Abstract
We have used single-particle reconstruction in cryo-electron microscopy to determine a structure of the Thermus thermophilus ribosome in which the ternary complex of elongation factor Tu (EF-Tu), tRNA and guanine nucleotide has been trapped on the ribosome using the antibiotic kirromycin. This represents the state in the decoding process just after codon recognition by tRNA and the resulting GTP hydrolysis by EF-Tu, but before the release of EF-Tu from the ribosome. Progress in sample purification and image processing made it possible to reach a resolution of 6.4 Å. Secondary structure elements in tRNA, EF-Tu and the ribosome, and even GDP and kirromycin, could all be visualized directly. The structure reveals a complex conformational rearrangement of the tRNA in the A/T state and the interactions with the functionally important switch regions of EF-Tu crucial to GTP hydrolysis. Thus, the structure provides insights into the molecular mechanism of signalling codon recognition from the decoding centre of the 30S subunit to the GTPase centre of EF-Tu.
Keywords: cryo-electron microscopy, elongation factor, GTPase, ribosome, translation
Introduction
Ribosomes are large macromolecular machines that translate the genetic message to make proteins. Although the ribosome itself is at the core of this process, several protein factors have important functions in each of the four broad stages of translation, that is, initiation, elongation, termination and ribosome recycling (Ramakrishnan, 2002; Frank and Spahn, 2006). Many of these factors are GTPases, among them initiation factor IF2, elongation factors EF-Tu (ternary complex of elongation factor Tu), EF-G and release factor RF3. Thus, GTP hydrolysis by protein factors is an essential part of translation in each of the stages of translation.
Among the GTPase translation factors, EF-Tu has an essential function in tRNA selection during decoding. EF-Tu in the GTP-bound form has high affinity for aminoacyl tRNAs (aa-tRNAs). The ternary complex of EF-Tu, aa-tRNA and GTP binds to the ribosomal A-site in the initial step of decoding. A cognate interaction between the codon on mRNA and anticodon on tRNA in the A-site leads to GTP hydrolysis by EF-Tu and subsequent dissociation of EF-Tu•GDP from the ribosome.
The structures of isolated EF-Tu in complex with GDP or the GTP analogue GDPNP, as well as its ternary complex with GTP and aa-tRNA, have all been characterized in molecular detail (for review, see Hilgenfeld, 1995; Andersen et al, 2003). The hydrolysis of GTP and the release of the gamma phosphate induce a major rearrangement of the universally conserved switch I (effector loop) and switch II regions, which in turn results in a gross conformational change in EF-Tu. The global change also results in a strong reduction in affinity for aa-tRNA of the GDP-bound form. How the ribosome induces GTP hydrolysis by EF-Tu is a major question in translation.
Kinetic, structural, genetic and biochemical data over the last decade have established that decoding is a complicated multistep process in which an active role is played by the ribosome itself (Rodnina and Wintermeyer, 2001; Ogle and Ramakrishnan, 2005; Frank and Spahn, 2006; Korostelev et al, 2008; Marshall et al, 2008). Kinetic work shows that a rapid, reversible, initial binding of the ternary complex is followed by a slower codon recognition step (Rodnina and Wintermeyer, 2001; Marshall et al, 2008). Codon recognition by cognate (but not near-cognate) tRNA leads to an acceleration of GTPase activation and GTP hydrolysis. This suggests that binding of cognate tRNA induces conformational changes in the ribosomal complex required for GTP hydrolysis by EF-Tu. GTP hydrolysis is followed by EF-Tu release and movement (accommodation) of the tRNA into the peptidyl transferase centre, after which peptide bond formation occurs rapidly. The antibiotic kirromycin traps the process after GTP hydrolysis but before EF-Tu release.
A number of biochemical and structural studies can be correlated directly with the kinetic data. During codon recognition, the correctness of the codon–anticodon interaction is checked by interactions of three conserved bases of 16S RNA at the decoding centre of the 30S subunit with the minor groove of the codon–anticodon base pairs (Ogle et al, 2001). Successful codon–anticodon recognition by the 30S subunit leads to a conformational change in the 30S subunit that would move the shoulder of the subunit closer to EF-Tu (Ogle et al, 2002). This led to the view that the additional energy obtained from the minor groove interactions of the ribosome with cognate tRNA at the decoding centre led to a domain closure of the ribosome essential for GTP hydrolysis by EF-Tu and subsequent steps of successful decoding (Ogle et al, 2002; Ogle and Ramakrishnan, 2005).
Early chemical footprinting studies suggested the existence of an A/T state as long as EF-Tu was present on the ribosome, in which the aminoacyl end of the incoming tRNA cannot enter the peptidyl transferase centre until GTP hydrolysis occurs and EF-Tu has been released, thus ensuring that decoding has taken place prior to peptide bond formation (Moazed and Noller, 1989). Single-particle cryo-EM studies of an EF-Tu ribosome complex stalled with kirromycin (similar to the one reported here) have directly revealed the existence of the A/T state (Stark et al, 1997). In this A/T state, the aminoacyl end of the tRNA is still in contact with its binding pocket in EF-Tu and thus unable to enter the peptidyl transferase centre, but the anticodon of the tRNA is in the decoding centre of the 30S subunit. Later higher resolution studies of the same complex (Stark et al, 2002; Valle et al, 2002, 2003) showed that the A/T state was characterized by a distorted tRNA as well as changes in the conformation of EF-Tu and the ribosome. Indeed, distortions of the tRNA during decoding have long been suggested as important in decoding (Yarus and Smith, 1995).
Although cryo-EM has provided important structural information about the kirromycin-stalled decoding complex, previous cryo-EM maps have been obtained at low to intermediate resolution (20–10 Å). At this resolution, detailed interpretation of maps in molecular terms is problematic; indeed, there are several discrepancies in the interpretation of previous cryo-EM maps of this complex (Stark et al, 1997, 2002; Valle et al, 2002, 2003). At about 7 Å, it becomes possible to directly visualize double helices of RNA as well as alpha helices of proteins, thus making placement of individual domains far more accurate and allowing a more detailed analysis of molecular interactions and conformational changes, as demonstrated by Petry et al (2005) in their interpretation of X-ray crystallographic maps of the ribosome. However, X-ray crystallography of functional ribosomal complexes is still very challenging. Despite considerable progress in the structural analysis of several ribosomal complexes at atomic or near-atomic resolution, no crystal structure of a GTPase factor bound to the ribosome has yet been reported.
