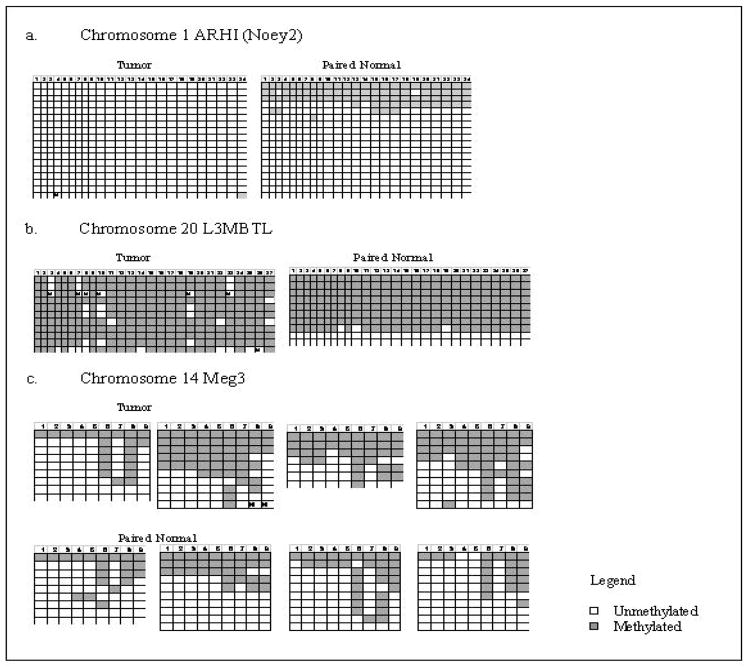
Figure 5. Bisulfite sequencing data for chromosomes 1, 14 and 20.
Each row represents an individual clone and each square a CpG dinucleotide. Open squares indicate unmethylated CpG and filled squares methylated CpG.
(a) Chromosome 1 - tumor retained the unmethylated allele while both alleles are present in the paired normal
(b) Chromosome 20 - tumor retained the methylated allele while corresponding normal demonstrates presence of both alleles. A tumor- normal pair that did not show uniparentalism was also analyzed and showed a 50% pattern in both tumor and paired normal (data not included).
Chromosome 14 - four tumor and paired normal samples are represented in the upper and lower panel respectively. Tumor samples demonstrate a partial methylation pattern while paired normal samples include both unmethylated and methylated alleles.
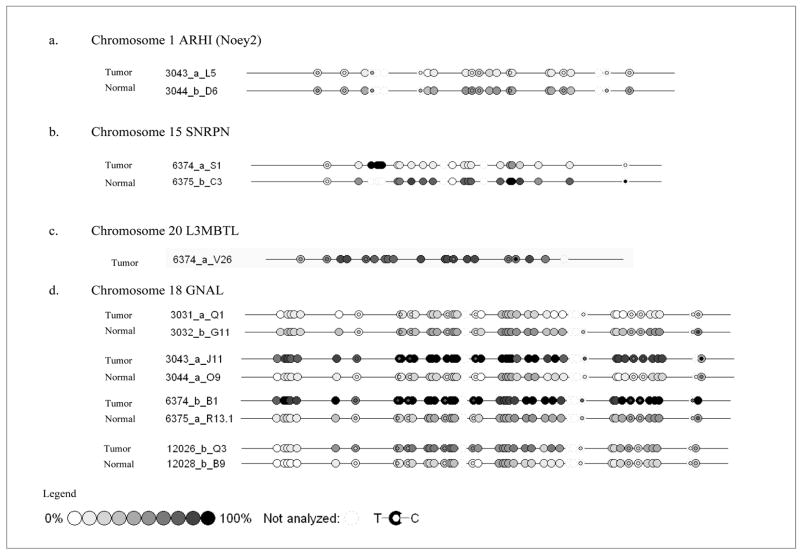
Figure 6. Mass-array Sequenom data for chromosomes 1, 15, 18 and 20.
Each circle indicates a CpG dinucleotide, unmethylated CpG is white, 50% methylation as expected for an imprinted gene is gray and 100% methylation is indicated by black. Controls for Mass-Arrray Sequenom included 0% to100% gradient, and ACF-paired normal DNA as control normal colon DNA. Tumor and paired-normal were run in duplicates when possible to confirm results. Paired normal for chromosome 20 did not give any result.
