Abstract
Colchicine and nocodazole, both established microtubule disruptors, are useful tools to investigate cytoskeletal-dependent signaling cascades and the associated downstream transcriptional targets. Since cytoskeletal events impact pathophysiologic consequences in the vascular system, the signaling requirements underlying colchicine-stimulated expression of PAI-1 and CTGF, two prominent cell deformation-sensitive fibrosis-initiating proteins, were evaluated in vascular smooth muscle cells. Microtubule disruption rapidly induced EGFR transactivation (at the src kinase-sensitive EGFRY845 site) in a ROS-dependent manner. Genetic deficiency of EGFR, inhibition of EGFR signaling with AG1478 or introduction of a kinase-deficient EGFR construct effectively blocked colchicine-stimulated PAI-1 and CTGF expression. MEK/ERK involvement downstream of ROS generation was critical for PAI-1, but not CTGF, expression following cytoskeletal perturbation suggesting bifurcation of signaling pathways downstream of EGFR activation. Colchicine also stimulated SMAD2/3 phosphorylation by a Rho/ROCK-dependent mechanism independent of TGF-β1 release or receptor activity. Rho/ROCK signaling initiated by tubulin network collapse was required for both CTGF and PAI-1 induction. Colchicine-initiated SMAD3 phosphorylation, however, was essential for PAI-1, but not CTGF, expression further highlighting divergence of signaling events downstream of Rho/ROCK that mediate microtubule deformation-associated changes in profibrotic gene transcription.
Keywords: PAI-1, CTGF, Microtubule Cytoskeleton, Colchicine, EGFR, SMAD, Rho/ROCK
INTRODUCTION
The microtubule cytoskeleton, a central regulator of cell shape, transduces intracellular signals in response to cues from the extracellular matrix, cell-cell interactions and growth factors [1]. The tubulin network sequesters, by direct or indirect interactions, various effector elements (e.g., SMADs, Rho-GEFs) modulating their activity and subcellular trafficking [2–7]. Not surprisingly, therefore, microtubule disruption impacts events that regulate the expression of downstream, cell deformation-sensitive, genes. Cytoskeletal network reorganization, however, is associated with changes in a relatively modest complement of genes, unlike the global transcriptional reprogramming induced by serum growth factors [e.g., 8,9], with PAI-1 [10], uPA [11,12], TGF-β1 [13], CTGF [14,15], COX-2 [16], VEGF [17] and several MMPs [18–20] prominent among this restricted subset of cell shape-sensitive genes.
The microtubule-disrupting drugs colchicine and nocodazole have emerged as useful reagents for the identification of “deformation” sensitive genetic responses [21,22]. Several of the downstream targets activated by microtubule disassembly are important regulators of tissue fibrosis (e.g. collagen, PAI-1, CTGF). Clarification of signaling pathways associated with disruption of the tubulin-based cytoskeleton, therefore, may well have specific clinical implications. This is particularly true in the vascular system where cytoskeletal alterations commonly accompany ischemia/reperfusion injury, arterial tensional adaptations and wound repair as well as the response to microtubule-targeting drugs. PAI-1 induction is evident in each setting [23–26], warranting further definition of mechanisms activated by cytoskeletal remodeling that affect regulation of profibrotic genes.
This report details events initiated by tubulin network alterations leading to PAI-1 and CTGF expression in VSMC. EGFR transactivation, involving EGFRY845 site phosphorylation, and Rho/ROCK signaling were essential elements in the microtubule deformation-sensitive PAI-1 and CTGF expression control pathways. MEK/ERK1/2 involvement downstream of the EGFR was critical for PAI-1, but not CTGF, induction. Colchicine treatment also stimulated delayed SMAD2/3 phosphorylation independent of autocrine TGF-β1 release or TGF-β1 receptor activity but requiring Rho/ROCK signaling. Induced SMAD3 activity by microtubule disruption, furthermore, was required for PAI-1 expression but dispensable for CTGF induction. These data indicate that multiple pathways are engaged as a consequence of cytoskeletal disruption and suggest, furthermore, that transcriptional controls on individual microtubule deformation-sensitive genes are complex and likely different.
MATERIALS AND METHODS
Cell culture
R22 aortic smooth muscle cells as well as EGFR wild-type (EGFR+/+) and null (EGFR−/−) MEFs were grown to near confluence in DMEM/10% FBS [27,28] then maintained in serum-free medium for 2–3 days prior to addition of colchicine (1–10 μM), nocodazole (0.5–5 μM), TGF-β1 (0.1 to 1 ng/ml) or EGF (10 ng/ml). Primary VSMC were propagated in F-12/DMEM supplemented with 10% FBS and serum-deprived for 1–2 days before incubation with colchicine or growth factors. Pretreatment with the pharmacologic inhibitors AG1478 (EGFR), C3 transferase (Rho), Y-27632 (p160ROCK), SIS3 (SMAD3), PD98059 or U0126 (MEK) as well as the ROS inhibitors (NAC and DPI) is described in the text.
Adenoviral constructs
Cells were infected (MOI=50–100) with the adenoviral GFP-tagged kinase-dead EGFR1 mutant EGFRK721A, wild-type EGFR1 or control-GFP expression constructs in low-serum medium for 48 hours as described [28] then stimulated with EGF(10 ng/ml), TGF- β1 (1ng/ml) or colchicine (10 μM) for 4 hours prior to cell extraction. Infectivity was >90% (assessed by GFP fluorescence microscopy).
Western analysis
R22 cells and primary VSMC were solubilized at 4°C in lysis buffer (0.5% deoxycholate, 0.1% SDS, 50 mM HEPES, pH 7.5, 1% Triton X-100, 1% NP-40, 150 mM NaCl, 50 mM NaF, 1mM vanadate, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 1mM PMSF) and extracts clarified at 14,000 ×g for 15 minutes. MEFs were disrupted in 4% SDS/PBS for 10 minutes, lysates vortexed briefly, boiled for 5 minutes then centrifuged at 14,000 rpm for 15 minutes. Aliquots (30–50 μg cellular protein) were electrophoretically-separated, transferred to nitrocellulose, membranes blocked in 5% milk in 0.05% Triton-X 100/PBS, incubated overnight with antibodies to PAI-1, EGFR, pERK1/2, ERK2, pSMAD2Ser465/467, pSMAD3Ser423/425, SMAD2/3, RhoA, EGFRpY845, pp60c-src-pY416; anti-phospho-tyrosine (4G10), or TGF-βRI in blocking buffer and washed three times in 0.05% Triton-X 100/PBS prior to incubation with secondary antibodies [28]. Immunoreactive proteins were visualized with ECL reagent and quantitated by densitometry.
Immunofluorescence microscopy
For visualization of microtubule organization, cells were fixed in 3.7% paraformaldehyde, permeabilized in 0.1% Triton X-100/PBS for 10 minutes, blocked in 2% BSA for 20 minutes then incubated with mouse monoclonal antibodies to tubulin for one hour and then with appropriate Alexa fluor 568 conjugated antibodies to mouse IgG. Following a final series of PBS rinses, coverslips were mounted with Vectashield reagent containing DAPI (Vector Laboratories) to visualize nuclei. Immunocytochemical localization of PAI-1 used specific antibodies and the same protocol for tubulin imaging described above.
Rho GTP assay
PBS-washed cells were extracted (in 25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol containing leupeptin and 1 mM sodium orthovanadate) by constant agitation for 15 minutes at 4°C. Clarified lysates (600 μg protein) were incubated with Rhotekin RBD-agarose beads for 45 minutes at 4°C. Active (i.e., Rhotekin-bound) Rho and total Rho levels (GTP-Rho + GDP-Rho) were determined by western blotting with RhoA antibodies.
Measurement of ROS
Serum-deprived VSMC were stimulated with colchicine for various lengths of time (1–120 minutes) then incubated with DCF-DA (Molecular Probes; 10 μM) in 37°C in the dark for 10 minutes. Following a PBS rinse, cells were scraped in 1 ml of 0.05% Triton X-100/PBS for fluorometer measurements (excitation at 488 nm; detection at 515 nm) and H2O2 levels normalized to cell number.