We present here a cryo-EM map of the Thermus thermophilus 70S ribosome bound to the ternary complex (EF-Tu•Phe-tRNA•GTP, stalled with kirromycin) at a resolution of 6.4 Å at 0.5 cutoff of the Fourier shell correlation (FSC) criterion, with significant information extending to 4.7 Å, as determined by the 3σ criterion (Orlova et al, 1997). At this resolution, we can directly visualize not only secondary structure elements such as α-helices as well as the RNA backbone but also even ligands such as GDP and kirromycin. Moreover, the crystal structure of the T. thermophilus ribosome at 2.8-Å resolution (Selmer et al, 2006) greatly facilitated interpretation of the cryo-EM map in molecular terms, which revealed important molecular interactions and conformational changes. The structure reveals a complex rearrangement of the tRNA and provides a detailed view of the interaction of the GTPase centre of EF-Tu and especially the switch regions with the ribosome. The structure provides insights into how successful recognition of the cognate codon is signalled from the decoding centre of the 30S subunit to the GTPase centre of EF-Tu about 80 Å away, and leads to a model of how the ribosome might stimulate the GTPase activity of translational GTPases.
Results and discussion
Intrinsic conformational heterogeneity of the ribosomal decoding complex and progression to higher resolution
A general problem with ribosomal complexes is achieving equal stoichiometry of all the components. To avoid this problem and generate the most homogeneous complex possible, we used an affinity tag method previously described for the crystallography of release factor complexes (Petry et al, 2005). In this procedure, ternary complex containing His-tagged EF-Tu was bound to programmed ribosomes containing tRNAfMet in the P-site in the presence of kirromycin. The entire ribosomal complex was affinity purified on a nickel column using the His-tag on EF-Tu. As previously demonstrated for release factors, only the ribosome complex, which had EF-Tu and tRNA, that was cognate for the A-site codon was retained on the nickel column.
However, although the specimen behaved quite well during initial phases of image reconstruction and always exhibited strong density for the ternary complex, the maps calculated from the full data set showed indications of conformational heterogeneity as we progressed to sub-nanometer resolution. For example, the L7/L12 stalk base region of the ribosome was strongly disordered and had a fragmented appearance. To obtain a more homogeneous population of particle images, we classified the data set (586 329 particle images) using an unsupervised ‘3D K-means procedure' (Penczek et al, 2006) and obtained a major sub-population (I) consisting of 55% of the particles (Supplementary Figure S1). Interestingly, cryo-EM maps calculated from all of the minor sub-populations of particle images exhibit density for the ternary complex (Supplementary Figures S1 and S2). The behaviour of the data set is thus different from those of unpurified ribosomal complexes, where a classification procedure resulted in a significant fraction of ribosomes lacking the ligand (for example, Valle et al, 2002; Penczek et al, 2006; Connell et al, 2007) and directly corroborates the success of our affinity purification using the His-tagged EF-Tu.
The 70S•tRNA•EF-Tu•GDP•kirromycin complex at 6.4-Å resolution
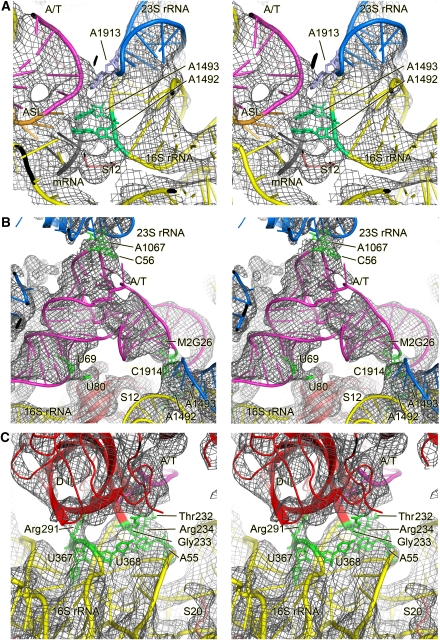
The major sub-population, consisting of 323 688 particles, was used to obtain an improved cryo-EM map (Figure 1A; Supplementary Movie 1) at the unprecedented resolution of 6.4 Å by the conservative 0.5 cutoff of the FSC criterion (Supplementary Figure S3). As would be expected from the increased resolution, the map reveals significantly more details than previous maps of the ribosomal decoding complex at low to intermediate resolution (Stark et al, 1997, 2002; Valle et al, 2002, 2003) or even previous cryo-EM maps of ribosomal complexes at 7 to 8-Å resolution (Schüler et al, 2006; Connell et al, 2007). Not only are α-helical secondary structure elements of proteins clearly resolved but also density for unstructured peptide chains corresponding to the extended tails of ribosomal proteins (Figure 2; Supplementary Figure S4). Distinct density is present even for the low-molecular ligands GDP (Figure 2B) and the antibiotic kirromycin (Figure 2C). However, β-sheets are not fully resolved: they appear as flat surfaces and only occasionally does the density indicate a separation of strands. This improved cryo-EM map enables a more detailed analysis of the interactions of EF-Tu and tRNA with the ribosome.
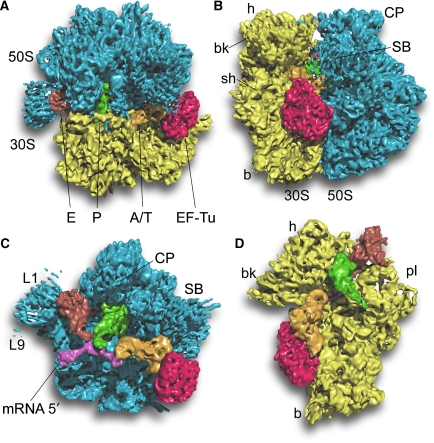
Figure 1.
Overview of the 70S•EF-Tu•Phe-tRNA•GDP•kirromycin complex. A surface representation of the cryo-EM map is shown (A) from the top; (B) from the L7/L12 side; (C) from the 30S side, with 30S removed and (D) from the 50S side, with 50S removed. The components are coloured distinctly (30S subunit, yellow; 50S subunit, blue; EF-Tu, red; A/T-tRNA, orange; P-tRNA, green; E-tRNA, brown; mRNA, pink).