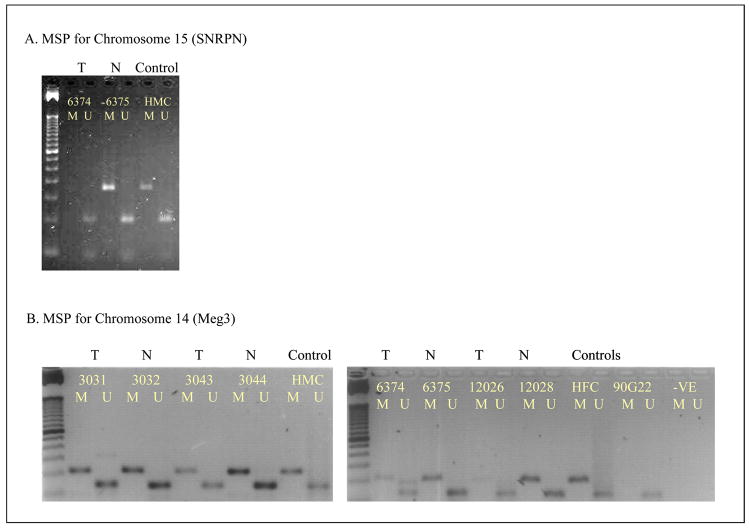
Chromosome 15, includes the SNRPN cluster of imprinted genes, showed whole chromosome uniparentalism and retained the unmethylated allele as detected both by methylation specific PCR (fig 4a) and by mass-array analysis (fig 6b). Both uniparental chromosome 1 and chromosome 15 included only a single tumor from our set of 13 tumors and both retained the unmethylated allele. In contrast, analysis of chromosome 20 that included the L3MBTL imprinted gene indicated the retention of the methylated allele by bisulfite sequencing and mass-array (Fig 5b and Fig 6c).
Figure 4.
Methylation specific PCR for tumors with uniparental chromosome 14 (Meg3) and chromosome 15 (SNRPN). T and N indicate tumor and paired normal DNA. M indicates methylation specific PCR product, U indicates unmethylated PCR product. Control DNA - HFC and HMC are pooled human lymphocyte DNA; RP11-90G22 is a BAC clone that includes Meg3. The left lane shows an Invitrogen 50 bp DNA ladder electrophoretic standard.
Our analyses of chromosomes 1, 15 and 20, included one tumor that showed partial uniparentalism in the case of chromosome 1 and one tumor showing whole chromosome uniparentalism in the cases of chromosome 15 and chromosome 20. Our analyses included three tumors; two tumors retained the unmethylated allele and one tumor the methylated allele, indicating a lack of selection for imprint status.
To determine if the allele retention patterns are locus-specific, we also analyzed four tumors showing whole chromosome uniparentalism of chromosomes 14 and 18. Sequencing data for chromosome 14 showed a partial methylation pattern (fig 5c), while MSP showed the presence of both unmethylated and methylated allele (fig 4b). Previously published data analyzing the Meg3 promoter upstream region does not indicate the expected 50% methylation pattern at all CpG dinucleotides in normal tissue, with a select few CpG dinucleotides exhibiting a hemi-methylated state; these reports describe a consolidated average for the sequencing data of 5 to 10 individual clones [25,26]. We were thus unable to compare our data with a precise picture of the methylation pattern for a particular allele. Our data may represent the methylated allele for reasons discussed above or be indicative of aberrant methylation at the locus, a result of the tumor environment.
Chromosome 18 includes GNAL, an imprinted gene that contains two alternative promoters (Golf and XLGolf) both reported to be differentially methylated in human brain tissue [27]. However our Mass-array analyses indicate that only the downstream Golf promoter is imprinted in colon tissue, while the upstream XLGolf promoter is unmethylation (suppl. fig 1c & d). At the imprinted Golf promoter three tumors retained a methylated allele and one tumor retained the unmethylated allele (Fig 6d). These data do not support a specific selection based on imprint status on chromosome 18, even at an individual locus (Table 1).
4. Discussion
Our results analyzing colon carcinoma DNAs demonstrate the frequent conversion of heterozygous genotypes to homozygous genotypes without change in copy number, indicative of uniparental disomy. We identified both segmental and whole chromosome uniparentalism in five of thirteen tumors studied on SNP microarrays. Four of these five tumors displayed coordinate uniparentalism for chromosome 14 and chromosome 18. Since such uniparentalism was observed in tumor DNA but not in corresponding normal tissue DNA from each patient, uniparentalism must be arising during the course of tumor progression. We further evaluated the role imprinting status plays in the selection of chromosomes during the conversion of the heterozygous genotype to the homozygous genotype, and found the imprinted state is not selected for and does not affect which allele is retained and reduplicated by the uniparentalism.