RESULTS
Microtubule collapse induces PAI-1 expression, src activation and EGFRY845 phosphorylation
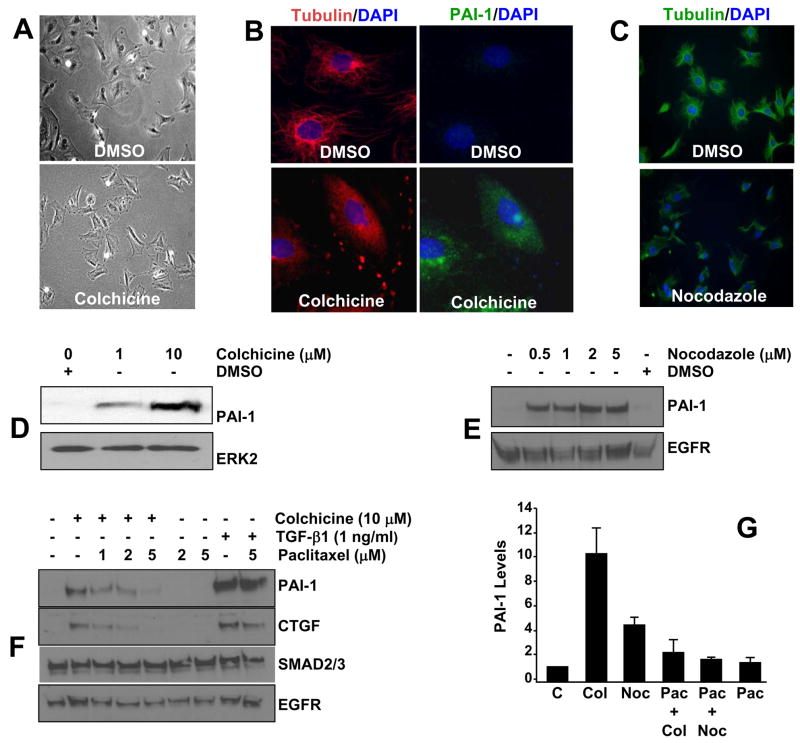
Deformation of the VSMC microtubule network with colchicine (Figure 1A,B) or nocodazole (Figure1C) induced a striking elevation in PAI-1 expression (Figure 1D,E). Stabilization of the tubulin cytoskeleton by pre-incubation with Paclitaxel (taxol; particularly at 5 μM) suppressed colchicine- and nocodazole-induced, but not TGF-β1-dependent, PAI-1 (and CTGF) induction (Figure 1 F,G) suggesting that microtubule disruption, rather than just drug exposure, is the basis for the associated increase in PAI-1/CTGF levels.
Figure 1. PAI-1 expression in response to microtubule disruption.
Exposure of quiescent VSMC to colchicine (10 μM in A,B) or nocodazole (2 μM in C) for 4 hours alters cell morphology (A), due to disruption of the tubulin network (B,C), and results in concentration-dependent increases PAI-1 levels (B,D,E). A 1 hour pre-incubation with the microtubule-stabilizing drug Paclitaxel, particularly at 2 and 5 μM, effectively suppressed colchicine-induced (10 μM) (F,G) or nocodazole-stimulated (1 μM) (G), but not TGF-β1-initiated (F), PAI-1 and CTGF expression. ERK2 (D), EGFR and SMAD2/3 (E,F) provided loading controls. Exposure to colchicine and nocodazole (or DMSO vehicle) and TGF-β1, in all cases, was for 4 hours. In (G), C = control (drug-free), Col = colchicine, Noc = nocodazole, Pac = paclitaxel; data plotted is the mean±SD of triplicate western assessments of PAI-1 levels.
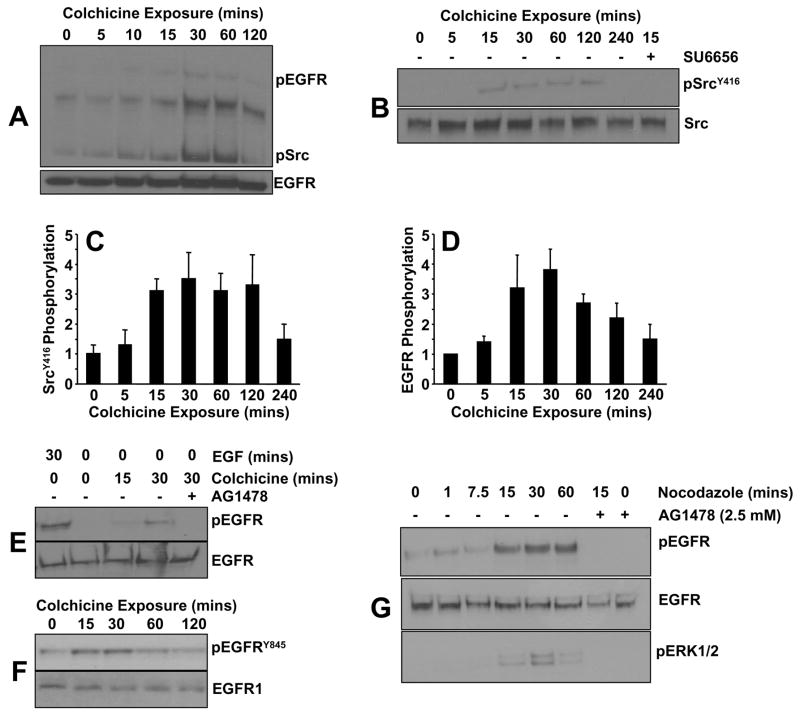
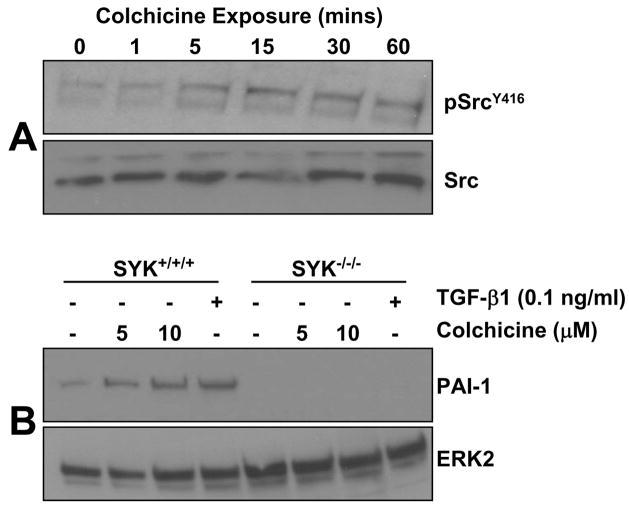
Previous studies implicated src family non-receptor tyrosine kinases, and specifically pp60c-src, as signaling intermediates in response to microtubule disassembly [10,15]. Analysis of protein phosphorylation patterns (assessed with the phosphotyrosine-specific antibody 4G10) as a function of time of colchicine addition disclosed a rapid increase in src kinase phosphorylation within 10 minutes of drug exposure (Figure 2A). The sensitivity of the colchicine-induced kinase activity to the src family inhibitor SU6656 (Figure 2B) is consistent with the prior identification of the involved kinase as pp60c-src [10]. Maximal srcY416 site phosphorylation occurred approximately 30 minutes after colchicine addition remaining elevated for at least 2 hours (Figure 2C). A pattern of phosphorylation similar to that of pp60c-src was also evident for a protein of approximately 170 kDa (Figure 2A) which co-migrated with the phosphorylated form of the EGFR (Figure 2A,D). While the elevated pEGFR levels associated with colchicine exposure are less than what commonly accompanies stimulation by the physiologic ligand EGF (Figure 2D), the kinetics of EGFR phosphorylation closely paralleled that of src kinase (Figure 2C,E). Consistent with src kinase involvement, the EGFRY845 site (a pp60c-src target residue; [29]) was significantly increased in response to microtubule disruption (Figure 2F). Consistent with the role of Src family members in EGFRY845 phosphorylation in VSMC, colchicine also activated (within 15 minutes) c-Src kinase activity (measured by SrcY416 phosphorylation) in mouse embtyonic fibroblasts (MEFs) (Figure 3A). Importantly, colchicine failed to stimulate PAI-1 expression in MEFs tripley-deficient in src family kinases (src, yes, fyn; SYF−/−/−) but effectively induced PAI-1 in wild type SYF+/+/+) fibroblasts confirming a participation of these non-receptor tyrosine kinases in PAI-1 gene induction by microtubule deformation (Figure 3B).