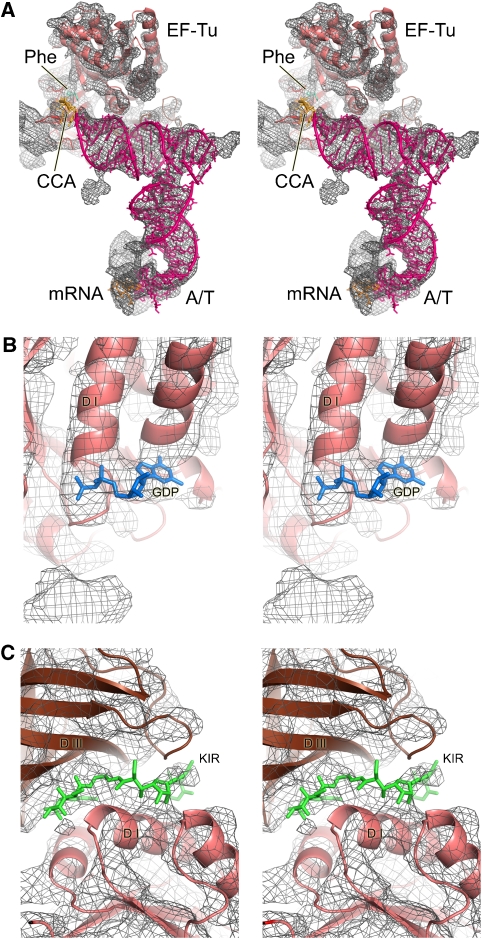
Figure 2.
Details seen in the electron density map at 5.7- to 6.4-Å resolution. (A) Overall structure of the ternary complex showing the interactions between the EF-Tu and A/T tRNA. The density for the ternary complex has been computationally separated using a mask generous enough to show the sites of interaction with the ribosome. At this resolution, secondary structure elements of RNA and protein are clearly distinguishable. (B) A region of EF-Tu showing additional density that corresponds to GDP. (C) Density for the low-molecular weight kirromycin seen between the domains of EF-Tu.
Overall, the conformation of the 70S•tRNA•EF-Tu•GDP•kirromycin complex is similar to the crystal structure of the 70S ribosome complex with mRNA and tRNAs. A good fit could be obtained by docking the atomic model of the T. thermophilus ribosome (Selmer et al, 2006) as three rigid bodies corresponding to the main body of the 50S subunit and the head and the body/platform domains of the 30S subunit. In addition, the L1 and L7/L12 stalks required further adjustment.
Apart from the ribosome itself, atomic models of mRNA and the P- and E-site tRNAs derived from the crystal structure of the 70S ribosome from T. thermophilus in complex with mRNA and tRNAs (Selmer et al, 2006) could also readily be docked into the cryo-EM map. A molecular model for the ribosome-bound ternary complex was derived from the X-ray structures of the ribosome-bound anticodon stem loop of the A-site tRNA (Yusupov et al, 2001; Selmer et al, 2006), the ternary complex (Nissen et al, 1995) and EF-Tu•GDP in complex with methylkirromycin (also called aurodox; Vogeley et al, 2001) as detailed below. Thus, we could derive a nearly complete molecular model for the kirromycin-stalled ribosomal decoding complex.
A complex rearrangement in the A/T tRNA
It is clear that the tRNA in the A/T state has to be deformed in comparison to the accommodated A-site tRNA to allow codon–anticodon interaction in the decoding centre to occur while the acceptor stem and the aminoacyl-end are still bound to EF-Tu. A distortion of the anticodon loop was proposed to account for the difference in orientation of accommodated A tRNA and A/T tRNA in one cryo-EM study (Stark et al, 2002). In other cryo-EM studies, a kink-like deformation of the A/T tRNA was proposed that occurs at the junction between the anticodon and D stems (Valle et al, 2002, 2003). No deformation of the anticodon loop can be observed in our present cryo-EM map, and the X-ray structures of the anticodon stem loops together with the A-site mRNA codon (Selmer et al, 2006) can be readily fit into the cryo-EM density map (Figure 2A). A kink-like deformation of the A/T tRNA at the junction between the anticodon and D stems can be clearly seen in our map in good overall agreement with earlier observations (Valle et al, 2003).
However, the exact nature of the tRNA deformation could not be determined precisely at the resolution of the previous cryo-EM studies, and the deformation was modelled as smoothly distributed over the four base pairs surrounding the junction (Valle et al, 2003). In contrast, in our present map, we do not observe a significant change of the upper base pairs of the anticodon stem or the lower base pairs of the D stem. Moreover, it was not possible to fit the density by docking the T-acceptor arm together with the D stem and D loop of the tRNA as a single rigid body. Independent rigid-body docking of the T-acceptor arm and the D stem (together with parts of the D loop and the variable loop) leads to an improved fit for the A/T tRNA (Figures 2 and 3). As a result, the D loop is rotated relative to the T-acceptor arm such that the angle between both arms of the L-shaped tRNA is opened in the A/T tRNA compared with the canonical tRNA structure. This rotation of the D stem is expected to have an effect on the conformation of the D loop, which stabilizes the relative positioning of the T stem loop and D stem by a tertiary interaction with the T loop. Indeed, the present cryo-EM density accounts for G18 and G19, which directly interact with the T loop, but the linker regions including positions 16, 17 and 20 appear to be disordered. This observed change in the D loop conformation explains a long-standing observation from kinetic data on decoding (Rodnina et al, 1994), which used a tRNA with proflavin at positions 16 and 17 as a reporter. The fluorescence from this reporter showed a marked temporal increase during decoding, consistent with the observed disorder in the D loop.
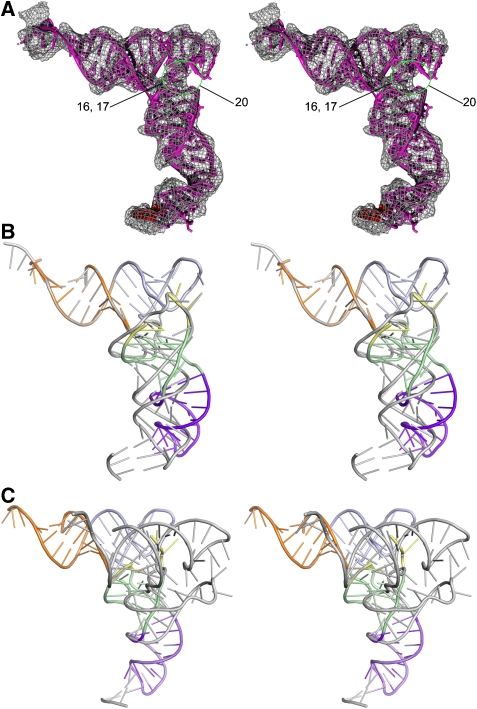
Figure 3.