Sporadic colon cancers occur as a result of numerous alterations of the genome, now including the generation of loss of heterozygosity without corresponding change in chromosomal copy number. Other recent studies have also employed SNP microarrays, with uniparentalism detected in pancreatic endocrine tumors [28], hepatocellular carcinoma [29], basal cell carcinomas [30], neuroblastomas [31], breast cancer [32], various myeloproliferative syndromes [33–36] and lymphomas [37–39].
Three recent reports indicate the presence of uniparental chromosomes in colon carcinomas, although none have looked at the imprint status of these chromosomes [40–42]. Gaasenbeek et al. examined 45 cell lines with array comparative genomic hybridization and SNP arrays to identify regions of uniparental chromosomes, although no patient samples were analyzed [40]. An analysis of 15 adenocarcinomas by Andersen et al identified uniparental regions on chromosome 3, 8,13,15 and 20, although in these analyses copy number was determined from a set of 113 unrelated germ line samples [41]. In contrast, our analyses examined SNP microarray data for tumor and paired-normal DNA to determine LOH and copy number, and identified a number of whole and segmental chromosomes displaying uniparentalism. In a third study analyzing colon cell lines, Melcher et al proposed that the formation of uniparental chromosomes is an early event in tumorigenesis, because such chromosomes contain genes that have been associated with early events of a linear model of colon cancer progression [42]. In contrast, we have used SNP microarray analysis on four aberrant crypt foci with high genomic instability as determined by inter- (simple sequence repeat) PCR, and found no uniparental chromosomes or subchromosomal regions; this suggests that the onset of the uniparental form of instability occurs during progression at the adenoma stage or later [43].
Potential roles for uniparental chromosomes in cancer progression include both the revealing of recessive somatic mutations and the disruption of imprinted genes. Loss of imprinting has been implicated in various cancers [14–16]. We found no selection bias toward either the methylated or unmethylated imprinted state during the generation of uniparental chromosomes indicating it is a random process in this regard.
Sporadic colon cancer progression is generally believed to result from destabilization of the genome, followed by ongoing selection for tumor-advantageous mutations. This destabilization has been demonstrated to include several processes including single base alterations, large amplifications or deletions, reciprocal and non-reciprocal translocations, and epigenetic changes and based on this work and work by others uniparentalism. Our results reinforce the necessity to view sporadic colon cancer as the result of several genomic instabilities together effecting diverse genes and pathways, producing both the inter-tumor and intra-tumor heterogeneity that typifies carcinomas. And it is these heterogeneities that in turn have so frustrated our efforts at curing colorectal cancer by non-surgical means.
Supplementary Material
Table 2.
Imprint status at chromosomes showing partial (chromosome1) and whole (chromosome 14, 15, 18 and 20) chromosome uniparentalism. Data based on methylation specific PCR (MSP), bisulfite sequencing and Sequenom MassArray analyses.
| CHROMOSOME | IMPRINTED GENE | ALLELE RETAINED |
|---|---|---|
| 1 | ARHI (Noey2) | Unmethylated |
| 14 | Meg3 | Partial methylation |
| Partial methylation | ||
| Partial methylation | ||
| Partial methylation | ||
|
| ||
| 15 | SNRPN | Unmethylated |
|
| ||
| 18 | GNAL (Golf) | Unmethylated |
| Methylated | ||
| Methylated | ||
| Methylated | ||
|
| ||
| 20 | L3MBTL | Methylated |
NORMAL IMPRINT PATTERNS:
Meg3: methylated (M) – paternal, unmethylated (UM) - maternal
SNRPN: methylated (M) – maternal, unmethylated (UM) - paternal
L3MBTL: methylated (M) – maternal, unmethylated (UM) –paternal
Parental source of expression unknown for ARHI and GNAL.