Figure 2. Colchicine exposure stimulates EFGR/c-Src kinase phosphorylation.
Western analysis using phospho-tyrosine (4G10) and c-src kinase pY416 residue- specific antibodies indicated that both c-src kinase (A–C) and the EGFR (A,D) are tyrosine phosphorylated following colchicine addition. While the magnitude of EGFR tyrosine phosphorylation initiated by colchicine is significantly less than that induced by the physiologic ligand EGF (E), phosphorylation of the EGFRY845 residue (an established Src kinase target) is a rapid response to cytoskeletal deformation in VSMC (F). Similar to colchicine, nocodazole treatment also results in EGFR and ERK1/2 activation suggesting that stimulation of downsteam signaling is a general consequence of microtubule disruption (G).
Figure 3. Role of Src family kinases in cell deformation induced PAI-1 induction.
Colchcine treatment induced a rapid increase (within 15 minutes) in Src kinase activation (as assessed by phosphorylation at the SrcY416 site) without alterations in total amounts c-Src protein (A). PAI-1 induction was clearly evident in colchicine-stimulated wild-type mouse embryonic fibroblasts (SYF+/+/+ MEFs) but not in Src, Yes and Fyn triple-null cells (SYK−/−/− MEFs) (B). Such genetic approaches validate a crucial function of Src non-receptor tyrosine kinases in PAI-1 expression in response to microtubule deformation.
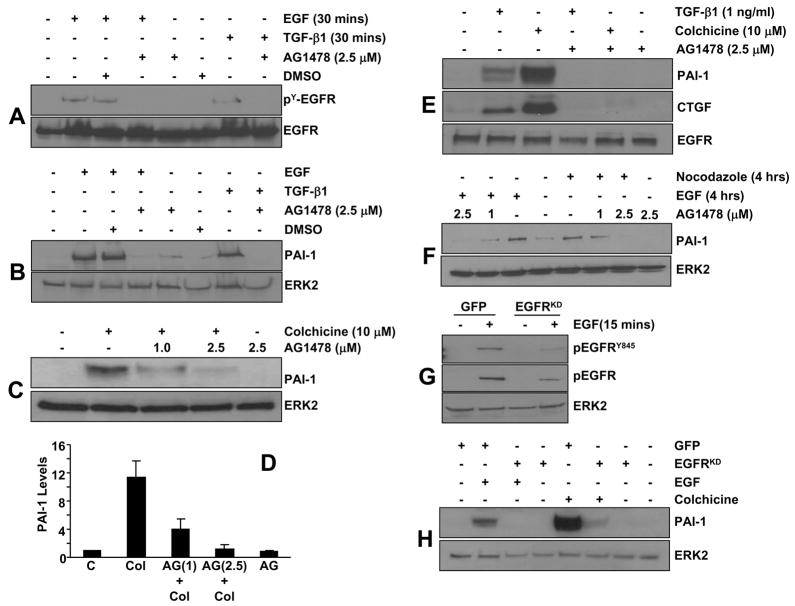
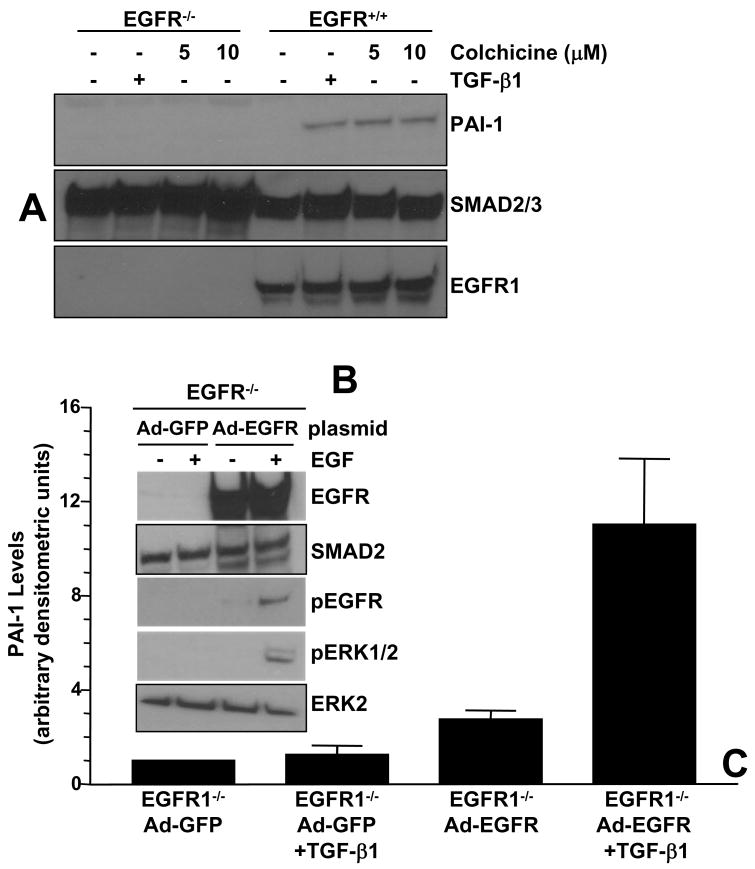
Preincubation of VSMC with the EGFR kinase inhibitor AG1478 effectively blocked EGFR tyrosine phosphorylation (Figure 4A) and downstream PAI-1 expression (Figure 4B) in response to EGF as well as TGF-β1. Colchicine-stimulated PAI-1 (Figure 4C,D) and CTGF (Figure 4E) induction was similarly sensitive to AG1478 pretreatment, implicating EGFR signaling in PAI-1/CTGF gene control following microtubule disruption. Nocodazole, moreover, similarly stimulated EGFR activation and AG1487 pretreatment virtually eliminated PAI-1 induction by nocodazole in a dose dependent fashion (Figure 4F). Consistent with the pharmacologic blockade, adenoviral delivery of the kinase-dead GFP-EGFRK721A mutant (EGFRKD), but not a GFP-control construct, significantly attenuated (by >75%) EGF-directed EGFR phosphorylation overall and, particularly, at the Y845 residue (Figure 4G). EGFRK721A also completely inhibited both EGF- and colchicine-stimulated PAI-1 induction (Figure 4H). While all three stimuli (EGF, TGF-β1, colchicine) markedly increased PAI-1 expression in EGFR1 wild-type (EGFR1+/+) fibroblasts, they were completely ineffective in EGFR1-deficient (EGFR1−/−) MEFs (e.g., Figure 5A). PAI-1 responsiveness, moreover, was restored in EGFR−/− cells engineered to re-express a wild-type EGFR1 construct (Figure 5B,C) suggesting that an EGFR1 with intact kinase activity is necessary for induced PAI-1 expression.
Figure 4. Involvement of EGFR in colchicine-mediated PAI-1 and CTGF expression.