Distortions in the tRNA during decoding. (A) A molecular model for the A/T-tRNA derived by fitting various segments of the tRNA separately into the electron density. The region of disorder in the D loop is highlighted (green circle, missing nucleotides are labelled) and is consistent with fluorescence changes in a reporter in this loop (see text for details). (B, C) Superposition of the distorted A/T-tRNA (coloured ribbon: acceptor stem, orange; T stem and T loop, cyan; D stem with variable loop, green; D loop, yellow; anticodon stem loop, magenta) and canonical tRNA (grey ribbon). The superpositions were carried out using the acceptor arm (B) and the anticodon stem loop (C), and show that in addition to the kink between the D stem and anticodon stem loop, there is also a rotation of the D loop and stem relative to the acceptor arm.
The opening of the tRNA at the junction between the D stem and T-acceptor arm moves the anticodon loop away from the position of the mRNA codon. To facilitate codon–anticodon interactions by counteracting this movement, the kink between the D and anticodon stems has to be even larger than previously proposed (Valle et al, 2003). Indeed, if we superpose atomic models for the tRNAs at the D stem, the distance between G34 of the anticodon loop between the A/A tRNA (Yusupov et al, 2001) and the previous model of the A/T tRNA (Valle et al, 2003) is 12.5 Å, and this is increased to 18.0 Å for the present structure of the A/T tRNA. The exact conformation of the A/T tRNA at the kink cannot be described in atomic terms even at the improved resolution of the current cryo-EM map, but it is in good agreement with local conformational changes of the unpaired nucleotides between the anticodon and D stems, for example, G26 and G45, which was suggested as a conformational hinge point when the structure of tRNA was originally determined (Robertus et al, 1974). The nearby Hirsh suppressor mutation at position 24 in the D stem (Hirsh, 1971) was recently shown to increase the rate of GTP hydrolysis in decoding (Cochella and Green, 2005). Presumably mutating this residue alters the properties of the neighbouring residues, facilitating a hinge movement in tRNA, thus allowing the transition state to be reached even for incorrect tRNAs.
The angle of twist between the acceptor stem and T stem of the tRNA is greater in the ternary complex with a GTP analogue than it is in free tRNA (Nissen et al, 1995). In the presence of antibiotics enacyloxin IIa or kirromycin, this twist is partially relieved and the conformation of the tRNA within the ternary complex is more similar to the conformation of free tRNA (Parmeggiani et al, 2006). The untwisted T-acceptor arm (without the 3′-CCA end, which was fit together with EF-Tu) from the ribosome-bound A/A tRNA (Yusupov et al, 2001) fits slightly better into the cryo-EM map than the one from the kirromycin-containing ternary complex (PDB identifier 1OB2; cross-correlation coefficient of 0.80 versus 0.78) and we used it for our final model of the A/T tRNA (Figures 2, 3 and 4). This difference is small and may not be significant at the present resolution of our cryo-EM map, but the larger twist present in the ternary complex in the absence of antibiotics (Nissen et al, 1995) is relieved during the events of ribosomal decoding and a conformational change of the T-acceptor arm of tRNA might be part of the mechanism leading to EF-Tu dissociation from the tRNA.
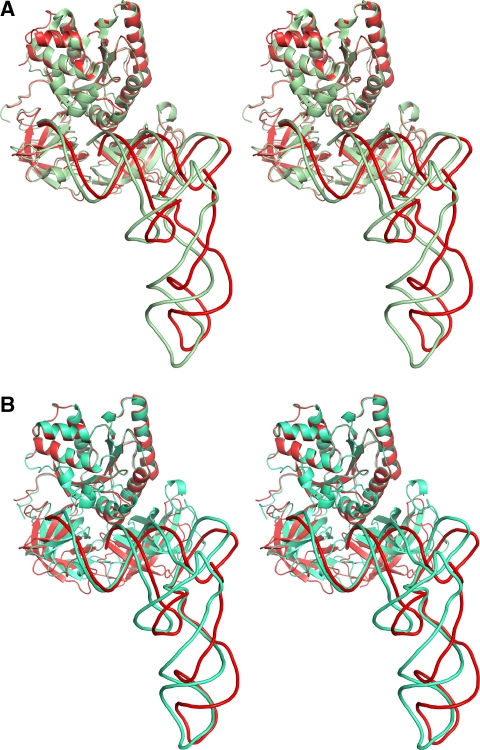
Figure 4.
Structures of the ternary complex on and off the ribosome. Superposition of the molecular model for the ribosome-bound ternary complex (red ribbons) with (A) the X-ray structure of the kirromycin-containing ternary complex (PDB identifier 1OB2, green ribbons) or (B) the X-ray structure of the ternary complex (Nissen et al, 1995) (turquoise ribbons). The ternary complexes are aligned at domain I (G domain) of EF-Tu.
In summary, if we consider the X-ray structure of the ternary complex to represent the structure of unbound ternary complex in solution (Nissen et al, 1995) and if we assume that the present model of the kirromycin-stalled ribosome-bound ternary complex represents a state that follows but is close to the transition state for GTP hydrolysis during decoding, the tRNA undergoes conformational rearrangements that are far more complex than previously anticipated (Figures 3 and 4): (i) the twist between the T and acceptor stems is relieved, (ii) the region between the T-acceptor arm and D stem is opened, (iii) accompanied by a partial unfolding of the D loop and (iv) a kink is introduced at the junction between the D and anticodon stems. These rearrangements that occur throughout the whole tRNA molecule are consistent with a direct role of tRNA in signalling successful codon recognition to the GTPase centre on EF-Tu (Piepenburg et al, 2000).