Acknowledgments
This work was supported by the NIH grant R01CA74018. Nicholas Petrelli provided the surgical specimens, and pathological examinations were done by Janet Winston. Michelle Zanche of the University of Rochester provided valuable help with the SNP microarrays. Valuable discussions with Michael Higgins led to the analysis of imprint status in the uniparental chromosomes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balmain A. Cancer genetics: from Boveri and Mendel to microarrays. Nature Rev Cancer. 2001;1:77–82. doi: 10.1038/35094086. [DOI] [PubMed] [Google Scholar]
- 2.Boveri T. The Origin of Malignant Tumors. 1929. [Google Scholar]
- 3.Anderson GR, Stoler DL, Brenner BM. Cancer: the evolved consequence of a destabilized genome. BioEssays. 2001;23:1037–46. doi: 10.1002/bies.1149. [DOI] [PubMed] [Google Scholar]
- 4.Rajagopalan H, Lengauer C. Aneuploidy and Cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 6.Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–43. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- 7.Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–59. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 8.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 9.da Rocha ST, Ferguson-Smith AC. Genomic Imprinting. Curr Biol. 2004;14:R646–9. doi: 10.1016/j.cub.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Holmes R, Soloway PD. Regulation of imprinted DNA methylation. Cytogenet Genome Res. 2006;113:122–9. doi: 10.1159/000090823. [DOI] [PubMed] [Google Scholar]
- 11.Bartos JD, Gaile DP, McQuaid DE, Conroy JM, Darbary H, Nowak NJ, Block A, Petrelli NJ, Mittelman A, Stoler DL, Anderson GR. aCGH local copy number aberrations associated with overall copy number genomic instability in colorectal cancer: coordinate involvement of the regions including BCR and ABL. Mutat Res. 2007;615:1–11. doi: 10.1016/j.mrfmmm.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson GR, Brenner BM, Swede H, Chen N, Henry WM, Conroy JM, Karpenko MJ, Issa J-P, Bartos JD, Brunelle JK, Jahreis GP, Kahlenberg MS, Basik M, Sait S, Rodriguez-Bigas MA, Nowak NJ, Petrelli NJ, Shows TB, Stoler DL. Intrachromosomal Genomic Instability in Human Sporadic Colorectal Cancer Measured by Genome-Wide Allelotyping and Inter-(Simple Sequence Repeat) PCR. Cancer Res. 2001;61:8274–83. [PubMed] [Google Scholar]
- 13.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. BioEssays. 2000;22:452–9. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 14.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 15.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–8. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- 16.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–9. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 17.Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA. 1999;96:8064–9. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoler D, Chen N, Basik M, Kahlenberg M, Rodriguez-Bigas M, Petrelli N, Anderson G. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci U S A. 1999 Dec 21;96(26):15121–6. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basik M, Stoler D, Kontzoglou K, Rodriguez-Bigas M, Petrelli N, Anderson G, Greider CW. Genomic instability in sporadic colorectal cancer quantitated by inter-simple sequence repeat PCR analysis. Genes Chromosomes Cancer. 1997;18:19–29. [PubMed] [Google Scholar]
- 20.Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, Yang G, Kennedy GC, Webster TA, Cawley S, Walsh PS, Jones KW, Fodor SP, Mei R. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–11. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 21.Liu WM, Di X, Yang G, Matsuzaki H, Huang J, Mei R, Ryder TB, Webster TA, Dong S, Liu G, Jones KW, Kennedy GC, Kulp D. Algorithms for large-scale genotyping microarrays. Bioinformatics. 2003;19:2397–403. doi: 10.1093/bioinformatics/btg332. [DOI] [PubMed] [Google Scholar]
- 22.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001:29. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy SK, Wylie AA, Coveler KJ, Cotter PD, Papenhausen PR, Sutton VR, Shaffer LG, Jirtle RL. Epigenetic detection of human chromosome 14 uniparental disomy. Hum Mutat. 2003;22:92–7. doi: 10.1002/humu.10237. [DOI] [PubMed] [Google Scholar]
- 24.Swede H, Bartos JD, Chen N, Shaukat A, Dutt SS, McQuaid DA, Natarajan N, Rodriguez-Bigas MA, Nowak NJ, Wiseman SM, Alrawi S, Brenner BM, Petrelli NJ, Cummings KM, Stoler DL, Anderson GR. Genomic profiles of colorectal cancers differ based on patient smoking status. Cancer Genet Cytogenet. 2006;168:98–104. doi: 10.1016/j.cancergencyto.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, Grundy R, Ferguson-Smith AC, Maher ER. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br J Cancer. 2005;92:1574–80. doi: 10.1038/sj.bjc.6602478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90:2179–86. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 27.Corradi JP, Ravyn V, Robbins AK, Hagan KW, Peters MF, Bostwick R, Buono RJ, Berrettini WH, Furlong ST. Alternative transcripts and evidence of imprinting of GNAL on 18p11.2. Mol Psychiatry. 2005;10:1017–25. doi: 10.1038/sj.mp.4001713. [DOI] [PubMed] [Google Scholar]