AG1478 completely inhibited EGF-dependent (4G10 antibody-reactive) EGFR tyrosine phosphorylation as expected (A) and significantly attenuated PAI-1 expression (B) in VSMC. AG1478 pretreatment similarly blocked PAI-1 and CTGF induction in response a 4-hour exposure to either colchicine or TGF-β1 (C,D,E). Exposure to EGF and TGF-β1 for EGFR phosphorylation (A) and PAI-1 expression (B) assessments was for 30 minutes and 4 hours, respectively. DMSO solvent alone had no effect on either EGFR phosphorylation or PAI-1 induction (A,B). Data plotted in (D) is the mean±SD of triplicate western determination of PAI-1 levels in control (C) cultures, in colchicine (Col) or AG1478 (AG) treated VSMC and in cells pretreated with AG1478 (at 1 or 2.5 μM) prior to colchicine addition. As was the case for colchicine, nocodazole- mediated PAI-1 induction was also AG1478-sensitive (F). EGF-dependent EGFR phosphorylation, and specifically EGFRY845-induced phosphorylation, are significantly reduced following infection with a mutant GFP-EGFRK721A (kinase-dead; EGFRKD) adenoviral construct, compared to the EGFR phosphotyrosine levels in cells identically infected with the GFP-control virus (GFP), confirming function-blocking status of this mutant construct (G). Similar to the effective blockade of EGF-stimulated PAI-1 induction by the GFP-EGFRK721A construct, but not by GFP-control adenoviruses, colchicine-mediated PAI-1 expression is also completely eliminated by GFP-EGFRK721A viral infection confirming EGFR involvement in PAI-1 gene regulation.
Figure 5. EGFR1-null MEFs do not express PAI-1 in response to stimulation with colchicine or TGF-β1.
Colchicine-, as well as TGF-β1, -dependent increases in PAI-1 levels are evident in EGFR+/+ but not EGFR−/− MEFs (A). EGFR−/− cells were infected with adenoviruses bearing wild-type EGFR1 (Ad-EGFR) or control-GFP (Ad-GFP) constructs; expression of the introduced EGFR was confirmed by western blotting (B). Phosphorylation of the EGFR and ERK1/2 in response to a 15 minute EGF stimulation occurred in EGFR-reconstituted MEFs but not in cells infected with GFP-control adenoviruses (B). Similarly, recovery of TGF-β1-stimulated PAI-1 expression was obvious for EGFR-reconstituted MEFs and not in cells engineered to express GFP alone (C). Data plotted in (C) is the mean±SD of triplicate western assessments of cellular PAI-1 levels.
Differential MEK/ERK involvement in colchicine-induced PAI-1 and CTGF expression
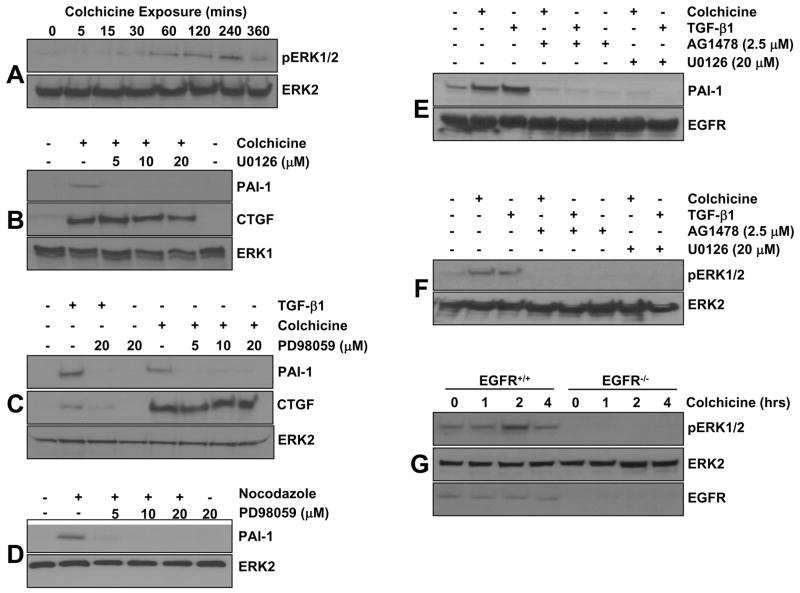
ERK1/2 activation is evident within 30 minutes and maximal at 2–4 hrs post-colchicine addition (Figure 6A), a time frame consistent with ERK involvement downstream of the EGFR, and sensitive to the two MEK inhibitors U0126 (Figure 6B) and PD98059 (Figure 6C). While the PAI-1 response to microtubule disruption requires MEK signaling, MEK kinase activity is dispensable for CTGF induction by colchicine in VSMC (Figures 6B,C) indicating a differential ERK1/2 requirement for PAI-1 vs. CTGF expression in response to cytoskeltal deformation. Nocodazole treatment similarly stimulated ERK1/2 phophorylation (Figure 2G) which is also required for PAI-1 induction by this microtubule disrupting drug (Figure 6D). Since AG1478 pretreatment inhibits colchicine or nocodazole-mediated PAI-1 induction (Figure 4C–F, 6E), it was necessary to assess the potential upstream involvement of the EGFR in signal propagation to ERK1/2. Increased ERK1/2 phosphorylation (Figure 6F) and PAI-1 expression (Figure 6E) following colchicine, nocodazole or TGF-β1 addition is virtually ablated by pretreatment of VSMC with AG1478 as well as U0126 (Figure 6E,F). Colchicine stimulates ERK1/2 phosphorylation, moreover, in EGFR+/+ but not EGFR−/− MEFs (Figure 6G), confirming the necessity for a functional EGFR1→ERK1/2 signaling axis for PAI-1 induction in response to microtubule disruption.
Figure 6. Role of the EGFR→MEK/ERK pathway in colchicine-induced PAI-1, but not CTGF, expression.
ERK1/2 is activated within 15–30 minutes of colchicine exposure with maximal phosphorylation evident at approximately 4 hours (A). PAI-1 induction by microtubule deformation, as well as by TGF-β1, is effectively ablated by pretreatment with the MEK inhibitors U0126 (B) or PD98059 (C). Colchicine-stimulated CTGF expression, in contrast, is insensitive to MEK inhibitors even at the highest concentration of U0126 or PD98059 (20 μM) (B,C). Nocodazole-mediated PAI-1 induction has similar MEK signaling requirements as PD98059 pretreatment virtually eliminates this response (D). PAI-1 expression (E) and ERK1/2 phosphorylation (F) in response to colchicine or TGF-β1 is also sensitive to preincubation of VSMC with the specific EGFR inhibitor AG1478. Colchicine-induced ERK1/2 phosphorylation, moreover, is evident in EGFR+/+ but not in EGFR−/− cells (G).
ROS involvement in EGFR activation and PAI-1 induction by microtubule disruption
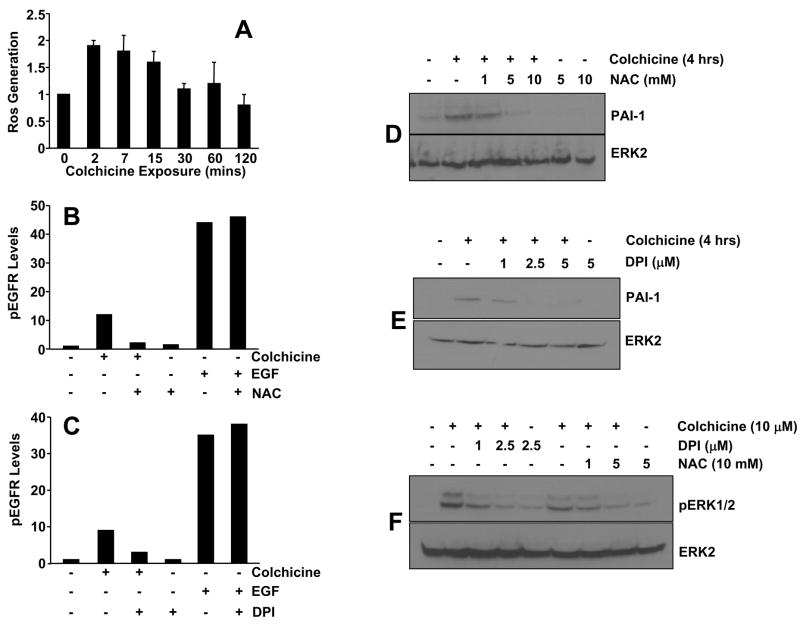
Colchicine addition rapidly generated (within 2 minutes) an increase in ROS (Figure 7A) that preceded the initial window of EGFR phosphorylation (15 to 30 minutes) (Figure 2A). NAC and DPI, established ROS scavengers, completely inhibited both colchicine-induced EGFR phosphorylation (Figure 7B,C) and PAI-1 expression (Figure 7D,E) suggesting a direct upstream signaling role of ROS in PAI-1 induction via EGFR transactivation. Indeed, ERK1/2 phosphorylation following microtubule disruption is effectively diminished by NAC or DPI pretreatment (Figure 7F), while neither inhibitor affected EGFR phosphorylation (Figure 7B,C) or ERK1/2 activation (not shown) in response to EGF.