The structure of ribosome-bound EF-Tu•GDP•kirromycin
Not surprisingly, EF-Tu on the ribosome has a distinct conformation that is different from the GTP- or GDP-bound forms of the isolated structure. The EF-Tu•GDP•methylkirromycin structure (Vogeley et al, 2001) could be docked readily into the cryo-EM density as a single rigid body (Figure 2) in contrast to the EF-Tu•GMPPNP structure (Berchtold et al, 1993) (Supplementary data). Methylkirromycin is functionally equivalent to kirromycin and prevents the large conformational change of EF-Tu upon GTP hydrolysis by gluing together domains I and III. However, the two structures are not equivalent: in the methylkirromycin structure, domains II and III are rotated relative to domain I by about 10° and 15°, respectively (Vogeley et al, 2001). The docking shows that the conformation of EF-Tu in complex with GDP and methylkirromycin (Vogeley et al, 2001) is very similar to the structure of EF-Tu in the kirromycin-stalled, ribosome-bound ternary complex. This result is at variance with the cryo-EM analysis of Stark et al (2002) on this complex, who could not obtain a good fit with the EF-Tu•GDP•methylkirromycin structure and instead proposed a large movement of domain III of EF-Tu. In contrast, Escherichia coli EF-Tu from the X-ray structure of the kirromycin-containing Phe-tRNAPhe•EF-Tu•GMPPNP•kirromycin complex (PDB code: 1OB2) could be docked as a single rigid body into the previous cryo-EM map of Valle et al (2003) and this is in agreement with our findings presented here, because the latter EF-Tu structure is very similar to the EF-Tu•GDP•methylkirromycin structure.
In agreement with lower resolution cryo-EM studies (Stark et al, 1997; Valle et al, 2003), the relative orientation of tRNA and EF-Tu is changed in the ribosome-bound ternary complex relative to the X-ray structures of ternary complex (Nissen et al, 1995) or the Phe-tRNAPhe•EF-Tu•GMPPNP•kirromycin complex (PDB code: 1OB2). The changes can be described as a small rotation of EF-Tu and a translational movement in the direction from the acceptor stem towards the T loop of the tRNA (Figure 4). This movement of EF-Tu is partially counteracted by the release of the twist between the acceptor and T stems of the tRNA that is present in the unbound ternary complex (Nissen et al, 1995). At the T stem, however, domain III of EF-Tu moves 5–6 Å relative to the tRNA while maintaining extensive interactions with it (Figure 2). We did not attempt to describe the underlying changes, which probably involve detailed changes in side chain contacts.
Interactions of the ribosome with the ternary complex and its implications for GTPase activation
A central unresolved question in decoding is how codon recognition in the decoding centre of the 30S subunit leads to GTP hydrolysis by EF-Tu over 80 Å away. The cryo-EM structure of the ternary complex bound to the ribosome presented here, by allowing a more detailed analysis in molecular terms, allows us to address this question. Kinetic data have shown that the structures of the transition state and the GDPNP complex are different (Rodnina et al, 1995). However, kirromycin-stalled complexes, such as the one analysed here, are very similar to the state at the moment of GTP hydrolysis, because kirromycin does not affect the rates of GTP hydrolysis and Pi release, but blocks the subsequent rearrangements of EF-Tu (Kothe and Rodnina, 2006).
At the decoding centre, the conformation of the tRNA anticodon stem loop and its interactions with the ribosome are indistinguishable from that of accommodated tRNA corresponding to previous crystallographic studies (Ogle et al, 2001; Yusupov et al, 2001; Selmer et al, 2006). In particular, the density clearly shows that A1492 and A1493 are in the flipped out conformation (Figure 5A) and make minor groove interactions with the codon–anticodon helix, as shown previously (Ogle et al, 2001). The density for G530 is also consistent with its role in minor groove interactions. These interactions were proposed to be specific for Watson–Crick geometry at the first two positions of the codon–anticodon helix, and thus provided the additional binding energy required to induce a conformational change of the 30S in a cognate tRNA-specific manner (Ogle et al, 2001, 2002). The conformational change consisted in particular of a movement of the shoulder domain of the 30S subunit towards the ternary complex, and such a movement was proposed to help stabilize the transition state for GTP hydrolysis by EF-Tu (Ogle et al, 2002).
Figure 5.
Interactions of the ribosome-bound ternary complex with elements of the ribosome. Stereo representation of the molecular model for the ternary complex and the interacting ribosomal elements (coloured ribbon) docked into the cryo-EM density (grey mesh). Important components or residues are labelled. (A) Details of the decoding centre showing that A1492 and A1493 of 16S RNA are extended out of helix 44 and making contact with the minor groove of the codon–anticodon helix along with G530. Also shown is the base of A1913 in helix 69 of 23S RNA, which is inserted between 16S and 23S RNA and appears to contact the tRNA around position 38 (Selmer et al, 2006). (B) Interactions of the A/T-tRNA with the ribosome outside the decoding region. Residues that are putatively involved in the interactions are highlighted in green. (C) Interaction of the shoulder of the 30S subunit with domain II of EF-Tu. As in (B), residues that are putatively involved in the interactions are highlighted in green. Colour code: blue, 50S subunit; yellow, 30S subunit; pink, A/T-site tRNA.
The interactions of the anticodon helix with the 16S rRNA are augmented by an interaction between helix 69 of 23S rRNA and the backbone of tRNA nucleotides 25 and 26. (Figure 5B). These interactions appear to be essentially unchanged compared with the interactions of the accommodated A/A tRNA (Yusupov et al, 2001; Selmer et al, 2006), although the axis of the anticodon helix of the A/T tRNA might be slightly rotated away from helix 69 of 23S RNA. The similarity of the interactions of helix 69 with accommodated A/A and A/T tRNA suggests that they are unlikely to actively facilitate the kinked conformation of the tRNA, as has been proposed (cf. Valle et al, 2003). This view would also be consistent with the observation that ribosomes lacking helix 69 translate mRNA with normal accuracy in vitro (Ali et al, 2006).