- 28.Nagano Y, Kim do H, Zhang L, White JA, Yao JC, Hamilton SR, Rashid A. Allelic alterations in pancreatic endocrine tumors identified by genome-wide single nucleotide polymorphism analysis. Endocr Relat Cancer. 2007;14:483–92. doi: 10.1677/ERC-06-0090. [DOI] [PubMed] [Google Scholar]
- 29.Midorikawa Y, Yamamoto S, Ishikawa S, Kamimura N, Igarashi H, Sugimura H, Makuuchi M, Aburatani H. Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene. 2006;25:5581–90. doi: 10.1038/sj.onc.1209537. [DOI] [PubMed] [Google Scholar]
- 30.Teh MT, Blaydon D, Chaplin T, Foot NJ, Skoulakis S, Raghavan M, Harwood CA, Proby CM, Philpott MP, Young BD, Kelsell DP. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 2005;65:8597–603. doi: 10.1158/0008-5472.CAN-05-0842. [DOI] [PubMed] [Google Scholar]
- 31.George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, Look AT, Maris JM. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS ONE. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murthy SK, DiFrancesco LM, Ogilvie RT, Demetrick DJ. Loss of heterozygosity associated with uniparental disomy in breast carcinoma. Mod Pathol. 2002;15:1241–50. doi: 10.1097/01.MP.0000032535.62750.D1. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S, Lillington D, Lister TA, Young BD. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–4. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 34.Flotho C, Steinemann D, Mullighan CG, Neale G, Mayer K, Kratz CP, Schlegelberger B, Downing JR, Niemeyer CM. Genome-wide single-nucleotide polymorphism analysis in juvenile myelomonocytic leukemia identifies uniparental disomy surrounding the NF1 locus in cases associated with neurofibromatosis but not in cases with mutant RAS or PTPN11. Oncogene. 2007;26:5816–21. doi: 10.1038/sj.onc.1210361. [DOI] [PubMed] [Google Scholar]
- 35.Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP. SNP Array Karyotyping Allows for the Detection of Uniparental Disomy and Cryptic Chromosomal Abnormalities in MDS/MPD-U and MPD. PLoS ONE. 2007;2:e1225. doi: 10.1371/journal.pone.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gondek LP, Tiu R, O’Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD and MDS-derived AML. Blood. 2008;111:1534–42. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielaender I, Martín-Subero JI, Wagner F, Martínez-Climent JA, Siebert R. Partial uniparental disomy: a recurrent genetic mechanism alternative to chromosomal deletion in malignant lymphoma. Leukemia. 2006;20:904–5. doi: 10.1038/sj.leu.2404173. [DOI] [PubMed] [Google Scholar]
- 38.Fitzgibbon J, Iqbal S, Davies A, O’shea D, Carlotti E, Chaplin T, Matthews J, Raghavan M, Norton A, Lister TA, Young BD. Genome-wide detection of recurring sites of uniparental disomy in follicular and transformed follicular lymphoma. Leukemia. 2007;21:1514–20. doi: 10.1038/sj.leu.2404696. [DOI] [PubMed] [Google Scholar]
- 39.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13:4777–85. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- 40.Gaasenbeek M, Howarth K, Rowan AJ, Gorman PA, Jones A, Chaplin T, Liu Y, Bicknell D, Davison EJ, Fiegler H, Carter NP, Roylance RR, Tomlinson IP. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex changes and multiple forms of chromosomal instability in colorectal cancers. Cancer Res. 2006;66:3471–9. doi: 10.1158/0008-5472.CAN-05-3285. [DOI] [PubMed] [Google Scholar]
- 41.Andersen CL, Wiuf C, Kruhøffer M, Korsgaard M, Laurberg S, Ørntoft TF. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. doi: 10.1093/carcin/bgl086. [DOI] [PubMed] [Google Scholar]
- 42.Melcher R, Al-Taie O, Kudlich T, Hartmann E, Maisch S, Steinlein C, Schmid M, Rosenwald A, Menzel T, Scheppach W, Luhrs H. SNP-Array genotyping and spectral karyotyping reveal uniparental disomy as early mutational event in MSS- and MSI-colorectal cancer cell lines. Cytogenet Genome Res. 2007;118:214–21. doi: 10.1159/000108303. [DOI] [PubMed] [Google Scholar]
- 43.Alrawi SJ, Carroll RE, Hill HC, Gibbs JF, Tan D, Brenner BM, Nowak NJ, Swede H, Stoler DL, Anderson GR. Genomic instability of human aberrant crypt foci measured by inter-(simple sequence repeat) PCR and array-CGH. Mutat Res. 2006;601:30–8. doi: 10.1016/j.mrfmmm.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Schofield PN, Joyce JA, Lam WK, Grandjean V, Ferguson-Smith A, Reik W, Maher ER. Genomic imprinting and cancer; new paradigms in the genetics of neoplasia. Toxicol Lett. 2001;120:151–60. doi: 10.1016/s0378-4274(01)00294-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.