Figure 7. ROS generation is required for EGFR transactivation and PAI-1 induction in response to microtubule deformation.
Reactive oxygen species (H2O2) are rapidly generated (within 2 minutes) following colchicine addition (A). Colchicine-stimulated EGFR phosphorylation, but not that in VSMC exposed to EGF, is effectively inhibited by a 30 minute pretreatment with the ROS scavengers NAC (B) or DPI (C). NAC and DPI suppressed both PAI-1 induction (D,E) and ERK1/2 activation (F), two critical downstream targets of EGFR signaling, following colchicine-initiated microtubule disruption.
SMAD2/3 activation in response to microtubule disruption is required for PAI-1, but not CTGF, induction
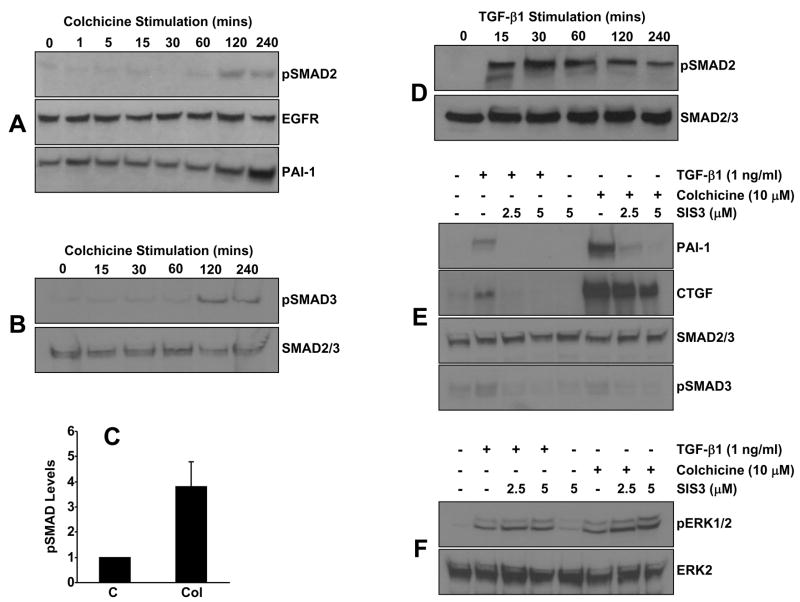
SMAD2/3 are phosphorylated in VSMCs within 1–2 hours post-colchicine stimulation (Figure 8A–C) in contrast to the rapid (within 15 minutes), and more robust, increase in pSMAD2 levels in response to TGF-β1 (Figure 8D). A specific inhibitor of SMAD3 (SIS3) [30]eliminated PAI-1 and CTGF induction by TGF-β1. SIS3 pretreatment also completely inhibited PAI-1, but only marginally CTGF, expression in colchicine-stimulated cells suggesting a differential involvement of SMAD3 in the activation of specific profibrogenic factors by cytoskeletal deformation (Figure 8E). Importantly, SIS3 did not affect ERK1/2 phosphorylation in either TGF-β1 or colchicine treated VSMC (Figure 8F) supporting both the specificity of SIS3 and the functionality of the EGFR→MEK/ERK pathway in the presence of SIS3. Clearly, ERK1/2 activation in the absence of SMAD3 signaling is not sufficient for PAI-1 induction in response to either TGF-β1 or colchicine.
Figure 8. SMAD3 signaling is required for PAI-1 but not CTGF induction by colchicine.
SMAD2 (A) and SMAD3 (B) are phosphorylated in VSMC with similar delayed kinetics (requiring 1 to 2 hours) upon colchicine addition. Overall pSMAD content increases by >4-fold in colchicine-stimulated cultures (C). Data plotted in (C) is the mean±SD of triplicate western determinations of cellular pSMAD2/3 levels; C = unstimulated quiescent VSMC, Col = colchicine-treated cells. These responses are in sharp contrast to the rapidity (within 15 minutes) and magnitude of TGF-β1-induced SMAD2 phosphorylation (D). Pretreatment with SIS3, a specific SMAD3 inhibitor, eliminated PAI-1 expression in response to both microtubule disruption and, as expected, TGF-β1 (E). Increased ERK1/2 phosphorylation in TGF-β1- and colchicine-stimulated VSMC was unaffected by SIS3 (F).
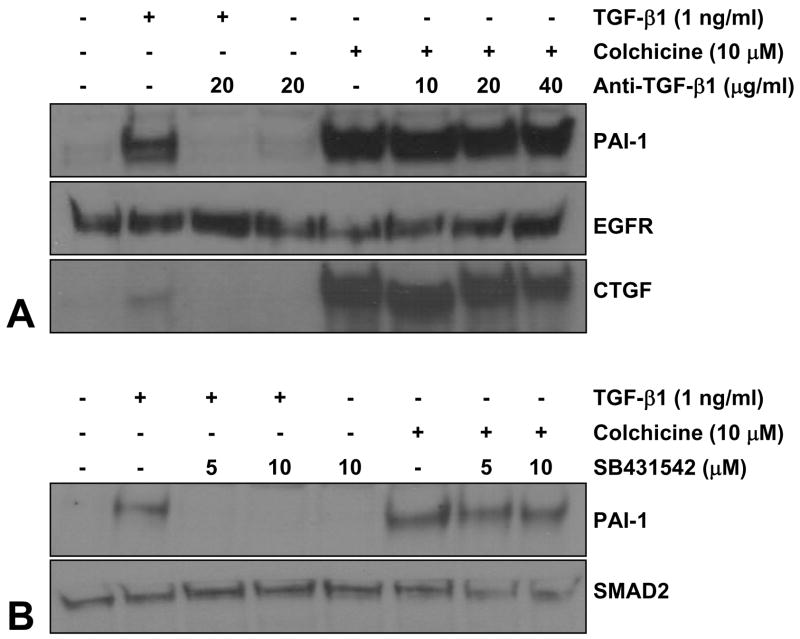
In view of the similarities in the repertoire of participating signaling intermediates (e.g., EGFR, c-src, Rho/Rock, MEK/ERK1/2) between the microtubule- and TGF-β1-initiated pathways of PAI-1 induction (this paper; [28]), and previous observations that TGF-β1 expression is also regulated by cytoskeletal deformation [13], it was important to determine if colchicine-stimulated PAI-1 expression was mediated by an autocrine TGF-β1 loop. Pretreatment with TGF-β1 neutralizing antibodies markedly decreased TGF-β1-induced PAI-1 and CTGF expression (as anticipated) confirming the relevance of this approach to block TGF-β1-initiated signaling (Figure 9A). Colchicine-stimulated PAI-1 or CTGF expression, in contrast, was unaffected by TGF-β1 neutralizing antibody blockade regardless of the pretreatment concentration suggesting that autocrine TGF-β1 loop and/or ligand-dependent TGF-βR activation is not likely involved in the microtubule deformation pathway leading to PAI-1 or CTGF induction (Figure 9A). Moreover, pretreatment with SB431542, a specific inhibitor of the TGF-β1 activin receptor-like kinases (ALKs) ALK-4 (activin type I receptor), ALK-5 (TGF-β type I receptor), and ALK-7 (nodal type I receptor) did not affect colchicine-stimulated PAI-1 expression while effectively eliminating both PAI-1 induction (Figure 9B) and SMAD2 phosphorylation (not shown) in response to TGF-β1.