Interestingly, we see that the shoulder domain of the 30S subunit, which was proposed to move towards the ternary complex upon cognate tRNA binding (Ogle et al, 2002), makes contact with the ternary complex in two locations (see Table I for a complete list of contacts). His80 of S12, which has been implicated by recent genetic evidence in the signalling of codon recognition to EF-Tu through the aa-tRNA (Gregory et al, 2009), is very close to U69 of the acceptor arm of tRNA, presumably making direct contact. The EF-Tu loop containing residues 232–235 in domain II is in close contact with the tertiary base pair between A55 and U368 and the backbone of helix h5 around U358 in the shoulder domain of 16S RNA (Figure 5C). The backbone of 16S rRNA helix h15 around U368 appears to be contacted by Arg291. Interestingly, the EF-Tu mutations both in E. coli G222D (G233 in T. thermophilus) and Salmonella typhimurium G280V (G292 in T. thermophilus) show impaired GTPase activation (Swart et al, 1987; Tubulekas and Hughes, 1993), suggesting that the contacts induced by cognate tRNA between the shoulder of the 30S subunit and the ternary complex are important for the stabilization of the transition state for GTP hydrolysis. Moreover, the contacts are not due to a movement of the 30S shoulder alone. Rather, the EF-Tu residues involved have moved by 4–5 Å relative to the GTP conformation.
Table 1.
Summary of contacts between the ribosome and ternary complexa
| Ligand | Approximate location | Approximate ribosomal location |
|---|---|---|
| A/T-site | U69 | Protein S12 His 80 |
| tRNA | C56 | 23S rRNA A1067 |
| C25, M2G26 | 23S rRNA A1913, A1914 | |
| OMG34 | 16S rRNA C1054 | |
| A36 | 16S rRNA G530 | |
| A36, YG37 | 16S rRNA A1492, A1493 | |
| EF-Tu | Domain II Arg 291 | 16S rRNA U368 |
| Domain II Thr 232–Arg 234 | 16S rRNA U358, U368, A55 | |
| Domain I, switch I (effector loop) | 16S rRNA A344 | |
| Domain I Pro114, His 19 (P-loop) | 23S rRNA G2661 (SRL) | |
| aContacts are inferred at locations where there is a strong fusion of density in the cryo-EM map. All listed contacts are clearly visible when the map is contoured at 3σ or lower. | ||
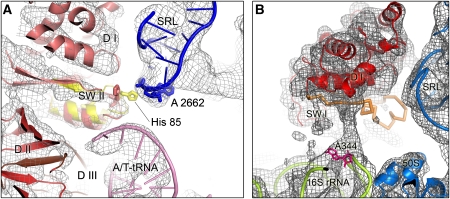
In the 50S subunit, the sarcin–ricin loop (SRL), a highly conserved loop of 23S RNA has long been known to be crucial for GTPase factor function (Hausner et al, 1987; Moazed et al, 1988). More recently, single-molecule FRET (smFRET) measurements have suggested that the SRL functions as a GTPase-activating factor, because cleavage of the SRL with the ribotoxin α-sarcin stalls decoding in a state similar to the GTPase-activating state, whereas the initial steps including codon recognition are hardly affected (Blanchard et al, 2004). The SRL interacts with the P-loop of domain I of EF-Tu (Table I) and may have an active function in inducing the GTPase conformation by manipulating the conformation of a switch region known to be important for GTP hydrolysis in GTPases generally. Interestingly, the cryo-EM map is more consistent with the conformation of the switch II region of the EF-Tu•GDP•methylkirromycin structure (Vogeley et al, 2001) than that in the ternary complex with GDPNP (Nissen et al, 1995). Our structure suggests that if EF-Tu were in its GTP-bound conformation, the catalytically important His85 (His84 in E. coli) of EF-Tu would sterically clash with the SRL (Figure 6A; Supplementary Movie 2). The rearrangement of switch II of EF-Tu could therefore be caused by an interaction of this residue with the SRL, suggesting a direct role of this important element in GTPase activation. The importance of His85 (His84 in E. coli) for ribosome-dependent GTPase activity in general (Scarano et al, 1995; Zeidler et al, 1995) and especially for the chemical step (Daviter et al, 2003) has been demonstrated by mutagenesis experiments. The position of the reoriented His85 is such that it could interact with the γ-phosphate, thereby stabilizing the transition state of GTPase reaction (Vogeley et al, 2001). In a previous study, close contacts were observed between EF-G•GMPPNP with the SRL in a 70S•EF-G•GMPPNP complex, but could not be analysed in detail to the limited resolution of the previous cryo-EM map (Connell et al, 2007). Nevertheless, it is consistent with an active role of the SRL in GTPase activation also for EF-G.
Figure 6.
Interactions of the switch regions of EF-Tu with the ribosome. (A) Close up on the switch II region (SW II) and the sarcin-ricin loop (SRL). The cryo-EM map is shown as grey mesh and models for the docked components as coloured ribbons. In addition, the structure of SW II from the EF-Tu•GMPPNP X-ray structure (PDB identifier 1EXM) is superposed (yellow ribbons) after the structure has been aligned onto the fitted EF-Tu•GDP•aurodox structure (red ribbons), which in turn was docked as a rigid body into the density map. Furthermore, the functionally important His85 (arrow) is highlighted in the docked X-ray structures of EF-Tu•GMPPNP and EF-Tu•GDP•aurodox (we note that we cannot observe the residue directly at the present resolution in the cryo-EM map), as well as A2662 of the SRL (pink residue) to indicate the possibility of a close contact between the SRL and His85 of EF-Tu•GMPPNP but not His85 of EF-Tu•GDP•aurodox. Abbreviations: D I, D II, D III: domains I–III of EF-Tu. (B) Region showing density extending from the switch I region of EF-Tu to nucleotide A344 in helix 14 in the shoulder of the 30S subunit. This switch undergoes a major rearrangement upon GTP hydrolysis and may be in an intermediate, partially disordered form in this structure, with the contact with the 30S subunit being potentially important.
The other switch region of EF-Tu, switch I, undergoes a dramatic rearrangement from a helical conformation to an extended beta hairpin upon GTP hydrolysis (Abel et al, 1996). In the structure here, the density for the switch I region is neither in the GTP nor the GDP form but is broken up (Figure 6B), suggesting that it is in a disordered, dynamic state during the switch from one form to the other. However, interestingly, there is a protrusion of density from the switch I region to nucleotide A344 of helix 14 in the shoulder of 16S RNA. This nucleotide has also been implicated in contacts with the switch I regions of EF-G (Connell et al, 2007) and EF4 (LepA) (Connell et al, 2008) and might therefore constitute a contact site for the switch regions of translational GTPases in general. The position of A344 is a function of the conformational state of the ribosome (for example, the state of the ratchet between the subunits); thus, although different factors are activated by different states of the ribosome, some of the crucial underlying interactions may be identical. In any case, the shoulder movement of the 30S subunit in response to codon recognition may thus have a direct role in the GTPase activity of EF-Tu in addition to stabilizing the GTPase-activated state more generally.
The conformation of the switch I region could also be manipulated by the ribosome through the tRNA. A hydrophobic gate formed by Val20 and Ile61 shields the γ-phosphate of the G-nucleotide from bulk solvent and it has been proposed that opening the gate could be part of the ribosome-mediated GTPase reaction (Berchtold et al, 1993). Interestingly, in the EF-Tu•GDP•methylkirromycin X-ray structure, which—as we have shown here—represents the GTPase-activated state, this gate is opened. Ile61 along with other residues of the switch I region is disordered and thus has moved away from Val20 (Vogeley et al, 2001). Although the GTP conformation of the switch I region appears to be destabilized in the GTPase-activated conformation of EF-Tu, one would not have expected this to happen in the context of a bound tRNA, because the switch I region is stabilized in the GTP conformation by interactions with the tRNA (Parmeggiani et al, 2006). However, in the ribosome-bound ternary complex (with regard to the free ternary complex) the tRNA is moved relative to EF-Tu by 5–7 Å (Stark et al, 1997; Valle et al, 2003). The apparent disorder of the switch I region in the GTPase-activated state on the ribosome could be caused by this tRNA movement, which in turn is probably facilitated by the interactions of the tRNA with the ribosome, especially in the decoding centre. Thus, our model suggests how both the ribosome and tRNA contribute to GTPase activation, in line with previous functional studies (Rodnina and Wintermeyer, 2001; Marshall et al, 2008).
Kinetic data show that codon recognition precedes GTPase activation (Rodnina et al, 1995; Blanchard et al, 2004). However, it is likely that codon recognition requires transient changes to the structure of the ternary complex that are stabilized by a cognate interaction. In the model proposed by Valle et al (2003), the distortion in the tRNA is essentially due to the localized kink, which is proposed to occur as the last step before codon recognition. However, a localized kink of the tRNA far away from EF-Tu can hardly explain how tRNA is actively participating in signalling codon recognition to EF-Tu. Moreover, the kink in their model would move the anticodon in a direction away from the codon. If the mRNA codon is placed in a position to interact with the kinked tRNA, the anticodon loop of the undistorted tRNA just before the formation of the kink would sterically clash with the codon.
In contrast, in our structural model, where the extent and direction of the kink are different due to additional changes elsewhere in the tRNA, the structures for both A/T and the undistorted tRNA can be superposed at the D stem without a sterical clash with the mRNA codon. Moreover, models where a major conformational change to a higher energy state is required before codon recognition (Frank et al, 2005) are difficult to reconcile with kinetic data showing that all steps before codon recognition are fast and reversible (Rodnina and Wintermeyer, 2001).
We therefore favour a model where the ternary complex approaches the ribosome in an essentially undistorted conformation. The X-ray structure of the ternary complex (Nissen et al, 1995) can be docked into the ribosome in such a way that major steric clashes are absent and that all the interaction sites of the ternary complex are relatively close to their corresponding interaction sites on the ribosome. In this hypothetical situation for the initial binding mode, the angle between the codon and anticodon is not optimal for codon–anticodon interaction, but local flexibility in tRNA and mRNA might allow transient probing of the mRNA codon by the anticodon loop of the tRNA. It is plausible that the first key event is then the stabilization of the correct codon–anticodon interaction by A-minor interactions with the 16S rRNA, and the closing of the 30S subunit (Ogle et al, 2001, 2002). The additional energy from the cognate interactions induces a domain closure in the 30S subunit, thus forcing the anticodon stem into an accommodated orientation (Ogle et al, 2002; Ogle and Ramakrishnan, 2005). Because EF-Tu and the acceptor stem of the tRNA are held by the ribosome and cannot follow the rotational movement of the anticodon stem, this will result in the kink at the D stem junction. The strain thus imposed can be partially relieved by the observed opening between the T and D stems of the tRNA and the relative movement of tRNA and EF-Tu.
In this view, it is not the anticodon stem loop but the elbow region of the tRNA that will undergo the largest movement during decoding. It therefore makes sense that the elbow region is not involved in contacts with the ribosome apart from the interaction with the flexible L7/L12 stalk base region. Our structure-based model is in excellent agreement with a model that involves two activation energy barriers during tRNA selection (Cochella et al, 2007) and with a model based on smFRET data (Blanchard et al, 2004). Blanchard et al (2004) could show that the elbow of the incoming tRNA moves towards the P-site from the codon recognition state of the decoding complex to the GTPase-activated state. Large and rare thermal fluctuations (Lee et al, 2007) might lead to the observed distortions in the structure of the ternary complex. As discussed above, the interactions of the ribosome with the distorted structure of the ternary complex will stabilize the conformation of EF-Tu in the state required for GTP hydrolysis. After GTPase activity and subsequent dissociation of EF-Tu•GDP, the tRNA can re-adopt the classical conformation during accommodation into the A/A state.
Conclusions
The work presented here reports several advances. Technically, it shows that the production of biochemically homogeneous samples combined with the ability to properly classify conformationally distinct subspecies can result in significantly improved cryo-EM maps of the ribosome complex. Such an improvement in turn allows us to interpret in much greater detail the interactions of tRNA and EF-Tu trapped at a point just after GTP hydrolysis during decoding, thus shedding light on this crucial event in tRNA selection.
The structure suggests that the distortion of tRNA observed previously was neither just due to a distortion of the anticodon loop nor a single kink above the anticodon stem. Rather, it is a complex and likely linked set of changes, all of which are required to stabilize the state required for GTP hydrolysis. These changes also mean that the tRNA itself has moved relative to EF-Tu. The detailed molecular interpretation also rationalizes the role of specific residues in EF-Tu that were known to affect GTP hydrolysis. It suggests a direct role for helix 14 of 16S rRNA (around A344) and the SRL (helix 95 of 23S rRNA) in the rearrangement of the switch I and II regions of EF-Tu, respectively. These results have a general implication for the activation of GTPases by the ribosome, which have a crucial function in every stage of translation.