Figure 9. Colchicine-mediated PAI-1 and CTGF induction occurs independently of autocrine TGF-β1 signaling or TGF-βRI activity.
Pretreatment of VSMC with TGF-β1 neutralizing antibodies (20 μg/ml) eliminated PAI-1 and CTGF expression in response to TGF-β1 (A). Colchicine-stimulated PAI-1/CTGF induction, in contrast, is not affected by TGF-β1 neutralizing antibodies regardless of the pretreatment concentration (10–40 μg/ml) (A). Incubation with the ALK4, 5, and 7 TGF-βRI kinase inhibitor SB431542 completely suppressed TGF-β1-initated SMAD2 phosphorlyation (not shown) and PAI-1 expression as expected (B). SB431542, however, did not block the PAI-1 response to colchicine-mediated microtubule network collapse (B).
SMAD2/3 activation and PAI-1/CTGF induction by microtubule alterations requires Rho-ROCK signaling
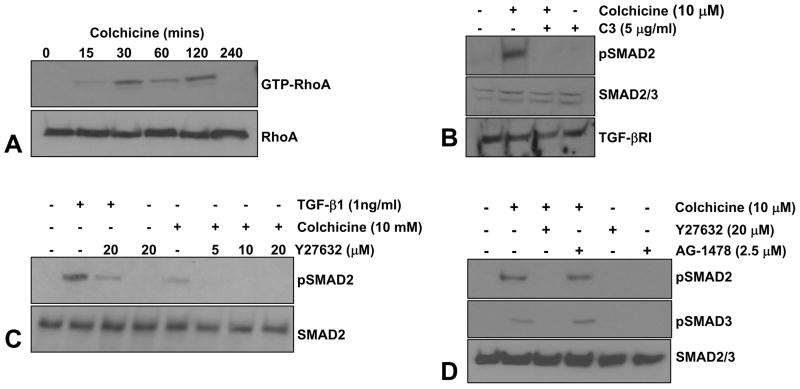
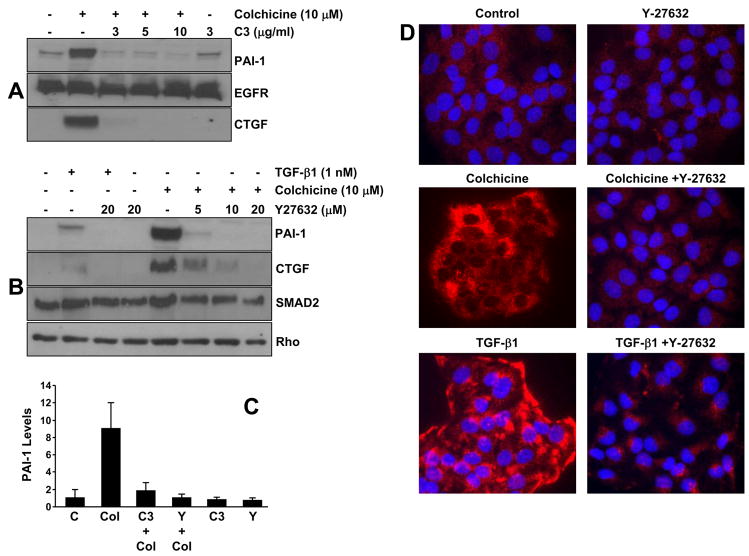
Colchicine treatment of quiescent R22 cells increased RhoA GTP loading (within 15–30 minutes) while total RhoA (GDP-and GTP-bound) levels remained unchanged (Figure 10A). Interference with either Rho or ROCK signaling by pretreatment with C3 transferase or Y-27632, respectively, inhibited SMAD2/3 phosphorylation in response to colchicine without altering either total SMAD2/3 or TGF-βRI receptor levels implicating an upstream role for Rho/ROCK as modulators of SMAD activation by tubulin network collapse (Figure 10B–D). The EGFR inhibitor, AG1478, in contrast, failed to affect colchicine-mediated pSMAD2/3 levels (Figure 10D). Prior incubation with C3 transferase and Y-27632 eliminated colchicine-stimulated PAI-1 as well as CTGF expression suggesting involvement of the Rho/ROCK system in both the microtubule disruption-sensitive and TGF-β1-mediated pathways leading to PAI-1 and CTGF induction (Figure 11A–C). Rho/ROCK-dependent PAI-1 expression upon stimulation with colchicine or TGF-β1 is not restricted to vascular smooth muscle cells as human (HaCaT) keratinocytes responded similarly (Figure 11D).
Figure 10. RhoA is activated by alterations in microtubule architecture and is required for SMAD phosphorylation.
Colchicine treatment of quiescent R22 VSMC rapidly stimulated RhoA GTPase loading (within 15–30 minutes) while total levels of cellular RhoA (both GDP- and GTP-bound) remain unaltered (A). To further assess the potential participation of the Rho pathway in colchicine-initiated SMAD activation, VSMC were incubated with specific inhibitors of Rho (C3 transferase) and its downstream effector ROCK (Y27632) prior to colchicine addition. Colchicine-mediated SMAD2 phosphorylation was blocked by both C3 transferase (B) and Y-27632 (C) suggesting an upstream role for activated Rho GTPase and ROCK in SMAD2 activation following microtubule disruption. AG1478, an inhibitor of EGFR signaling, however, has no effect on colchicine-induced SMAD2/3 phosphorylation (D). Total SMAD2/3 protein and TGF-βRI levels are not altered by inhibition of Rho, ROCK or EGFR signaling.
Figure 11. Rho/ROCK signaling is required for PAI-1/CTGF expression in response to both microtubule disruption and TGF-β1.
Pretreatment of R22 VSMC with the Rho or ROCK inhibitors C3 transferase and Y27632, respectively, effectively blocked both colchicine- (A,B) and TGF-β1- (B) stimuated PAI-1 and CTGF expression at all tested concentrations illustrating Rho signaling involvement in the microtubule deformation- and TGF-β1-mediated PAI-1 inductive responses. Data plotted in (C) is the mean±SD of triplicate western assessments of PAI-1 levels in control (C) R22 cultures, in colchicine (Col), C3 transferase (C3) and Y27632 (Y) treated VSMC as well as in R22 cells pretreated with C3 and Y prior to colchicine addition. Rho/ROCK-involvement in PAI-I expression in response to colchicine and TGF-β1 is not limited to VSMC as a similar dependency is also evident in immortalized human (HaCaT) keratinocytes (D).
DISCUSSION
Microtubule disruption, in colchicine or nocdazole-treated cells, is associated with transient EGFR phosphorylation, specifically at the EGFRY845 src family kinase target residue. Adenoviral delivery of a kinase dead GFP-EGFRK721A mutant, or preincubation with AG1478, similarly inhibited colchicine-dependent PAI-1 expression, supporting the necessity for a functional EGFR in signal propagation. Colchicine-stimulated ERK1/2 phosphorylation, in fact, is evident in EGFR+/+ but not EGFR−/− fibroblasts positioning the MEK-ERK cascade as a downstream EGFR target that mediates microtubule collapse-associated PAI-1 expression. ROS generation in colchicine-treated cells, moreover, appears crucial for EGFR transactivation as ROS inhibitors (e.g., NAC and DPI) ablate both EGFR phosphorylation and PAI-1 induction. EGFR transactivation by non-EGF initiators [28,31–35] and subsequent ERK1/2 phosphorylation, at least in response to microtubule disruption or changes in cellular tension, is dependent on ROS generation/signaling in sharp contrast to the native ligand (EGF) highlighting stimulus-specific modes of EGFR mobilization [17,35–37]. While ERK1/2 activation is required for PAI-1 induction, MEK inhibition with either U0126 or PD98059 suggests that these kinases are dispensable for colchicine-stimulated CTGF expression, consistent with previous observations in other cell types [14].
Cytoskeletal-deformation efficiently stimulated RhoA-GTP loading and Rho/ROCK signaling is critical for colchicine-induced PAI-1, as well as CTGF, expression (this paper, [14]). The Rho guanine exchange factors GEF-H1 and p190Rho-GEF interact with microtubules [2,4,38]. Microtubule disassembly, therefore, may lead to aberrant release of these GTP-exchange factors activating, thereby, Rho family members [4,38]. GEF-H1 suppression, in fact, retards nocodazole-induced Rho-GTP loading in HeLa cells [39], consistent with the observation that stabilization of the microtubule skeleton by Paclitaxel (Taxol) pretreatment prior to colchicine or nocodazole addition inhibits microtubule-induced PAI-1 and CTGF expression. Microtubules also anchor SMAD proteins likely to maintain these receptor-activated signaling effectors in an inactive configuration. Tubulin network collapse may release cytoskeletal restrains on SMAD2/3 trafficking, facilitating SMAD phosphorylation, nuclear translocation, and transcriptional activity [2,40]. Clearly, colchicine-mediated PAI-1 or CTGF induction does not involve an autocrine TGF-β1 loop regardless of commonalities in signaling elements (e.g., EGFR, Rho/ROCK, MEK-ERK1/2, SMAD2/3) that mediate TGF-β1- and colchicine-stimulated PAI-1 expression (this paper; [28]). Pretreatment with SB431542, a specific inhibitor of the TGF-βRI kinase, furthermore, has no effect on colchicine-mediated PAI-1 induction, but completely eliminates SMAD2/3 activation and the response of the PAI-1 gene to TGF-β1. These findings are not without precedent. It has become increasingly evident that SMAD signaling can be initiated by non-TGF-β superfamily ligands independent of the TGF-βRI [41]. Nocodazole-induced SMAD2 phsophorylation is also not dependent on TGF-βRI kinase activity [40]. Relative to the increased levels and kinetics of SMAD2/3 phosphorylation evident in colchicine-treated VSMC (this paper), the time course and amplitude of nocodazole-stimulated pSMAD2 is similarly delayed and significantly less, respectively, than that associated with TGF-β1 exposure. MPS1 (a kinase activated by microtubule disruption) may, in fact, phosphorylate SMAD2 and initiate transcriptional responses independent of TGF-βR involvement [40]. Most novel, however, is the identification of the upstream role of Rho/Rock, but not the EGFR pathway, as a modulator of SMAD2/3 phosphorylation induced by cytoskeletal deformation similar to Rho involvement in TGF-β1-stimulated SMAD2/3 activation ([28]; Figure 12). Importantly, activation of SMAD3 is crucial for PAI-1 expression by microtubule disruption suggesting the necessity of both Rho/ROCK/SMAD and EGFR/ERK signaling cascades in this inductive response.
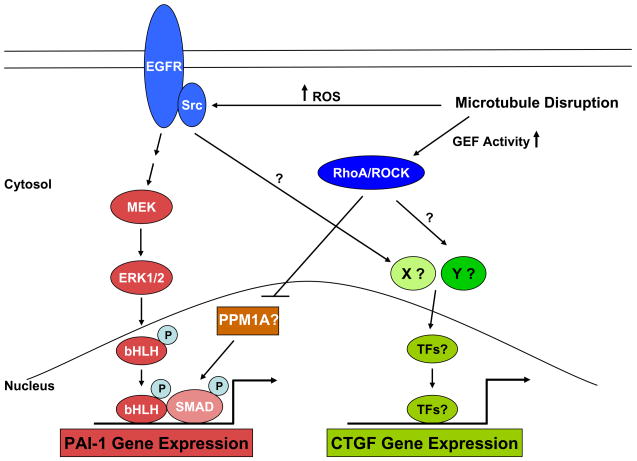
Figure 12. Alterations in the microtubule cytoskeleton initiates multiple signaling events that lead to PAI-1 expression.
Drug-initiated microtubule disassembly mobilizes several signaling pathways that impact PAI-1 transcription. Colchicine- induced disruption of microtubule cytoskeleton leads to the activation of EGFR and Src family kinases, perhaps as a consequence of an increase in the generation of ROS. pp60c-src-phosphorylated EGFR (at the Y845 site) signals to MEK-ERK initiating ERK interactions with specific transcription factors (TFs), including members of the bHLH-LZ family that are known to be MAP kinase targets [28] and which initiate PAI-1 gene expression via the ERK1/2 pathway [9]. Colchicine also stimulates SMAD2/3 phosphorylation (at the c-terminal end) independent of the TGF-β1-ligand dependent mechanisms (e.g., de novo synthesis or release of the active form of TGF-β1 from the matrix) and independent of TGF-β1 receptor activity. pSMAD2/3 may cooperate with pERK1/2-activated bHLH proteins to initiate PAI-1 transcription. Mobilization of the RhoA/ROCK pathway downstream of microtobule deformation appears necessary for sustaining SMAD phsophorylation and subsequent PAI-1 induction [28]. In this model, stimulation of the Rho/Rock pathway by a microtubule-dependent increase in Rho GEF function may regulate the activity of specific SMAD phosphatases (e.g., PPM1A) impacting, thereby, the duration of SMAD-dependent transcription of target genes such as PAI-1. MEK/ERK involvement downstream of ROS generation was critical for PAI-1, but not CTGF, expression following microtubule perturbation suggesting bifurcation of pathways downstream of EGFR activation. Colchicine-initiated SMAD3 phosphorylation, moeover, was essential for PAI-1, but not CTGF, expression further highlighting divergence of signaling events downstream of Rho/ROCK.
Acknowledgments
Supported by NIH grant GM57242.
Abbreviations
- PAI-1
plasminogen activator inhibitor-1
- CTGF
connective tissue growth factor
- EGFR
epidermal growth factor receptor
- ROS
reactive oxygen species
- ERK
extracellular signal-regulated kinases
- ROCK
Rho-associated, coiled-coil containing protein kinase
- TGF-β
transforming growth factor-β
- SMAD2/3
TGF-β receptor-activated signaling effectors
- GEF
guanine nucleotide exhange factor
- uPA
urokinase plasminogen activator
- COX-2
cyclooxygenase-2
- VEGF
vascular endothelial growth factor
- MMP
matrix metallotproteinase
- VSMC
vascular smooth muscle cells
- MEF
mouse embryonic fibroblasts
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- EGF
epidermal growth factor
- NAC
N-acetyl cysteine
- DPI
diphenyleneiodonium chloride
- DCF-DA
dichlorofluorescein diacetate
- GFP
green fluorescence protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fuchs E, Karakesisoglou I. Bridging cytoskeletal intersections. Genes Dev. 2001;15:1–14. doi: 10.1101/gad.861501. [DOI] [PubMed] [Google Scholar]
- 2.Dong C, Li Z, Alvarez R, Jr, Feng XH, Goldschmidt-Clermont PJ. Microtubule binding to Smads may regulate TGF beta activity. Mol Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 3.van Horck FP, Ahmadian MR, Haeusler LC, Moolenaar WH, Kranenburg O. Characterization of p190RhoGEF, a RhoA-specific guanine nucleotide exchange factor that interacts with microtubules. J Biol Chem. 2001;276:4948–56. doi: 10.1074/jbc.M003839200. [DOI] [PubMed] [Google Scholar]
- 4.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–85. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janmey PA. The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiol Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- 6.Forgacs G, Yook SH, Janmey PA, Jeong H, Burd CG. Role of the cytoskeleton in signaling networks. J Cell Sci. 2004;117:2769–2775. doi: 10.1242/jcs.01122. [DOI] [PubMed] [Google Scholar]
- 7.Rosette C, Karin M. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-κB. J Cell Biol. 1995;128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 9.Qi L, Higgins SP, Lu Q, Samarakoon R, Wilkins-Port CE, Ye Q, Higgins CE, Staiano-Coico L, Higgins PJ. SERPINE1 (PAI-1) Is a Prominent Member of the Early G0→G1 Transition “Wound Repair” Transcriptome in p53 Mutant Human Keratinocytes. J Invest Dermatol. 2008;128:749–753. doi: 10.1038/sj.jid.5701068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samarakoon R, Higgins PJ. MEK/ERK pathway mediates cell-shape-dependent plasminogen activator inhibitor type 1 gene expression upon drug-induced disruption of the microfilament and microtubule networks. J Cell Sci. 2002;115:3093–103. doi: 10.1242/jcs.115.15.3093. [DOI] [PubMed] [Google Scholar]
- 11.Irigoyen JP, Besser D, Nagamine Y. Cytoskeleton reorganization induces the urokinase-type plasminogen activator gene via the Ras/extracellular signal-regulated kinase (ERK) signaling pathway. J Biol Chem. 1997;272:1904–1909. doi: 10.1074/jbc.272.3.1904. [DOI] [PubMed] [Google Scholar]
- 12.Faisal A, Kleiner S, Nagamine Y. Non-redundant role of Shc in Erk activation by cytoskeletal reorganization. J Biol Chem. 2004;279:3202–11. doi: 10.1074/jbc.M310010200. [DOI] [PubMed] [Google Scholar]
- 13.Varedi M, Ghahary A, Scott PG, Tredget EE. Cytoskeleton regulates expression of genes for transforming growth factor-beta 1 and extracellular matrix proteins in dermal fibroblasts. J Cell Physiol. 1997;172:192–9. doi: 10.1002/(SICI)1097-4652(199708)172:2<192::AID-JCP6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 14.Ott C, Iwanciw D, Graness A, Giehl K, Goppelt-Struebe M. Modulation of the expression of connective tissue growth factor by alterations of the cytoskeleton. J Biol Chem. 2003;278:44305–11. doi: 10.1074/jbc.M309140200. [DOI] [PubMed] [Google Scholar]
- 15.Graness A, Cicha I, Goppelt-Struebe M. Contribution of Src-FAK signaling to the induction of connective tissue growth factor in renal fibroblasts. Kidney Int. 2006;69:1341–9. doi: 10.1038/sj.ki.5000296. [DOI] [PubMed] [Google Scholar]
- 16.Subbaramaiah K, Hart JC, Norton L, Dannenberg AJ. Microtubule-interfering agents stimulate the transcription of cyclooxygenase-2. Evidence for involvement of ERK1/2 and p38 mitogen-activated protein kinase cascades. J Biol Chem. 2000;275:14838–14845. doi: 10.1074/jbc.275.20.14838. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Yang JH, Huang H, Kennedy SP, Turi TG, Thompson JF, Libby P, Lee RT. Transcriptional profile of mechanically induced genes in human vascular smooth muscle cells. Circ Res. 1999;85:1118–1123. doi: 10.1161/01.res.85.12.1118. [DOI] [PubMed] [Google Scholar]
- 18.Ailenberg M, Silverman M. Cellular activation of mesangial gelatinase by cytochalasin D is accompanied by enhanced mRNA expression of both gelatinase A and its membrane-associated gelatinase A activator (MT-MMP) Biochem J. 1996;313:879–884. doi: 10.1042/bj3130879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unemori EN, Werb Z. Reorganization of polymerized actin: a possible trigger for induction of procollagenase in fibroblasts cultured in and on collagen gels. J Cell Biol. 1986;103:1021–1031. doi: 10.1083/jcb.103.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aggeler J, Frisch SM, Werb Z. Changes in cell shape correlate with collagenase gene expression in rabbit synovial fibroblasts. J Cell Biol. 1984;98:1662–1671. doi: 10.1083/jcb.98.5.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 22.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 23.Providence KM, Kutz SM, Staiano-Coico L, Higgins PJ. PAI-1 gene expression is regionally induced in wounded epithelial cell monolayers and required for injury repair. J Cell Physiol. 2000;182:269–80. doi: 10.1002/(SICI)1097-4652(200002)182:2<269::AID-JCP16>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Providence KM, Higgins PJ. PAI-1 expression is required for epithelial cell migration in two distinct phases of in vitro wound repair. J Cell Physiol. 2004;200:297–308. doi: 10.1002/jcp.20016. [DOI] [PubMed] [Google Scholar]
- 25.Pawar S, Kartha S, Toback FG. Differential gene expression in migrating renal epithelial cells after wounding. J Cell Physiol. 1995;165:556–65. doi: 10.1002/jcp.1041650314. [DOI] [PubMed] [Google Scholar]
- 26.Basile DP, Martin DR, Hammerman MR. Extracellular matrix-related genes in kidney after ischemic injury: potential role for TGF-β in repair. Am J Physiol. 1998;275:F894–903. doi: 10.1152/ajprenal.1998.275.6.F894. [DOI] [PubMed] [Google Scholar]
- 27.Samarakoon R, Higgins CE, Higgins SP, Kutz SM, Higgins PJ. Plasminogen activator inhibitor type-1 gene expression and induced migration in TGF-β1-stimulated smooth muscle cells is pp60c-src/MEK-dependent. J Cell Physiol. 2005;204:236–246. doi: 10.1002/jcp.20279. [DOI] [PubMed] [Google Scholar]
- 28.Samarakoon R, Higgins S, Higgins CE, Higgins PJ. TGF-β-induced plasminogen activator inhibitor-1 expression in vascular smooth muscle cells requires pp60c-src/EGFRY845 and Rho/Rock signaling. J Mol Cell Cardiol. 2008;44:527–538. doi: 10.1016/j.yjmcc.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 30.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced Extracellular matrix expression. Mol Pharmacol. 2006;69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 31.Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19:2013–2023. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Ushio-Fukai M, Griendling KK, Becker PL, Hilenski L, Halleran S, Alexander RW. Epidermal growth factor receptor transactivation by angiotensin II requires reactive oxygen species in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2001;21:489–495. doi: 10.1161/01.atv.21.4.489. [DOI] [PubMed] [Google Scholar]
- 33.Tschumperlin DJ, Dai G, Maly IV, Kikuchi T, Laiho LH, McVittie AK, Haley KJ, Lilly CM, So PT, Lauffenburger DA, Kamm RD, Drazen JM. Mechanotransduction through growth-factor shedding into the extracellular space. Nature. 2004;429:83–86. doi: 10.1038/nature02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wetzker R, Bohmer FD. Transactivation joins multiple tracks to the ERK/MAPK cascade. Nat Rev Mol Cell Biol. 2003;4:651–657. doi: 10.1038/nrm1173. [DOI] [PubMed] [Google Scholar]
- 35.Khan EM, Lanir R, Danielson AR, Goldkorn T. Epidermal growth factor receptor exposed to cigarette smoke is aberrantly activated and undergoes perinuclear trafficking. FASEB J. 2008;22:910–917. doi: 10.1096/fj.06-7729com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemarié CA, Tharaux PL, Esposito B, Tedgui A, Lehoux S. Transforming growth factor-alpha mediates nuclear factor kappaB activation in strained arteries. Circ Res. 2006;99:434–441. doi: 10.1161/01.RES.0000237388.89261.47. [DOI] [PubMed] [Google Scholar]
- 37.Lehoux S, Tedui A. Cellular mechanics and gene expression in blood vessels. J Biomech. 2003;36:631–643. doi: 10.1016/s0021-9290(02)00441-4. [DOI] [PubMed] [Google Scholar]
- 38.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 39.Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol Biol Cell. 2008;19:2147–2153. doi: 10.1091/mbc.E07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu S, Wang W, Clarke DC, Liu X. Activation of Mps1 promotes transforming growth factor-beta-independent Smad signaling. J Biol Chem. 2007;282:18327–18338. doi: 10.1074/jbc.M700636200. [DOI] [PubMed] [Google Scholar]
- 41.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]