Materials and methods
Preparation of the 70S•tRNA•EF-Tu•GDP•kirromycin complex
Ribosomes, tRNA and mRNA were prepared as described previously (Selmer et al, 2006). Ternary complex was formed in two steps, the formation of the binary EF-Tu-GTP complex, followed by the addition of Phe-tRNAPhe. Formation of the full ribosomal complex was achieved by forming two subcomplexes concurrently, the ternary complex and initiation complex, and then mixing the two. Ribosomal complexes containing His-tagged EF-Tu were purified as described previously for release factors (Petry et al, 2005).
Cryo-EM of the 70S•tRNA•EF-Tu•GDP•kirromycin complexes
Cryo-EM micrographs were collected on a Tecnai G2 Polara (FEI) and scanned on a D8200 Primscan drum scanner (Heidelberger Druckmaschinen). In total, 586 329 projection images were collected from 452 micrographs at a 0.6–4.0 μm defocus range and processed using SPIDER (Frank et al, 1996). Using a cryo-EM reconstruction of the vacant E. coli 70S as a reference volume for initial refinement and as a seed structure for subsequent multi-particle refinement (Penczek et al, 2006; Connell et al, 2008), the data set was split into five sub-populations. Subsequently, the data set of the major sub-population I (323 688 projection images) was further refined.
Rigid body docking of atomic models was first carried out manually using O (Jones and Kjeldgaard, 1997) and then automatically using the SITUS software package (Wriggers et al, 1999) or SPIDER. Final figures were prepared with UCSF Chimera (Pettersen et al, 2004) or Pymol (DeLano Scientific).
See the Supplementary data for a more thorough description of the methods employed.
Accession codes
The electron density map and model of the 70S•tRNA•EF-Tu•GDP•kirromycin complex have been deposited in the 3D-EM and PDB databases with the accession numbers EMD-10474 and PDBID 3FIN and 3FIC, respectively.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends
Acknowledgments
This study was supported by grants from the DFG (SFB 740 TP A3 and TP Z1, SP 1130/2-1), the VolkswagenStiftung, the European Union 3D-EM Network of Excellence (to CMTS), the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum) and US NIH Grant GM 60635 (to PAP). VR was supported by the Medical Research Council (UK), US NIH Grant GM 67624, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet foundation.
References
- Abel K, Yoder MD, Hilgenfeld R, Jurnak F (1996) An alpha to beta conformational switch in EF-Tu. Structure 4: 1153–1159 [DOI] [PubMed] [Google Scholar]
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF (2006) Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 [DOI] [PubMed] [Google Scholar]
- Andersen GR, Nissen P, Nyborg J (2003) Elongation factors in protein biosynthesis. Trends Biochem Sci 28: 434–441 [DOI] [PubMed] [Google Scholar]
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365: 126–132 [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD (2004) tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R (2007) Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R (2005) An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM (2007) Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell 25: 751–764 [DOI] [PubMed] [Google Scholar]
- Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM (2008) A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol 15: 910–915 [DOI] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV (2003) Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332: 689–699 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M (2005) The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962 [DOI] [PubMed] [Google Scholar]
- Frank J, Spahn CM (2006) The ribosome and the mechanism of protein synthesis. Rep Prog Phys 69: 1383–1417 [Google Scholar]
- Gregory ST, Carr JF, Dahlberg AE (2009) A signal relay between ribosomal protein S12 and elongation factor EF-Tu during decoding of mRNA. RNA 15: 208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner TP, Atmadja J, Nierhaus KH (1987) Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie 69: 911–923 [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R (1995) Regulatory GTPases. Curr Opin Struct Biol 5: 810–817 [DOI] [PubMed] [Google Scholar]
- Hirsh D (1971) Tryptophan transfer RNA as the UGA suppressor. J Mol Biol 58: 439–458 [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M (1997) Electron density map interpretation. Method Enzymol 277B: 173–207 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF (2008) Structural dynamics of the ribosome. Curr Opin Chem Biol 12: 674–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U, Rodnina MV (2006) Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry 45: 12767–12774 [DOI] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S (2007) The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci USA 104: 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD (2008) Translation at the single-molecule level. Annu Rev Biochem 77: 177–203 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364 [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Harris JR, Beckman E, Zemlin F, Markl J, van Heel M (1997) Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J Mol Biol 271: 417–437 [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, Krab IM, Watanabe T, Nielsen RC, Dahlberg C, Nyborg J, Nissen P (2006) Enacyloxin IIa pinpoints a binding pocket of elongation factor Tu for development of novel antibiotics. J Biol Chem 281: 2893–2900 [DOI] [PubMed] [Google Scholar]
- Penczek PA, Frank J, Spahn CM (2006) A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol 154: 184–194 [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FV IV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Pape T, Pleiss JA, Wintermeyer W, Uhlenbeck OC, Rodnina MV (2000) Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39: 1734–1738 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250: 546–551 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W (1995) Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J 14: 2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Wintermeyer W (1994) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33: 12267–12275 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Scarano G, Krab IM, Bocchini V, Parmeggiani A (1995) Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Lett 365: 214–218 [DOI] [PubMed] [Google Scholar]
- Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM (2006) Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M (1997) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389: 403–406 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M (2002) Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol 9: 849–854 [DOI] [PubMed] [Google Scholar]
- Swart GW, Parmeggiani A, Kraal B, Bosch L (1987) Effects of the mutation glycine-222—aspartic acid on the functions of elongation factor Tu. Biochemistry 26: 2047–2054 [DOI] [PubMed] [Google Scholar]
- Tubulekas I, Hughes D (1993) A single amino acid substitution in elongation factor Tu disrupts interaction between the ternary complex and the ribosome. J Bacteriol 175: 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R (2001) Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu.GDP and aurodox. J Biol Chem 276: 17149–17155 [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA (1999) Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 125: 185–195 [DOI] [PubMed] [Google Scholar]
- Yarus M, Smith D (1995) tRNA on the ribosome: a waggle theory. In tRNA: Structure, Biosynthesis and Function, Söll D, RajBhandary U (eds), pp 443–468. Washington: American Society for Microbiology Press [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zeidler W, Egle C, Ribeiro S, Wagner A, Katunin V, Kreutzer R, Rodnina M, Wintermeyer W, Sprinzl M (1995) Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur J Biochem 229: 596–604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends